Advantages and Challenges of Using ctDNA NGS to Assess the Presence of Minimal Residual Disease (MRD) in Solid Tumors
Abstract
:Simple Summary
Abstract
1. Introduction
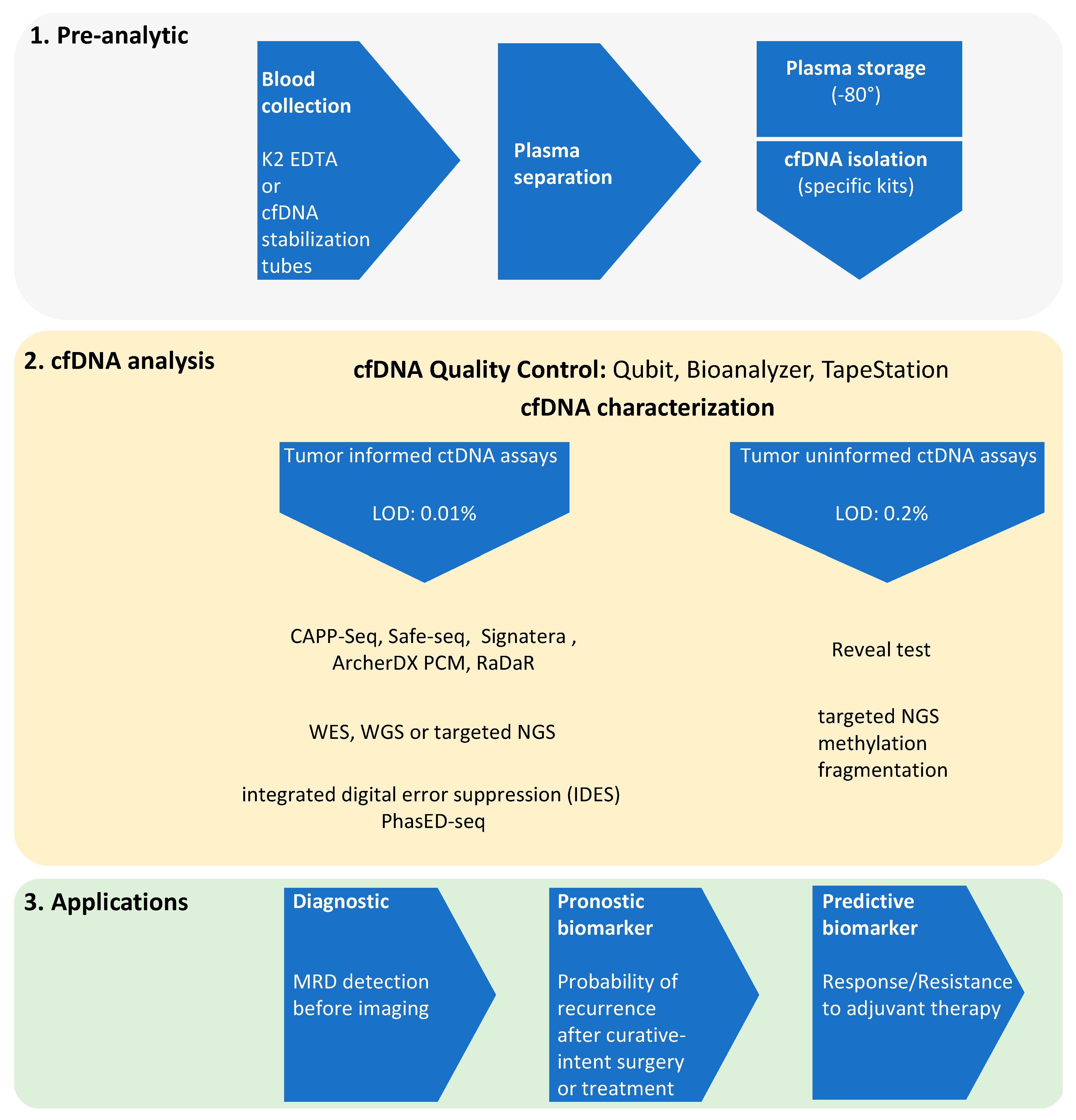
2. ctDNA NGS and MRD Evidence
2.1. Evidence of MRD in NSCLC
2.2. Evidence of MRD in CRC
2.3. MRD Evidence to Guide Immunotherapy
3. Challenges of the ctDNA-Based NGS Technology
3.1. False-Negativity Rate
3.2. False-Positivity Rate
3.3. Confounding Factor: CHIP
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Definitions
| MRD | minimal residual disease |
| NGS | next-generation sequencing |
| ctDNA | circulating tumor DNA |
| cfDNA | cell-free DNA |
| gDNA | genomic DNA |
| CRC | colorectal cancer |
| NSCLC | non-small-cell lung cancer |
| FDA | food and drug administration |
| VAF | variant allele frequency |
| CAPP-Seq | CAncer Personalized Profiling by deep Sequencing |
| IDES | integrated digital error suppression |
| PhasED-Seq | phased variant enrichment and detection sequencing |
| PPV | positive predictive value |
| SOC | standard of care |
| TKI | tyrosine kinase inhibitor |
| ICI | immune checkpoint inhibitor |
| PFS | progression free survival |
| OS | overall survival |
| TMB | tumor mutational burden |
| MSI | microsatellite instability |
| CRT | chemoradiotherapy |
| DCB | durable clinical benefit |
| CHIP | clonal hematopoiesis of undetermined potential |
| WBC | white blood cells |
| HRD | homologous recombination deficiency |
| CTC | circulating tumor cells |
| RFS | relapse-free survival |
References
- Thierry, A.R.; EI Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. NON-THEMATIC REVIEW. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Pisetsky, D.S.; Fairhurst, A.-M. The origin of extracellular DNA during the clearance of dead and dying cells. Autoimmunity 2009, 40, 281–284. [Google Scholar] [CrossRef]
- Tabernero, J.; Lenz, H.-J.; Siena, S.; Sobrero, A.; Falcone, A.; Ychou, M.; Humblet, Y.; Bouché, O.; Mineur, L.; Barone, C.; et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015, 16, 937–948. [Google Scholar] [CrossRef]
- Parseghian, C.M.; Loree, J.M.; Morris, V.K.; Liu, X.; Clifton, K.K.; Napolitano, S.; Henry, J.T.; Pereira, A.A.; Vilar, E.; Johnson, B.; et al. Anti-EGFR-resistant clones decay exponentially after progression: Implications for anti-EGFR re-challenge. Ann. Oncol. 2019, 30, 243–249. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Taniguchi, H.; Ikeda, M.; Bando, H.; Kato, K.; Morizane, C.; Esaki, T.; Komatsu, Y.; Kawamoto, Y.; Takahashi, N.; et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat. Med. 2020, 26, 1859–1864. [Google Scholar] [CrossRef]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. 2011, 108, 9530–9535. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.; Lindbjerg Andersen, C. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef] [Green Version]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Abbosh, C.; Frankell, A.; Garnett, A.; Harrison, T.; Weichert, M.; Licon, A.; Veeriah, S.; Daber, B.; Moreau, M.; Chesh, A.; et al. Abstract CT023: Phylogenetic tracking and minimal residual disease detection using ctDNA in early-stage NSCLC: A lung TRACERx study. Cancer Res. 2020, 80, CT023. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Schaffer, A. ctDNA Shows Promise as a Potential Biomarker for MRD in Locally Advanced NSCLC. Available online: https://www.onclive.com/view/ctdna-shows-promise-as-a-potential-biomarker-for-mrd-in-locally-advanced-nsclc (accessed on 30 August 2021).
- Kurtz, D.M.; Soo, J.; Alizadeh, A.A. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat. Biotechnol. 2021. [Google Scholar] [CrossRef]
- Parikh, A.; Van Seventer, E.; Siravegna, G.; Corcoran, R. Minimal Residual Disease Detection using a Plasma-only Circulating Tumor DNA Assay in Patients with Colorectal Cancer. Clin. Cancer Res. 2021. [Google Scholar] [CrossRef]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef] [Green Version]
- Azad, T.D.; Chaudhuri, A.A.; Fang, P.; Qiao, Y.; Esfahani, M.S.; Chabon, J.J.; Hamilton, E.G.; Yang, Y.D.; Lovejoy, A.; Aaron, M.; et al. Circulating tumor DNA analysis for detection of minimal residual disease after chemoradiotherapy for localized esophageal cancer. Gastroenterology 2020, 158, 494–505. [Google Scholar] [CrossRef]
- Garcia-Murillas, I.; Chopra, N.; Comino-Méndez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.F.; Farooq, M.; Cronin, P.A.; Anderson, K.S.; et al. Detection of residual disease after neoadjuvant therapy in breast cancer using personalized circulating tumor DNA analysis. bioRxiv 2018, 7392, 1–14. [Google Scholar] [CrossRef]
- Parsons, H.A.; Rhoades, J.; Reed, S.C.; Adalsteinsson, V.A. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin Cancer Res 2020, 26, 2556–2564. [Google Scholar] [CrossRef] [Green Version]
- Abbosh, C.; Birkbak, N.J.; Swanton, C. Early stage NSCLC—Challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 2018, 15, 577–586. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020, 580, 28. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Peterson, C.; Sriram, D.; Mahipal, A. Early stage colon cancer: Current treatment standards, evolving paradigms, and future directions. World J. Gastrointest. Oncol. 2020, 12, 808–832. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef] [Green Version]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019, 5, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Alberts, S.; Sargent, D.; Goldberg, R. Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: A randomized trial. JAMA 2012, 307, 1383–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taieb, J.; Tabernero, J.; Mini, E.; Subtil, F.; Folprecht, G.; Van Laethem, J.-L.; Thaler, J.; Bridgewater, J.; Petersen, L.N.; Blons, H.; et al. Oxaliplatin, fluorouracil, and leucovorin with or without cetuximab in patients with resected stage III colon cancer (PETACC-8): An open-label, randomised phase 3 trial. Lancet Oncol. 2014, 15, 862–873. [Google Scholar] [CrossRef]
- de Gramont, A.; Van Cutsem, E.; Schmoll, H.-J.; Tabernero, J.; Clarke, S.; Moore, M.J.; Cunningham, D.; Cartwright, T.H.; Hecht, J.R.; Rivera, F.; et al. Bevacizumab plus oxaliplatin-based chemotherapy as adjuvant treatment for colon cancer (AVANT): A phase 3 randomised controlled trial. Lancet Oncol. 2012, 13, 1225–1233. [Google Scholar] [CrossRef]
- Anandappa, G.; Starling, N.; Begum, R.; Bryant, A.; Sharma, S.; Renner, D.; Aresu, M.; Peckitt, C.; Sethi, H.; Feber, A.; et al. Minimal residual disease (MRD) detection with circulating tumor DNA (ctDNA) from personalized assays in stage II-III colorectal cancer patients in a U.K. multicenter prospective study (TRACC). J. Clin. Oncol. 2021, 39, 102. [Google Scholar] [CrossRef]
- Lianidou, E. Detection and relevance of epigenetic markers on ctDNA: Recent advances and future outlook. Mol. Oncol. 2021, 15, 1683. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, Y.; Mouliere, F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell 2019, 36, 350–368. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, T.V.; Tarazona, N.; Reinert, T.; Carbonell-Asins, J.A.; Roda, D.; Huerta, M.; Roselló, S.; Madsen, A.H.; Iversen, L.H.; Gotschalck, K.A.; et al. 420P Minimal residual disease detection and tracking tumour evolution using ctDNA in stage I-III colorectal cancer patients. Ann. Oncol. 2020, 31, S419–S420. [Google Scholar] [CrossRef]
- Hofman, P.; Heeke, S.; Alix-Panabières, C.; Pantel, K. Liquid biopsy in the era of immuno-oncology: Is it ready for prime-time use for cancer patients? Ann. Oncol. 2019, 30, 1448–1459. [Google Scholar] [CrossRef] [Green Version]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Guan, J.; Fu, Y. DNA Sensing in Mismatch Repair-Deficient Tumor Cells Is Essential for Anti-tumor Immunity. Cancer Cell 2021, 39, 96–108.e6. [Google Scholar] [CrossRef]
- Chan, T.; Yarchoan, M.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, J.; Cai, S.; Han, M.; Dong, H.; Zhao, J.; Zhu, B.; Wang, S.; Zhuo, M.; Sun, J.; et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients with Non-Small Cell Lung Cancer with Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019, 5, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Morrissey, M.P.; Hellmann, M.D. Use of circulating tumor DNA for cancer immunotherapy. Clin. Cancer Res. 2019, 25, 6909–6915. [Google Scholar] [CrossRef] [Green Version]
- Peters, S.; Cho, B.C.; Reinmuth, N.; Lee, K.H.; Luft, A.; Ahn, M.-J.; Baas, P.; Dols, M.C.; Smolin, A.; Vicente, D.; et al. Abstract CT074: Tumor mutational burden (TMB) as a biomarker of survival in metastatic non-small cell lung cancer (mNSCLC): Blood and tissue TMB analysis from MYSTIC, a Phase III study of first-line durvalumab ± tremelimumab vs chemotherapy. Cancer Res. 2019, 79, CT074. [Google Scholar] [CrossRef]
- Georgiadis, A.; Durham, J.N.; Keefer, L.A.; Bartlett, B.R.; Zielonka, M.; Murphy, D.; White, J.R.; Lu, S.; Verner, E.L.; Ruan, F.; et al. Noninvasive detection of microsatellite instabilit and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin. Cancer Res. 2019, 25, 7024–7034. [Google Scholar] [CrossRef] [Green Version]
- Willis, J.; Lefterova, M.I.; Artyomenko, A.; Kasi, P.M.; Nakamura, Y.; Mody, K.; Catenacci, D.V.T.; Fakih, M.; Barbacioru, C.; Zhao, J.; et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin. Cancer Res. 2019, 25, 7035–7045. [Google Scholar] [CrossRef]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24. [Google Scholar] [CrossRef]
- Janjigian, Y.; Lumish, M. ctDNA Analysis Is Feasible for Detection of Minimal Residual. Available online: https://www.esmo.org/oncology-news/ctdna-analysis-is-feasible-for-detection-of-minimal-residual-disease-after-treatment-across-a-variety-of-mmrd-tumour-types (accessed on 30 August 2021).
- Antonia, S.; Villegas, A.; Özgüroğlu, M. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018. [Google Scholar] [CrossRef]
- Food and Drug Agency. FDA approves durvalumab after chemoradiation for unresectable stage III NSCLC. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-durvalumab-after-chemoradiation-unresectable-stage-iii-nsclc (accessed on 30 August 2021).
- Moding, E.J.; Liu, Y.; Nabet, B.Y.; Chabon, J.J.; Chaudhuri, A.A.; Hui, A.B.; Bonilla, R.F.; Ko, R.B.; Yoo, C.H.; Gojenola, L.; et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat. Cancer 2020, 1, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Raja, R.; Kuziora, M.; Brohawn, P.; Ranade, K. Early Reduction in ctDNA Predicts Survival in Patients with Lung and Bladder Cancer Treated with Durvalumab. Clin. Cancer Res. 2018, 24, 6212–6222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anagnostou, V.; Forde, P.M.; White, J.R.; Niknafs, N.; Hruban, C.; Naidoo, J.; Marrone, K.; Ashok Sivakumar, I.K.; Bruhm, D.C.; Rosner, S.; et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non–small cell lung cancer. Cancer Res. 2019, 79, 1214–1225. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, S.B.; Narayan, A.; Kole, A.J.; Decker, R.H.; Teysir, J.; Carriero, N.J.; Lee, A.; Nemati, R.; Nath, S.K.; Mane, S.M.; et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 2018, 24, 1872–1880. [Google Scholar] [CrossRef] [Green Version]
- Hellmann, M.D.; Nabet, B.Y.; Rizvi, H.; Chaudhuri, A.A.; Wells, D.K.; Dunphy, M.P.S.; Chabon, J.J.; Liu, C.L.; Hui, A.B.; Arbour, K.C.; et al. Circulating Tumor DNA Analysis to Assess Risk of Progression after Long-term Response to PD-(L)1 Blockade in NSCLC. Clin. Cancer Res. 2020, 26, 2849–2858. [Google Scholar] [CrossRef] [Green Version]
- Nabet, B.Y.; Esfahani, M.S.; Moding, E.J.; Hamilton, E.G.; Chabon, J.J.; Rizvi, H.; Steen, C.B.; Chaudhuri, A.A.; Liu, C.L.; Hui, A.B.; et al. Noninvasive Early Identification of Therapeutic Benefit from Immune Checkpoint Inhibition. Cell 2020, 183, 363–376.e13. [Google Scholar] [CrossRef]
- Wu, T.D.; Madireddi, S.; de Almeida, P.E.; Banchereau, R.; Chen, Y.J.J.; Chitre, A.S.; Chiang, E.Y.; Iftikhar, H.; O’Gorman, W.E.; Au-Yeung, A.; et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020, 579, 274–278. [Google Scholar] [CrossRef]
- Valpione, S.; Galvani, E.; Tweedy, J.; Mundra, P.A.; Banyard, A.; Middlehurst, P.; Barry, J.; Mills, S.; Salih, Z.; Weightman, J.; et al. Immune awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat. Cancer 2020, 1, 210–221. [Google Scholar] [CrossRef]
- Fairfax, B.P.; Taylor, C.A.; Watson, R.A.; Nassiri, I.; Danielli, S.; Fang, H.; Mahé, E.A.; Cooper, R.; Woodcock, V.; Traill, Z.; et al. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat. Med. 2020, 26, 193–199. [Google Scholar] [CrossRef]
- Pantel, K.; Hayes, D. Disseminated breast tumour cells: Biological and clinical meaning. Nat. Rev. Clin. Oncol. 2018, 15, 129–131. [Google Scholar] [CrossRef]
- Vidal, J.; Muinelo, L.; Montagut, C. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, J.; Elez, E.; Vivancos, A. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann. Oncol. 2017, 28, 1294–1301. [Google Scholar] [CrossRef]
- Normanno, N.; Esposito Abate, R.; Lambiase, M.; Forgione, L.; Cardone, C.; Iannaccone, A.; Sacco, A.; Rachiglio, A.M.; Martinelli, E.; Rizzi, D.; et al. RAS testing of liquid biopsy correlates with the outcome of metastatic colorectal cancer patients treated with first-line FOLFIRI plus cetuximab in the CAPRI-GOIM trial. Ann. Oncol. 2018, 29, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Bachet, J.; Bouché, O.; Laurent-Puig, P. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: The AGEO RASANC prospective multicenter study. Ann. Oncol. 2018, 29, 1211–1219. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lo, S.N.; Wang, Y.; Li, L.; Christie, M.; Lee, M.; Wong, R.; Kosmider, S.; Skinner, I.; et al. Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer: Individual patient pooled analysis of three cohort studies. J. Cancer 2021, 148, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928. [Google Scholar] [CrossRef]
- Mansukhani, S.; Barber, L.; Starling, N.; Gerlinger, M. Ultra-Sensitive Mutation Detection and Genome-Wide DNA Copy Number Reconstruction by Error-Corrected Circulating Tumor DNA Sequencing. Clin. Chem. 2018, 64, 1626–1635. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, M.W.; Kennedy, S.R.; Salk, J.J.; Fox, E.J.; Hiatt, J.B.; Loeb, L.A. Detection of ultra-rare mutations by next-generation sequencing. PNAS 2012. [Google Scholar] [CrossRef] [Green Version]
- Dudley, J.; Schroers-Martin, J.; Alizadeh, A.; Diehn, M. Detection and Surveillance of Bladder Cancer Using Urine Tumor DNA. Cancer Discov. 2019, 9, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Phallen, J.; Sausen, M.; Velculescu, V. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, 403. [Google Scholar] [CrossRef] [Green Version]
- Strickler, J.H.; Loree, J.M.; Ahronian, L.G.; Parikh, A.R.; Andresson, A.; Pereira, L.; Mckinney, M.; Korn, W.M.; Atreya, C.E.; Banks, K.C.; et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2019, 8, 164–173. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Olsen, S.; Ku, B.M.; Lee, M.S.; Jung, H.A.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Park, K.; Choi, Y.-L.; et al. High concordance of actionable genomic alterations identified between circulating tumor DNA–based and tissue-based next-generation sequencing testing in advanced non–small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program. Cancer 2021, 127, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Lam, V.K.; Elamin, Y.Y.; Hong, L.; Colen, R.; Elshafeey, N.A.; Hassan, I.S.A.; Altan, M.; Blumenschein, G.R.; Rinsurongkawong, W.; et al. Clinical Outcomes in Non–Small-Cell Lung Cancer Patients Treated With EGFR-Tyrosine Kinase Inhibitors and Other Targeted Therapies Based on Tumor Versus Plasma Genomic Profiling. JCO Precis. Oncol. 2021, 1241–1249. [Google Scholar] [CrossRef]
- Chae, Y.K.; Davis, A.A.; Jain, S.; Santa-Maria, C.; Flaum, L.; Beaubier, N.; Platanias, L.C.; Gradishar, W.; Giles, F.J.; Cristofanilli, M. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol. Cancer Ther. 2017, 16, 1412–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieg-Bourne, C.C.; Okamura, R.; Kurzrock, R. Concordance between TP53 alterations in blood and tissue: Impact of time interval, biopsy site, cancer type and circulating tumor DNA burden. Mol. Oncol. 2020, 14, 1242–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pectasides, E.; Stachler, M.; Catenacci, D. Genomic Heterogeneity as a Barrier to Precision Medicine in Gastroesophageal Adenocarcinoma. Cancer Discov. 2018, 8, 37–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetson, D.; Ahmed, A.; Xu, X.; Nuttall, B.R.B.; Lubinski, T.J.; Johnson, J.H.; Barrett, J.C.; Dougherty, B.A. Orthogonal Comparison of Four Plasma NGS Tests With Tumor Suggests Technical Factors are a Major Source of Assay Discordance. JCO Precis. Oncol. 2019, 1–9. [Google Scholar] [CrossRef]
- Ng, S.; Chua, C.; Tan, I. Individualised multiplexed circulating tumour DNA assays for monitoring of tumour presence in patients after colorectal cancer surgery. Sci. Rep. 2017, 7, 40737. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.; Gibbs, P. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2018, 68, 663–671. [Google Scholar] [CrossRef]
- Loupakis, F.; Sharma, S.; Derouazi, M.; Murgioni, S.; Biason, P.; Rizzato, M.D.; Rasola, C.; Renner, D.; Shchegrova, S.; Malashevich, A.K.; et al. Detection of Molecular Residual Disease Using Personalized Circulating Tumor DNA Assay in Patients With Colorectal Cancer Undergoing Resection of Metastases. JCO Precis. Oncol. 2021, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.; Sun, K. Epigenetic Biomarkers in Cell-Free DNA and Applications in Liquid Biopsy. Genes 2019, 10, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quirico, L.; Orso, F. The power of microRNAs as diagnostic and prognostic biomarkers in liquid biopsies. Cancer Drug Resist. 2020, 3, 117–139. [Google Scholar] [CrossRef] [Green Version]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G. Clinical utility of circulating non-coding RNAs - an update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V. The biology, function, and biomedical applications of exosomes. Science 2020, 367. [Google Scholar] [CrossRef]
- Sol, N.; in’t Veld, S.G.J.G.; Vancura, A.; Tjerkstra, M.; Leurs, C.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Zwaan, K.; et al. Tumor-Educated Platelet RNA for the Detection and (Pseudo)progression Monitoring of Glioblastoma. Cell Reports Med. 2020, 1, 100101. [Google Scholar] [CrossRef]
- Keup, C.; Suryaprakash, V.; Hauch, S.; Storbeck, M.; Hahn, P.; Sprenger-Haussels, M.; Kolberg, H.-C.; Tewes, M.; Hoffmann, O.; Kimmig, R.; et al. Integrative statistical analyses of multiple liquid biopsy analytes in metastatic breast cancer. BMC 2021, 13, 85. [Google Scholar] [CrossRef]
- Steensma, D.; Bejar, R.; Ebert, B. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, M.; Kircher, M.; Shendure, J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lui, Y.Y.N.; Chik, K.-W.; Lo, Y.M.D. Predominant Hematopoietic Origin of Cell-Free DNA in Plasma and Serum after Sex-Mismatched Bone Marrow Transplantation. Available online: https://pubmed.ncbi.nlm.nih.gov/11861434/ (accessed on 26 August 2021).
| Assay | Company/Reference | Measure | Method | Tumor-Informed Approach |
|---|---|---|---|---|
| CAPP-Seq | Ref. [12] | large size gene panels | multiplex PCR amplification + ultradeep sequencing | yes |
| Safe-seq | Ref. [7] | medium size gene panels | deep sequencing with unique molecular identifier (UMI) barcoding | yes |
| Signatera | Natera | 16 somatic variants | multiplex PCR amplification + ultradeep sequencing | yes |
| PCM | ArcherDX | 28 genes | Anchored Multiplex PCR (AMP™) + ultradeep sequencing | yes |
| RaDaR | Inivata | 48 genes | multiplex PCR amplification + ultradeep sequencing | yes |
| Reveal | Guardant Health | somatic and epigenetic abberations | ultradeep sequencing + bioinformatic classifier to filter non-tumor derived variants | no |
| Entity | Intervention | Platform | Method | ctDNA Marker | Allocation | Acronym | NCT Number |
|---|---|---|---|---|---|---|---|
| NSCLC | adjuvant ChT or ICI | ArcherDx | PCM | somatic abberations | randomized | MERMAID-I | NCT04385368 |
| NSCLC | adjuvant ChT or ICI | ArcherDx | PCM | somatic abberations | randomized | MERMAID-II | NCT04642469 |
| NSCLC | adjuvant ChT or ICI | AVENIO | NGS | somatic abberations | non-randomized | NA | NCT04585490 |
| NSCLC | adjuvant ICI | AVENIO | NGS | somatic abberations | non-randomized | NA | NCT04585477 |
| NSCLC | adjuvant ChT or ICI | customized | CAPP-Seq | somatic abberations | non-randomized | NA | NCT04367311 |
| NSCLC | observational | customized | CAPP-Seq | somatic abberations | single group | TRACERx | NCT01888601 |
| NSCLC | adjuvant ChT or ICI | Inivata | RaDaR | somatic abberations | randomized | NA | NCT04966663 |
| CRC | adjuvant ChT | LUNAR | NGS | somatic and epigenetic abberations | randomized | COBRA | NCT04068103 |
| CRC | adjuvant ChT | LUNAR | NGS | somatic and epigenetic abberations | NA | PEGASUS | NCT04259944 |
| CRC | adjuvant ChT or ICI | LUNAR | NGS | somatic and epigenetic abberations | randomized | NA | NCT03803553 |
| CRC | observational | customized | NGS | NA | single group | IMPROVE | NCT03637686 |
| CRC | adjuvant ChT | customized | NGS | NA | randomized | IMPROVE-IT | NCT03748680 |
| CRC | adjuvant ChT | NA | Safe-SeqS | somatic abberations | randomized | DYNAMIC-II | ACTRN12615000381583 |
| CRC | adjuvant ChT | NA | Safe-SeqS | somatic abberations | randomized | DYNAMIC-III | ACTRN12617001566325 |
| CRC | adjuvant ChT | Ion Torrent PGM | NGS | somatic abberations | randomized | CIRCULATE | NCT04089631 |
| CRC | adjuvant ChT | Signatera | 16-plex PCR/NGS | somatic abberations | randomized | NA | NCT04920032 |
| CRC | personalized cancer vaccine | AVENIO | NGS | somatic abberations | randomized | NA | NCT04486378 |
| CRC | blood multi-analyte collection | LUNAR | NGS | somatic and epigenetic abberations | single group | MiRDA-C | NCT04739072 |
| CRC | observational | Signatera | 16-plex PCR/NGS | somatic abberations | single group | TRACC | NCT04050345 |
| CRC | observational | ProBio Trial | NGS | GW CNVs, MSI, and hypermutation | single group | CITCCA | NCT04726800 |
| CRC | observational | Signatera | 16-plex PCR/NGS | somatic abberations | single group | BESPOKE | NCT04264702 |
| MSI-H | ICI | F1, G360 or MSK-ACCESS | NGS | somatic abberations | single group | NA | NCT03832569 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larribère, L.; Martens, U.M. Advantages and Challenges of Using ctDNA NGS to Assess the Presence of Minimal Residual Disease (MRD) in Solid Tumors. Cancers 2021, 13, 5698. https://doi.org/10.3390/cancers13225698
Larribère L, Martens UM. Advantages and Challenges of Using ctDNA NGS to Assess the Presence of Minimal Residual Disease (MRD) in Solid Tumors. Cancers. 2021; 13(22):5698. https://doi.org/10.3390/cancers13225698
Chicago/Turabian StyleLarribère, Lionel, and Uwe M. Martens. 2021. "Advantages and Challenges of Using ctDNA NGS to Assess the Presence of Minimal Residual Disease (MRD) in Solid Tumors" Cancers 13, no. 22: 5698. https://doi.org/10.3390/cancers13225698
APA StyleLarribère, L., & Martens, U. M. (2021). Advantages and Challenges of Using ctDNA NGS to Assess the Presence of Minimal Residual Disease (MRD) in Solid Tumors. Cancers, 13(22), 5698. https://doi.org/10.3390/cancers13225698





