Uveal Melanoma Metastasis
Abstract
:Simple Summary
Abstract
1. Introduction
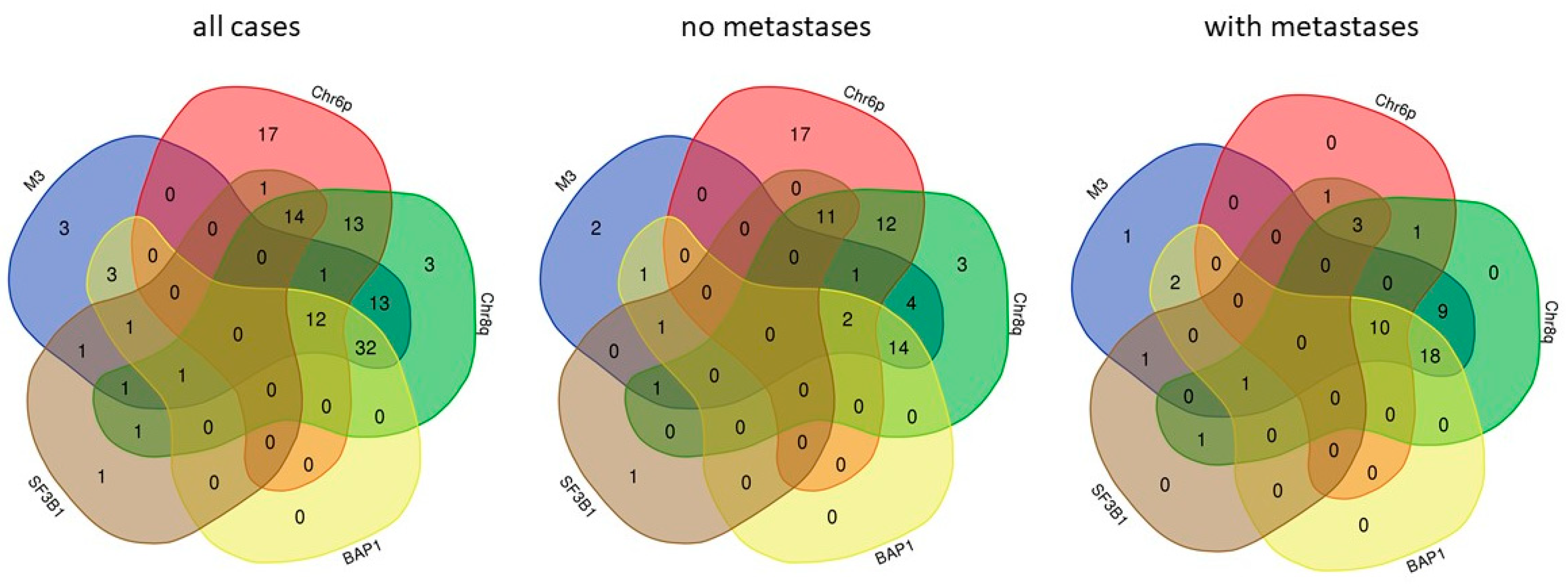
2. Metastasis Associated Molecular Characteristics of UM
3. Pathophysiology of UM Metastasis
4. Therapy of UM Metastases
5. UM Metastasis Models
6. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal melanoma. Nat. Rev. Dis. Prim. 2020, 6, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.; Gangemi, R.; Piaggio, F.; Angelini, G.; Barisione, G.; Ferrini, S.; Pfeffer, U. The biology of uveal melanoma. Cancer Metastasis Rev. 2017, 36, 109–140. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.D.; Topham, A. Incidence of uveal melanoma in the United States: 1973–1997. Ophthalmology 2003, 110, 956–961. [Google Scholar] [CrossRef]
- Virgili, G.; Gatta, G.; Ciccolallo, L.; Capocaccia, R.; Biggeri, A.; Crocetti, E.; Lutz, J.M.; Paci, E. Incidence of Uveal Melanoma in Europe. Ophthalmology 2007, 114, 2309–2315.e2. [Google Scholar] [CrossRef] [PubMed]
- Nayman, T.; Bostan, C.; Logan, P.; Burnier, M.N. Uveal Melanoma Risk Factors: A Systematic Review of Meta-Analyses. Curr. Eye Res. 2017, 42, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Lutz, J.M.; Cree, I.; Sabroe, S.; Kvist, T.K.; Clausen, L.B.; Afonso, N.; Ahrens, W.; Ballard, T.J.; Bell, J.; Cyr, D.; et al. Occupational risks for uveal melanoma results from a case-control study in nine European countries. Cancer Causes Control 2005, 16, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Shah, C.P.; Weis, E.; Lajous, M.; Shields, J.A.; Shields, C.L. Intermittent and chronic ultraviolet light exposure and uveal melanoma: A meta-analysis. Ophthalmology 2005, 112, 1599–1607. [Google Scholar] [CrossRef]
- Ferguson, R.; Vogelsang, M.; Ucisik-Akkaya, E.; Rai, K.; Pilarski, R.; Martinez, C.N.; Rendleman, J.; Kazlow, E.; Nagdimov, K.; Osman, I.; et al. Genetic markers of pigmentation are novel risk loci for uveal melanoma. Sci. Rep. 2016, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Belmar-Lopez, C.; Mancheno-Corvo, P.; Saornil, M.A.; Baril, P.; Vassaux, G.; Quintanilla, M.; Martin-Duque, P. Uveal vs. cutaneous melanoma. Origins and causes of the differences. Clin. Transl. Oncol. 2008, 10, 137–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Kooij, M.K.; Speetjens, F.M.; van der Burg, S.H.; Kapiteijn, E. Uveal versus cutaneous melanoma; same origin, very distinct tumor types. Cancers 2019, 11, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigel, D.S. Cutaneous ultraviolet exposure and its relationship to the development of skin cancer. J. Am. Acad. Dermatol. 2008, 58, S129–S132. [Google Scholar] [CrossRef] [PubMed]
- Balazs, E.A. Studies on the structure of the vitreous body. I. The absorption of ultraviolet light. Am. J. Ophthalmol. 1954, 38, 21–28. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piaggio, F.; Tozzo, V.; Bernardi, C.; Croce, M.; Puzone, R.; Viaggi, S.; Patrone, S.; Barla, A.; Coviello, D.; Jager, M.J.; et al. Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma. Cancers 2019, 11, 1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kujala, E.; Mäkitie, T.; Kivelä, T. Very Long-Term Prognosis of Patients with Malignant Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef] [Green Version]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220. [Google Scholar] [CrossRef] [Green Version]
- Roelofsen, C.D.M.; Wierenga, A.P.A.; van Duinen, S.; Verdijk, R.M.; Bleeker, J.; Marinkovic, M.; Luyten, G.P.M.; Jager, M.J. Five Decades of Enucleations for Uveal Melanoma in One Center: More Tumors with High Risk Factors, No Improvement in Survival over Time. Ocul. Oncol. Pathol. 2021, 7, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Croce, M.; Ferrini, S.; Pfeffer, U.; Gangemi, R. Targeted therapy of uveal melanoma: Recent failures and new perspectives. Cancers 2019, 11, 846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, A.; Heinzerling, L.; Bhardwaj, N.; Friedlander, P. Current melanoma treatments: Where do we stand? Cancers 2021, 13, 221. [Google Scholar] [CrossRef]
- Virgil, H. FDA Accepts Biologics License Application for Tebentafusp in Metastatic Uveal Melanoma. Available online: https://www.cancernetwork.com/view/fda-accepts-biologics-license-application-for-tebentafusp-in-metastatic-uveal-melanoma (accessed on 19 October 2021).
- Damato, B.E.; Dukes, J.; Goodall, H.; Carvajal, R.D. Tebentafusp: T cell redirection for the treatment of metastatic uveal melanoma. Cancers 2019, 11, 971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nathan, P.; Hassel, J.C.; Rutkowski, P.; Baurain, J.-F.; Butler, M.O.; Schlaak, M.; Sullivan, R.J.; Ochsenreither, S.; Dummer, R.; Kirkwood, J.M.; et al. Overall Survival Benefit with Tebentafusp in Metastatic Uveal Melanoma. N. Engl. J. Med. 2021, 385, 1196–1206. [Google Scholar] [CrossRef]
- Pelster, M.S.; Gruschkus, S.K.; Bassett, R.; Gombos, D.S.; Shephard, M.; Posada, L.; Glover, M.S.; Simien, R.; Diab, A.; Hwu, P.; et al. Nivolumab and Ipilimumab in Metastatic Uveal Melanoma: Results From a Single-Arm Phase II Study. J. Clin. Oncol. 2021, 39, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Vidal, C.; Fernandez-Diaz, D.; Fernandez-Marta, B.; Lago-Baameiro, N.; Pardo, M.; Silva, P.; Paniagua, L.; Blanco-Teijeiro, M.J.; Piñeiro, A.; Bande, M. Treatment of Metastatic Uveal Melanoma: Systematic Review. Cancers 2020, 12, 2557. [Google Scholar] [CrossRef]
- Rodrigues, M.; Mobuchon, L.; Houy, A.; Fiévet, A.; Gardrat, S.; Barnhill, R.L.; Popova, T.; Servois, V.; Rampanou, A.; Mouton, A.; et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat. Commun. 2018, 9, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Johansson, P.A.; Stark, A.; Palmer, J.M.; Bigby, K.; Brooks, K.; Rolfe, O.; Pritchard, A.L.; Whitehead, K.; Warrier, S.; Glasson, W.; et al. Prolonged stable disease in a uveal melanoma patient with germline MBD4 nonsense mutation treated with pembrolizumab and ipilimumab. Immunogenetics 2019, 71, 511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marseglia, M.; Amaro, A.; Solari, N.; Gangemi, R.; Croce, E.; Tanda, E.T.; Spagnolo, F.; Filaci, G.; Pfeffer, U.; Croce, M. How to make immunotherapy an effective therapeutic choice for uveal melanoma. Cancers 2021, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.A.; Rodriguez, D.A.; Kurtenbach, S.; Kuznetsov, J.N.; Sanchez, M.I.; Decatur, C.L.; Snyder, H.; Feun, L.G.; Livingstone, A.S.; Harbour, J.W. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef] [Green Version]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in Uveal Melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, A.R.; Ceraudo, E.; Sher, J.J.; Guan, Y.; Shoushtari, A.N.; Chang, M.T.; Zhang, J.Q.; Walczak, E.G.; Kazmi, M.A.; Taylor, B.S.; et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016, 48, 675–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, P.; Aoude, L.G.; Wadt, K.; Glasson, W.J.; Warrier, S.K.; Hewitt, A.W.; Kiilgaard, J.F.; Heegaard, S.; Isaacs, T.; Franchina, M.; et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2016, 7, 4624–4631. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Weng, L.; Bastian, B.C.; Chen, X. Functional characterization of uveal melanoma oncogenes. Oncogene 2021, 40, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Degese, M.S.; Iglesias-Bartolome, R.; Vaque, J.P.; Molinolo, A.A.; Rodrigues, M.; Zaidi, M.R.; Ksander, B.R.; Merlino, G.; Sodhi, A.; et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a Trio-regulated Rho GTPase Signaling Circuitry. Cancer Cell 2014, 25, 831–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, Q.; Dang, K.; Ma, S.; Cotton, J.L.; Yang, S.; Zhu, L.J.; Deng, A.C.; Ip, Y.T.; Johnson, R.L.; et al. YAP/TAZ Activation Drives Uveal Melanoma Initiation and Progression. Cell Rep. 2019, 29, 3200–3211. [Google Scholar] [CrossRef] [Green Version]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emley, A.; Nguyen, L.P.; Yang, S.; Mahalingam, M. Somatic mutations in GNAQ in amelanotic/hypomelanotic blue nevi. Hum. Pathol. 2011, 42, 136–140. [Google Scholar] [CrossRef]
- Livingstone, E.; Zaremba, A.; Horn, S.; Ugurel, S.; Casalini, B.; Schlaak, M.; Hassel, J.C.; Herbst, R.; Utikal, J.S.; Weide, B.; et al. GNAQ and GNA11 mutant nonuveal melanoma: A subtype distinct from both cutaneous and uveal melanoma. Br. J. Dermatol. 2020, 183, 928–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küsters-Vandevelde, H.V.N.; Klaasen, A.; Küsters, B.; Groenen, P.J.T.A.; Van Engen-Van Grunsven, I.A.C.H.; Van Dijk, M.R.C.F.; Reifenberger, G.; Wesseling, P.; Blokx, W.A.M. Activating mutations of the GNAQ gene: A frequent event in primary melanocytic neoplasms of the central nervous system. Acta Neuropathol. 2010, 119, 317–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dratviman-Storobinsky, O.; Cohen, Y.; Frenkel, S.; Pe’er, J.; Goldenberg-Cohen, N. Lack of Oncogenic GNAQ Mutations in Melanocytic Lesions of the Conjunctiva as Compared to Uveal Melanoma. Investig. Opthalmology Vis. Sci. 2010, 51, 6180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dono, M.; Angelini, G.; Cecconi, M.; Amaro, A.; Esposito, A.I.I.; Mirisola, V.; Maric, I.; Lanza, F.; Nasciuti, F.; Viaggi, S.; et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: Detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br. J. Cancer 2014, 110, 1058–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.O.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.; Maßhöfer, L.; Temming, P.; Rahmann, S.; Metz, C.; Bornfeld, N.; Van De Nes, J.; Hitpass, L.K.; Hinnebusch, A.G.; Horsthemke, B.; et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013, 45, 933–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harbour, J.W.; Roberson, E.D.O.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Liu-Smith, F.; Lu, Y. Opposite roles of BAP1 in overall survival of uveal melanoma and cutaneous melanoma. J. Clin. Med. 2020, 9, 411. [Google Scholar] [CrossRef] [Green Version]
- Royer-Bertrand, B.; Torsello, M.; Rimoldi, D.; El Zaoui, I.; Cisarova, K.; Pescini-Gobert, R.; Raynaud, F.; Zografos, L.; Schalenbourg, A.; Speiser, D.; et al. Comprehensive Genetic Landscape of Uveal Melanoma by Whole-Genome Sequencing. Am. J. Hum. Genet. 2016, 99, 1190–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yavuzyigitoglu, S.; Koopmans, A.E.; Verdijk, R.M.; Vaarwater, J.; Eussen, B.; Van Bodegom, A.; Paridaens, D.; Kiliç, E.; De Klein, A. Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology 2016, 123, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Luscan, A.; Just, P.A.; Briand, A.; Burin Des Roziers, C.; Goussard, P.; Nitschké, P.; Vidaud, M.; Avril, M.F.; Terris, B.; Pasmant, E. Uveal melanoma hepatic metastases mutation spectrum analysis using targeted next-generation sequencing of 400 cancer genes. Br. J. Ophthalmol. 2015, 99, 437–439. [Google Scholar] [CrossRef] [Green Version]
- Shain, A.H.; Bagger, M.M.; Yu, R.; Chang, D.; Liu, S.; Vemula, S.; Weier, J.F.; Wadt, K.; Heegaard, S.; Bastian, B.C.; et al. The genetic evolution of metastatic uveal melanoma. Nat. Genet. 2019, 51, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Beasley, A.B.; Ardakani, N.M.; Denisenko, E.; Calapre, L.; Jones, M.; Wood, B.A.; Warburton, L.; Forrest, A.R.R.; Gray, E.S. Intra- and intertumoral heterogeneity of liver metastases in a patient with uveal melanoma revealed by single-cell RNA sequencing. Mol. Case Stud. 2021, 7, a006111. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.A.; Yang, J.; Durante, M.A.; Shoushtari, A.N.; Moschos, S.J.; Wrzeszczynski, K.O.; Harbour, J.W.; Carvajal, R.D. Multiregional genetic evolution of metastatic uveal melanoma. Npj Genomic Med. 2021, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sottoriva, A.; Kang, H.; Ma, Z.; Graham, T.A.; Salomon, M.P.; Zhao, J.; Marjoram, P.; Siegmund, K.; Press, M.F.; Shibata, D.; et al. A big bang model of human colorectal tumor growth. Nat. Genet. 2015, 47, 209–216. [Google Scholar] [CrossRef]
- Amaro, A.; Chiara, S.; Pfeffer, U. Molecular evolution of colorectal cancer: From multistep carcinogenesis to the big bang. Cancer Metastasis Rev. 2016, 35, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Prescher, G.; Bornfeld, N.; Hirche, H.; Horsthemke, B.; Jöckel, K.H.; Becher, R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet 1996, 347, 1222–1225. [Google Scholar] [CrossRef] [PubMed]
- Prescher, G.; Bornfeld, N.; Becher, R. Two subclones in a case of uveal melanoma. Relevance of monosomy 3 and multiplication of chromosome 8q. Cancer Genet. Cytogenet. 1994, 77, 144–146. [Google Scholar] [CrossRef]
- Thornton, S.; Coupland, S.E.; Olohan, L.; Sibbring, J.S.; Kenny, J.G.; Hertz-Fowler, C.; Liu, X.; Haldenby, S.; Heimann, H.; Hussain, R.; et al. Targeted next-generation sequencing of 117 routine clinical samples provides further insights into the molecular landscape of uveal melanoma. Cancers 2020, 12, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehlers, J.P.; Worley, L.; Onken, M.D.; Harbour, J.W. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin. Cancer Res. 2005, 11, 3609–3613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piaggio, F.; Tozzo, V.; Bernardi, C.; Croce, M.; Lo Sicco, C.; Gangemi, R.; Puzone, R.; Viaggi, S.; Patrone, S.; Barla, A.; et al. How many mutations does it take to make a uveal melanoma? Arch. Clin. Exp. Ophthalmol. 2019, 1, 6–11. [Google Scholar]
- White, V.A.; Chambers, J.D.; Courtright, P.D.; Chang, W.Y.; Horsman, D.E. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer 1998, 83, 354–359. [Google Scholar] [CrossRef]
- Ehlers, J.P.; Worley, L.; Onken, M.D.; Harbour, J.W. Integrative genomic analysis of aneuploidy in uveal melanoma. Clin. Cancer Res. 2008, 14, 115–122. [Google Scholar] [CrossRef] [Green Version]
- van Weeghel, C.; Wierenga, A.P.A.A.P.A.; Versluis, M.; van Hall, T.; van der Velden, P.A.P.A.; Kroes, W.G.M.W.G.M.; Pfeffer, U.; Luyten, G.P.M.G.P.M.; Jager, M.J.M.J. Do GNAQ and GNA11 differentially affect inflammation and HLA expression in uveal melanoma? Cancers 2019, 11, 1127. [Google Scholar] [CrossRef] [Green Version]
- Volpi, E.V.; Chevret, E.; Jones, T.; Vatcheva, R.; Williamson, J.; Beck, S.; Campbell, R.D.; Goldsworthy, M.; Powis, S.H.; Ragoussis, J.; et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000, 113, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.; Parodi, F.; Diedrich, K.; Angelini, G.; Götz, C.; Viaggi, S.; Maric, I.; Coviello, D.; Pistillo, M.P.; Morabito, A.; et al. Analysis of the expression and single-nucleotide variant frequencies of the butyrophilin-like 2 gene in patients with uveal melanoma. JAMA Ophthalmol. 2016, 134, 1125–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jager, M.J.; Hurks, H.M.; Levitskaya, J.; Kiessling, R. HLA expression in uveal melanoma: There is no rule without some exception. Hum. Immunol. 2002, 63, 444–451. [Google Scholar] [CrossRef]
- Niederkorn, J.Y. Ocular immune privilege and ocular melanoma: Parallel universes or immunological plagiarism? Front. Immunol. 2012, 3, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souri, Z. Regulation of Inflammation in Uveal Melanoma; Leiden University Scholarly Publications: Leiden, The Netherlands, 2021. [Google Scholar]
- Bronkhorst, I.H.G.; Jager, M.J. Inflammation in uveal melanoma. Eye 2013, 27, 217–223. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, M.; Uschmajew, A.; Amaro, A.; Pfeffer, U. Data Fusion Techniques for the Integration of Multi-Domain Genomic Data from Uveal Melanoma. Cancers 2019, 11, 434. [Google Scholar] [CrossRef] [Green Version]
- Baradaran, P.C.; Kozovska, Z.; Furdova, A.; Smolkova, B. Targeting epigenetic modifications in uveal melanoma. Int. J. Mol. Sci. 2020, 21, 1–21. [Google Scholar]
- Field, M.G.; Kuznetsov, J.N.; Bussies, P.L.; Cai, L.Z.; Alawa, K.A.; Decatur, C.L.; Kurtenbach, S.; Harbour, J.W. BAP1 loss is associated with DNA methylomic repatterning in highly aggressive class 2 uveal melanomas. Clin. Cancer Res. 2019, 25, 5663–5673. [Google Scholar] [CrossRef] [Green Version]
- Landreville, S.; Agapova, O.A.; Matatall, K.A.; Kneass, Z.T.; Onken, M.D.; Lee, R.S.; Bowcock, A.M.; Harbour, J.W. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin. Cancer Res. 2012, 18, 408–416. [Google Scholar] [CrossRef] [Green Version]
- Souri, Z.; Jochemsen, A.G.; Versluis, M.; Wierenga, A.P.A.; Nemati, F.; van der Velden, P.A.; Kroes, W.G.M.; Verdijk, R.M.; Luyten, G.P.M.; Jager, M.J. HDAC Inhibition Increases HLA Class I Expression in Uveal Melanoma. Cancers 2020, 12, 3690. [Google Scholar] [CrossRef] [PubMed]
- Lafave, L.M.; Béguelin, W.; Koche, R.; Teater, M.; Spitzer, B.; Chramiec, A.; Papalexi, E.; Keller, M.D.; Hricik, T.; Konstantinoff, K.; et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat. Med. 2015, 21, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- Schoumacher, M.; Le Corre, S.; Houy, A.; Mulugeta, E.; Stern, M.H.; Roman-Roman, S.; Margueron, R. Uveal Melanoma Cells Are Resistant to EZH2 Inhibition Regardless of BAP1 Status. Nat. Med. 2016, 22, 577–578. [Google Scholar]
- Bakhoum, M.F.; Francis, J.H.; Agustinus, A.; Earlie, E.M.; Di Bona, M.; Abramson, D.H.; Duran, M.; Masilionis, I.; Molina, E.; Shoushtari, A.N.; et al. Loss of polycomb repressive complex 1 activity and chromosomal instability drive uveal melanoma progression. Nat. Commun. 2021, 12, 5402. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Dopierala, J.; Klaasen, A.; van Dijk, M.; Sibbring, J.; Coupland, S.E. Multiplex ligation-dependent probe amplification of uveal melanoma: Correlation with metastatic death. Investig. Ophthalmol. Vis. Sci. 2009, 50, 3048–3055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onken, M.D.; Worley, L.A.; Tuscan, M.D.; Harbour, J.W. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J. Mol. Diagnostics 2010, 12, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.D.; Chao, D.L.; Feuer, W.; Schiffman, J.; Char, D.H.; Harbour, J.W. Prognostic implications of tumor diameter in association with gene expression profile for uveal melanoma. JAMA Ophthalmol. 2016, 134, 734–740. [Google Scholar] [CrossRef] [Green Version]
- Eskelin, S.; Pyrhönen, S.; Summanen, P.; Hahka-Kemppinen, M.; Kivelä, T. Tumor doubling times in metastatic malignant melanoma of the uvea: Tumor progression before and after treatment. Ophthalmology 2000, 107, 1443–1449. [Google Scholar] [CrossRef]
- Uner, O.E.; See, T.R.O.; Szalai, E.; Grossniklaus, H.E.; Stålhammar, G. Estimation of the timing of BAP1 mutation in uveal melanoma progression. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Onken, M.D.; Blumer, K.J.; Cooper, J.A. Uveal melanoma cells use ameboid and mesenchymal mechanisms of cell motility crossing the endothelium. Mol. Biol. Cell 2021, 32, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Roszik, J.; Gombos, D.; Upshaw, J.; Sarli, V.; Meas, S.; Lucci, A.; Hall, C.; Patel, S. Pilot study of circulating tumor cells in early-stage and metastatic uveal melanoma. Cancers 2019, 11, 856. [Google Scholar] [CrossRef] [Green Version]
- Blanco, P.L.; Lim, L.A.; Miyamoto, C.; Burnier, M.N. Uveal melanoma dormancy: An acceptable clinical endpoint? Melanoma Res. 2012, 22, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Albini, A.; Mirisola, V.; Pfeffer, U. Metastasis signatures: Genes regulating tumor-microenvironment interactions predict metastatic behavior. Cancer Metastasis Rev 2008, 27, 75–83. [Google Scholar] [CrossRef]
- Mielgo, A.; Schmid, M.C. Liver tropism in cancer: The hepatic metastatic niche. Cold Spring Harb. Perspect. Med. 2020, 10, a037259. [Google Scholar] [CrossRef]
- Bol, K.F.; Mensink, H.W.; Aarntzen, E.H.J.G.; Schreibelt, G.; Keunen, J.E.E.; Coulie, P.G.; De Klein, A.; Punt, C.J.A.; Paridaens, D.; Figdor, C.G.; et al. Long overall survival after dendritic cell vaccination in metastatic uveal Melanoma Patients. Am. J. Ophthalmol. 2014, 158, 939–947. [Google Scholar] [CrossRef] [Green Version]
- Angi, M.; Kalirai, H.; Prendergast, S.; Simpson, D.; Hammond, D.E.; Madigan, M.C.; Beynon, R.J.; Coupland, S.E. In-depth proteomic profiling of the uveal melanoma secretome. Oncotarget 2016, 7, 49623–49635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niederkorn, J.Y. Immune escape mechanisms of intraocular tumors. Prog. Retin. Eye Res. 2009, 28, 329–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossniklaus, H.E.; Zhang, Q.; You, S.; McCarthy, C.; Heegaard, S.; Coupland, S.E. Metastatic ocular melanoma to the liver exhibits infiltrative and nodular growth patterns. Hum. Pathol. 2016, 57, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babchia, N.; Landreville, S.; Clément, B.; Coulouarn, C.; Mouriaux, F. The bidirectional crosstalk between metastatic uveal melanoma cells and hepatic stellate cells engenders an inflammatory microenvironment. Exp. Eye Res. 2019, 181, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Luyten, G.P.M.; Van Der Spek, C.W.; Brand, I.; Sintnicolaas, K.; De Waard-Siebinga, I.; Jager, M.J.; De Jong, P.T.V.M.; Schrier, P.I.; Luider, T.M. Expression of MAGE, gp100 and tyrosinase genes in uveal melanoma cell lines. Melanoma Res. 1998, 8, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Kan-Mitchell, J.; Liggett, P.E.; Harel, W.; Steinman, L.; Nitta, T.; Oksenberg, J.R.; Posner, M.R.; Mitchell, M.S. Lymphocytes Cytotoxic to Uveal and Skin Melanoma Cells from Peripheral Blood of Ocular Melanoma Patients. Cancer Immunol. Immunother. 1991, 33, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Ksander, B.R.; Geer, D.C.; Chen, P.W.; Salgaller, M.L.; Rubsamen, P.; Murray, T.G. Uveal melanomas contain antigenically specific and non-specific infiltrating lymphocytes. Curr. Eye Res. 1998, 17, 165–173. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, P.G.; Saban, D.R.; Dando, S.J. Immune cells in the retina and choroid: Two different tissue environments that require different defenses and surveillance. Prog. Retin. Eye Res. 2019, 70, 85–98. [Google Scholar] [CrossRef]
- Taylor, A.W.; Alard, P.; Yee, D.G.; Streilein, J.W. Aqueous humor induces transforming growth factor-β (TGF-β)-producing regulatory T-cells. Curr. Eye Res. 1997, 16, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Forrester, J.V.; Xu, H. Good news-bad news: The Yin and Yang of immune privilege in the eye. Front. Immunol. 2012, 3, 338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferguson, T.A.; Griffith, T.S. The role of Fas ligand and TNF-related apoptosis-inducing ligand (TRAIL) in the ocular immune response. Chem. Immunol. Allergy 2007, 92, 140–154. [Google Scholar] [CrossRef] [PubMed]
- Vendomèle, J.; Khebizi, Q.; Fisson, S. Cellular and molecular mechanisms of anterior chamber-associated immune deviation (ACAID): What we have learned from knockout mice. Front. Immunol. 2017, 8, 1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apte, R.S.; Sinha, D.; Mayhew, E.; Wistow, G.J.; Niederkorn, J.Y. Cutting edge: Role of macrophage migration inhibitory factor in inhibiting NK cell activity and preserving immune privilege. J. Immunol. 1998, 160, 5693–5696. [Google Scholar] [PubMed]
- Yoshida, M.; Takeuchi, M.; Streilein, J.W. Participation of pigment epithelium of iris and ciliary body in ocular immune privilege. 1. Inhibition of T-cell activation in vitro by direct cell- to-cell contact. Investig. Ophthalmol. Vis. Sci. 2000, 41, 811–821. [Google Scholar]
- Repp, A.C.; Mayhew, E.S.; Apte, S.; Niederkorn, J.Y. Human Uveal Melanoma Cells Produce Macrophage Migration-Inhibitory Factor to Prevent Lysis by NK Cells. J. Immunol. 2000, 165, 710–715. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.W.; Mellon, J.K.; Mayhew, E.; Wang, S.; He, Y.G.; Hogan, N.; Niederkorn, J.Y. Uveal melanoma expression of indoleamine 2,3-deoxygenase: Establishment of an immune privileged environment by tryptophan depletion. Exp. Eye Res. 2007, 85, 617–625. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Li, H.; Chen, P.W.; Alizadeh, H.; He, Y.; Hogan, R.N.; Niederkorn, J.Y. PD-L1 expression on human ocular cells and its possible role in regulating immune-mediated ocular inflammation. Investig. Ophthalmol. Vis. Sci. 2009, 50, 273–280. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.C.; Mehdizadeh, S.; Smith, J.; Young, A.; Mufazalov, I.A.; Mowery, C.T.; Daud, A.; Bluestone, J.A. Regulatory T cell control of systemic immunity and immunotherapy response in liver metastasis. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.R.; Kalirai, H.; Sacco, J.J.; Azevedo, R.A.; Duckworth, A.; Slupsky, J.R.; Coulson, J.M.; Coupland, S.E. Loss of BAP1 expression is associated with an immunosuppressive microenvironment in uveal melanoma, with implications for immunotherapy development. J. Pathol. 2020, 250, 420–439. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Bollin, K.; De MacEdo, M.P.; Carapeto, F.; Kim, K.B.; Roszik, J.; Wani, K.M.; Reuben, A.; Reddy, S.T.; Williams, M.D.; et al. Immune profiling of uveal melanoma identifies a potential signature associated with response to immunotherapy. J. Immunother. Cancer 2020, 8, 960. [Google Scholar] [CrossRef]
- Krishna, Y.; Acha-Sagredo, A.; Sabat-Pośpiech, D.; Kipling, N.; Clarke, K.; Figueiredo, C.R.; Kalirai, H.; Coupland, S.E. Transcriptome profiling reveals new insights into the immune microenvironment and upregulation of novel biomarkers in metastatic uveal melanoma. Cancers 2020, 12, 2832. [Google Scholar] [CrossRef] [PubMed]
- Krishna, Y.; McCarthy, C.; Kalirai, H.; Coupland, S.E. Inflammatory cell infiltrates in advanced metastatic uveal melanoma. Hum. Pathol. 2017, 66, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Dallinga, J.W.; Moonen, E.J.; Dumoulin, J.C.; Evers, J.L.; Geraedts, J.P.; Kleinjans, J.C. Decreased human semen quality and organochlorine compounds in blood. Hum Reprod 2002, 17, 1973–1979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, J.; Siarov, J.; Kiffin, R.; Mölne, J.; Mattsson, J.; Naredi, P.; Olofsson Bagge, R.; Martner, A.; Lindnér, P. Presence of tumor-infiltrating CD8+ T cells and macrophages correlates to longer overall survival in patients undergoing isolated hepatic perfusion for uveal melanoma liver metastasis. Oncoimmunology 2020, 9, 1854519. [Google Scholar] [CrossRef]
- Lachota, M.; Lennikov, A.; Malmberg, K.J.; Zagozdzon, R. Bioinformatic Analysis Reveals Central Role for Tumor-Infiltrating Immune Cells in Uveal Melanoma Progression. J. Immunol. Res. 2021, 9920234. [Google Scholar] [CrossRef] [PubMed]
- Tosi, A.; Cappellesso, R.; Dei Tos, A.P.; Rossi, V.; Aliberti, C.; Pigozzo, J.; Fabozzi, A.; Sbaraglia, M.; Blandamura, S.; Del Bianco, P.; et al. The immune cell landscape of metastatic uveal melanoma correlates with overall survival. J. Exp. Clin. Cancer Res. 2021, 40, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Schinzari, G.; Rossi, E.; Cassano, A.; Dadduzio, V.; Quirino, M.; Pagliara, M.; Blasi, M.A.; Barone, C. Cisplatin, dacarbazine and vinblastine as first line chemotherapy for liver metastatic uveal melanoma in the era of immunotherapy: A single institution phase II study. Melanoma Res. 2017, 27, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Seedor, R.S.; Eschelman, D.J.; Gonsalves, C.F.; Adamo, R.D.; Orloff, M.; Amjad, A.; Sharpe-Mills, E.; Chervoneva, I.; Shields, C.L.; Shields, J.A.; et al. An outcome assessment of a single institution’s longitudinal experience with uveal melanoma patients with liver metastasis. Cancers 2020, 12, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowcroft, A.; Loveday, B.P.T.; Thomson, B.N.J.; Banting, S.; Knowles, B. Systematic review of liver directed therapy for uveal melanoma hepatic metastases. Hpb 2020, 22, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Mariani, P.; Piperno-Neumann, S.; Servois, V.; Berry, M.G.; Dorval, T.; Plancher, C.; Couturier, J.; Levy-Gabriel, C.; Lumbroso-Le Rouic, L.; Desjardins, L.; et al. Surgical management of liver metastases from uveal melanoma: 16 years’ experience at the Institut Curie. Eur. J. Surg. Oncol. 2009, 35, 1192–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, K.; Sullivan, K.; Berd, D.; Mastrangelo, M.J.; Shields, C.L.; Shields, J.A.; Sato, T. Chemoembolization of the hepatic artery with BCNU for metastatic uveal melanoma: Results of a phase II study. Melanoma Res. 2005, 15, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Edelhauser, G.; Schicher, N.; Berzaczy, D.; Beitzke, D.; Höeller, C.; Lammer, J.; Funovics, M. Fotemustine chemoembolization of hepatic metastases from uveal melanoma: A retrospective single-center analysis. Am. J. Roentgenol. 2012, 199, 1387–1392. [Google Scholar] [CrossRef]
- Gonsalves, C.F.; Eschelman, D.J.; Sullivan, K.L.; Anne, P.R.; Doyle, L.; Sato, T. Radioembolization as salvage therapy for hepatic metastasis of uveal melanoma: A single-institution experience. Am. J. Roentgenol. 2011, 196, 468–473. [Google Scholar] [CrossRef]
- Eldredge-Hindy, H.; Ohri, N.; Anne, P.R.; Eschelman, D.; Gonsalves, C.; Intenzo, C.; Bar-Ad, V.; Dicker, A.; Doyle, L.; Li, J.; et al. Yttrium-90 microsphere brachytherapy for liver metastases from uveal melanoma clinical outcomes and the predictive value of fluorodeoxyglucose positron emission tomography. Am. J. Clin. Oncol. Cancer Clin. Trials 2016, 39, 189–195. [Google Scholar] [CrossRef]
- Ben-Shabat, I.; Belgrano, V.; Hansson, C.; Olofsson Bagge, R. The effect of perfusate buffering on toxicity and response in isolated hepatic perfusion for uveal melanoma liver metastases. Int. J. Hyperth. 2017, 33, 483–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varghese, S.; Xu, H.; Bartlett, D.; Hughes, M.; Pingpank, J.F.; Beresnev, T.; Alexander, H.R. Isolated hepatic perfusion with high-dose melphalan results in immediate alterations in tumor gene expression in patients with metastatic ocular melanoma. Ann. Surg. Oncol. 2010, 17, 1870–1877. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, M.E.; Terai, M.; Eschelman, D.J.; Gonsalves, C.F.; Chervoneva, I.; Shields, J.A.; Shields, C.L.; Yamamoto, A.; Sullivan, K.L.; Laudadio, M.; et al. Double-blinded, randomized phase II study using embolization with or without granulocyte-macrophage colony-stimulating factor in uveal melanoma with hepatic metastases. J. Vasc. Interv. Radiol. 2015, 26, 523–532.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spagnolo, F.; Grosso, M.; Picasso, V.; Tornari, E.; Pesce, M.; Queirolo, P. Treatment of metastatic uveal melanoma with intravenous fotemustine. Melanoma Res. 2013, 23, 196–198. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.H.; Boru, G.; Massengill, J.; Salem, M.M.; Davidorf, F.H. MET oncogene inhibition as a potential target of therapy for uveal melanomas. Investig. Ophthalmol. Vis. Sci. 2010, 51, 3333–3339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luke, J.J.; Olson, D.J.; Allred, J.B.; Strand, C.A.; Bao, R.; Zha, Y.; Carll, T.; Labadie, B.W.; Bastos, B.R.; Butler, M.O.; et al. Randomized Phase II trial and tumor mutational spectrum analysis from cabozantinib versus chemotherapy in metastatic uveal melanoma (Alliance A091201). Clin. Cancer Res. 2020, 26, 804–811. [Google Scholar] [CrossRef] [Green Version]
- Carvajal, R.D.; Sosman, J.A.; Quevedo, J.F.; Milhem, M.M.; Joshua, A.M.; Kudchadkar, R.R.; Linette, G.P.; Gajewski, T.F.; Lutzky, J.; Lawson, D.H.; et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: A randomized clinical trial. JAMA J. Am. Med. Assoc. 2014, 311, 2397–2405. [Google Scholar] [CrossRef] [PubMed]
- Mahipal, A.; Tijani, L.; Chan, K.; Laudadio, M.; Mastrangelo, M.J.; Sato, T. A pilot study of sunitinib malate in patients with metastatic uveal melanoma. Melanoma Res. 2012, 22, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Piperno-Neumann, S.; Diallo, A.; Etienne-Grimaldi, M.; Bidard, F.; Rodrigues, M.; Plancher, C.; Mariani, P.; Cassoux, N.; Decaudin, D.; Asselain, B.; et al. Phase II Trial of Bevacizumab in Combination With Temozolomide as First-Line Treatment in Patients With Metastatic Uveal Melanoma. Oncologist 2016, 21, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvajal, R.D.; Piperno-Neumann, S.; Kapiteijn, E.; Chapman, P.B.; Frank, S.; Joshua, A.M.; Piulats, J.M.; Wolter, P.; Cocquyt, V.; Chmielowski, B.; et al. Selumetinib in combination with dacarbazine in patients with metastatic uveal melanoma: A Phase III, Multicenter, Randomized Trial (SUMIT). J. Clin. Oncol. 2018, 36, 1232–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyrhönen, S.; Hahka-Kemppinen, M.; Muhonen, T.; Nikkanen, V.; Eskelin, S.; Summanen, P.; Tarkkanen, A.; Kivelä, T. Chemoimmunotherapy with bleomycin, vincristine, lomustine, dacarbazine (BOLD), and human leukocyte interferon for metastatic uveal melanoma. Cancer 2002, 95, 2366–2372. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Schinzari, G.; Maiorano, B.A.; Indellicati, G.; Di Stefani, A.; Pagliara, M.M.; Fragomeni, S.M.; De Luca, E.V.; Sammarco, M.G.; Garganese, G.; et al. Efficacy of immune checkpoint inhibitors in different types of melanoma. Hum. Vaccines Immunother. 2020, 11, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Maio, M.; Danielli, R.; Chiarion-Sileni, V.; Pigozzo, J.; Parmiani, G.; Ridolfi, R.; De Rosa, F.; Del Vecchio, M.; Di Guardo, L.; Queirolo, P.; et al. Efficacy and safety of ipilimumab in patients with pre-treated, uveal melanoma. Ann. Oncol. 2013, 24, 2911–2915. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Pagliara, M.M.; Orteschi, D.; Dosa, T.; Sammarco, M.G.; Caputo, C.G.; Petrone, G.; Rindi, G.; Zollino, M.; Blasi, M.A.; et al. Pembrolizumab as first-line treatment for metastatic uveal melanoma. Cancer Immunol. Immunother. 2019, 68, 1179–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piulats, J.M.; Espinosa, E.; de la Cruz Merino, L.; Varela, M.; Alonso Carrión, L.; Martín-Algarra, S.; López Castro, R.; Curiel, T.; Rodríguez-Abreu, D.; Redrado, M.; et al. Nivolumab Plus Ipilimumab for Treatment-Naïve Metastatic Uveal Melanoma: An Open-Label, Multicenter, Phase II Trial by the Spanish Multidisciplinary Melanoma Group (GEM-1402). J. Clin. Oncol. 2021, 39, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.; Moreno, R.; Gil-Martin, M.; Cascallo, M.; De Olza, M.O.; Cuadra, C.; Piulats, J.M.; Navarro, V.; Domenech, M.; Alemany, R.; et al. A Phase 1 trial of oncolytic adenovirus ICOVIR-5 administered intravenously to cutaneous and uveal melanoma patients. Hum. Gene Ther. 2019, 30, 352–364. [Google Scholar] [CrossRef]
- Ny, L.; Jespersen, H.; Karlsson, J.; Alsén, S.; Filges, S.; All-Eriksson, C.; Andersson, B.; Carneiro, A.; Helgadottir, H.; Levin, M.; et al. The PEMDAC phase 2 study of pembrolizumab and entinostat in patients with metastatic uveal melanoma. Nat. Commun. 2021, 12, 1–10. [Google Scholar] [CrossRef]
- Sussman, T.A.; Funchain, P.; Singh, A. Clinical Trials in Metastatic Uveal Melanoma: Current Status. Ocul. Oncol. Pathol. 2020, 6, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Orloff, M. Clinical trials in metastatic uveal melanoma: Immunotherapy. Ocul. Oncol. Pathol. 2021, 7, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Farach-Carson, M.C.; Jia, X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014, 32, 1256–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornabaio, G.; Barnhill, R.L.; Lugassy, C.; Bentolila, L.A.; Cassoux, N.; Roman-Roman, S.; Alsafadi, S.; Del Bene, F. Angiotropism and extravascular migratory metastasis in cutaneous and uveal melanoma progression in a zebrafish model. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fiorentzis, M.; Viestenz, A.; Siebolts, U.; Seitz, B.; Coupland, S.E.; Heinzelmann, J. The potential use of electrochemotherapy in the treatment of uveal melanoma: In vitro results in 3D tumor cultures and in vivo results in a chick embryo model. Cancers 2019, 11, 1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, J.; Emmons, M.F.; Faião-Flores, F.; Aplin, A.E.; Harbour, J.W.; Licht, J.D.; Wink, M.R.; Smalley, K.S.M. Decitabine limits escape from MEK inhibition in uveal melanoma. Pigment Cell Melanoma Res. 2020, 33, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Jager, M.J. Animal Eye Models for Uveal Melanoma. Ocul. Oncol. Pathol. 2015, 1, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, A.R.; Ran, L.; Guan, Y.; Sher, J.J.; Hitchman, T.D.; Zhang, J.Q.; Hwang, C.; Walzak, E.G.; Shoushtari, A.N.; Monette, S.; et al. GNA11 Q209L Mouse Model Reveals RasGRP3 as an Essential Signaling Node in Uveal Melanoma. Cell Rep. 2018, 22, 2455–2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.Y.; Deatherage, D.E.; Rodriguez, B.A.; Liyanarachchi, S.; Weng, Y.I.; Zuo, T.; Liu, J.; Cheng, A.S.; Huang, T.H. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res 2009, 69, 5936–5945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, J.R.; Yoo, J.H.; Shin, D.; Odelberg, S.J. Mouse models of uveal melanoma: Strengths, weaknesses, and future directions. Pigment Cell Melanoma Res. 2020, 33, 264–278. [Google Scholar] [CrossRef]
- Huang, J.L.Y.; Urtatiz, O.; Van Raamsdonk, C.D. Oncogenic G protein GNAQ induces uveal melanoma and intravasation in mice. Cancer Res. 2015, 75, 3384–3397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hitchman, T.D.; Bayshtok, G.; Ceraudo, E.; Moore, A.R.; Lee, C.; Jia, R.; Wang, N.; Pachai, M.R.; Shoushtari, A.N.; Francis, J.H.; et al. Combined Inhibition of Gaq and MEK Enhances Therapeutic Efficacy in Uveal Melanoma. Clin. Cancer Res. 2021, 27, 1476–1490. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.; Nilsson, L.M.; Mitra, S.; Alsén, S.; Shelke, G.V.; Sah, V.R.; Forsberg, E.M.V.; Stierner, U.; All-Eriksson, C.; Einarsdottir, B.; et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [Green Version]
- van der Ent, W.; Burrello, C.; Teunisse, A.F.A.S.; Ksander, B.R.; van der Velden, P.A.; Jager, M.J.; Jochemsen, A.G.; Snaar-Jagalska, B.E. Modeling of human uveal melanoma in zebrafish xenograft embryos. Invest. Ophthalmol. Vis. Sci. 2014, 55, 6612–6622. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.E.; Henle, A.M.; Amsterdam, A.; Hagen, H.R.; Lees, J.A. Uveal melanoma driver mutations in GNAQ/11 yield numerous changes in melanocyte biology. Pigment Cell Melanoma Res. 2018, 31, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Pandiani, C.; Strub, T.; Nottet, N.; Cheli, Y.; Gambi, G.; Bille, K.; Husser, C.; Dalmasso, M.; Béranger, G.; Lassalle, S.; et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ. 2021, 28, 1990–2000. [Google Scholar] [CrossRef]
- Lapadula, D.; Farias, E.; Randolph, C.E.; Purwin, T.J.; McGrath, D.; Charpentier, T.H.; Zhang, L.; Wu, S.; Terai, M.; Sato, T.; et al. Effects of oncogenic Gαq and Gα11 inhibition by FR900359 in uveal melanoma. Mol. Cancer Res. 2019, 17, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Schrage, R.; Schmitz, A.L.; Gaffal, E.; Annala, S.; Kehraus, S.; Wenzel, D.; Büllesbach, K.M.; Bald, T.; Inoue, A.; Shinjo, Y.; et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun. 2015, 6, 10156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onken, M.D.; Makepeace, C.M.; Kaltenbronn, K.M.; Kanai, S.M.; Todd, T.D.; Wang, S.; Broekelmann, T.J.; Rao, P.K.; Cooper, J.A.; Blumer, K.J. Targeting nucleotide exchange to inhibit constitutively active G protein a subunits in cancer cells. Sci. Signal. 2018, 11, eaao6852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proietti, I.; Skroza, N.; Bernardini, N.; Tolino, E.; Balduzzi, V.; Marchesiello, A.; Michelini, S.; Volpe, S.; Mambrin, A.; Mangino, G.; et al. Mechanisms of acquired BRAF inhibitor resistance in melanoma: A systematic review. Cancers 2020, 12, 2801. [Google Scholar] [CrossRef]
- Lutzky, J.; Feun, L.G.; Magallanes, N.; Kwon, D.; Harbour, J.W. NCT04552223: A phase II study of nivolumab plus BMS-986016 (relatlimab) in patients with metastatic uveal melanoma (UM) (CA224-094). J. Clin. Oncol. 2021, 39, TPS9590. [Google Scholar] [CrossRef]
- Di Giacomo, A.M.; Covre, A.; Finotello, F.; Rieder, D.; Danielli, R.; Sigalotti, L.; Giannarelli, D.; Petitprez, F.; Lacroix, L.; Valente, M.; et al. Guadecitabine plus ipilimumab in unresectable melanoma: The NIBIT-M4 clinical trial. Clin. Cancer Res. 2019, 25, 7351–7362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Author | Treatment | Setting | N. pts | Objective Response (%) | PFS (m) | OS (m) |
|---|---|---|---|---|---|---|
| Spagnolo, F. et al. [124] | fotemustine | Naïve | 25 | 8 | - | 13.9 |
| Schinzari, G. et al. [113] | cisplatin + dacarbazine + vinblastine | Naïve | 25 | 20 | 5.5 | 13.0 |
| Luke, J.J. et al. [126] | cabozantinib | Pretreated | 31 | 0 | 2 | 6.4 |
| Carvajal, R.D. et al. [127] | selumetinib | Naïve and pretreated | 69 | 14 | 4 | 11.8 |
| Mahipal, A. et al. [128] | sunitinib | Naïve and pretreated | 20 | 5 | 4.2 | 8.2 |
| Piperno-Neumann, S. et al. [129] | bevacizumab + temozolomide | Naïve | 36 | 0 | 3 | 10 |
| Carvajal, R.D. et al. [130] | selumetinib + dacarbazine | Naïve | 97 | 3 | 2.8 | - |
| Pyrhönen, S. et al. [131] | bleomycin + vincristine + lomustine + dacarbazine + IFN | Naïve and pretreated | 22 | 15 | 4 | 12 |
| Maio, M. et al. [133] | ipilimumab | pretreated | 84 | 5 | 3.6 | 6.0 |
| Rossi, E. et al. [134] | pembrolizumab | Naïve | 17 | 11.7 | 3.8 | nr |
| Piulats, J.M. et al. [136] | ipilimumab + nivoluimab | Naïve | 52 | 11.5 | 3 | 12.7 |
| Bol, K.F. et al. [86] | dendritic cell vaccination | Naïve and pretreated | 14 | - | - | 19.2 |
| Nathan P. et al. [22] | tebentafusp | Naïve | 252 | 9 | 3.3 | 21.7 |
| Ny, L. et al. [137] | pembrolizumab + etinostat | Naïve and pretreated | 29 | 14 | 2.1 | 13.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, E.; Croce, M.; Reggiani, F.; Schinzari, G.; Ambrosio, M.; Gangemi, R.; Tortora, G.; Pfeffer, U.; Amaro, A. Uveal Melanoma Metastasis. Cancers 2021, 13, 5684. https://doi.org/10.3390/cancers13225684
Rossi E, Croce M, Reggiani F, Schinzari G, Ambrosio M, Gangemi R, Tortora G, Pfeffer U, Amaro A. Uveal Melanoma Metastasis. Cancers. 2021; 13(22):5684. https://doi.org/10.3390/cancers13225684
Chicago/Turabian StyleRossi, Ernesto, Michela Croce, Francesco Reggiani, Giovanni Schinzari, Marianna Ambrosio, Rosaria Gangemi, Giampaolo Tortora, Ulrich Pfeffer, and Adriana Amaro. 2021. "Uveal Melanoma Metastasis" Cancers 13, no. 22: 5684. https://doi.org/10.3390/cancers13225684
APA StyleRossi, E., Croce, M., Reggiani, F., Schinzari, G., Ambrosio, M., Gangemi, R., Tortora, G., Pfeffer, U., & Amaro, A. (2021). Uveal Melanoma Metastasis. Cancers, 13(22), 5684. https://doi.org/10.3390/cancers13225684








