Identification of Risk Loci for Radiotoxicity in Prostate Cancer by Comprehensive Genotyping of TGFB1 and TGFBR1
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
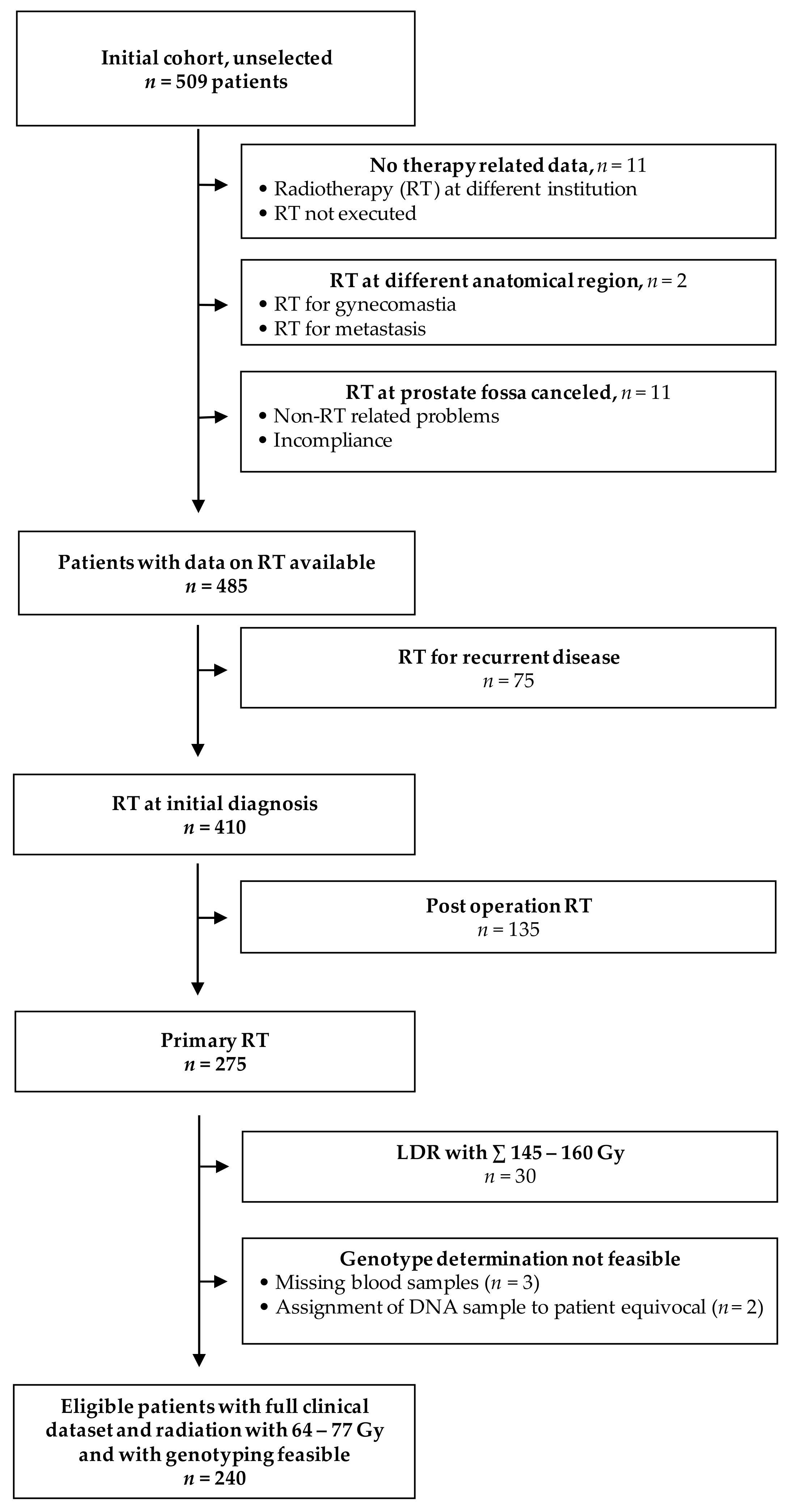
2.1. Patient Cohort
2.2. Administration of Radiotherapy
2.3. Scoring of Radiation Toxicity
2.4. Selection and Typing of Genetic Polymorphisms
2.5. Assessment of DNA Repair Capacity
3. Results
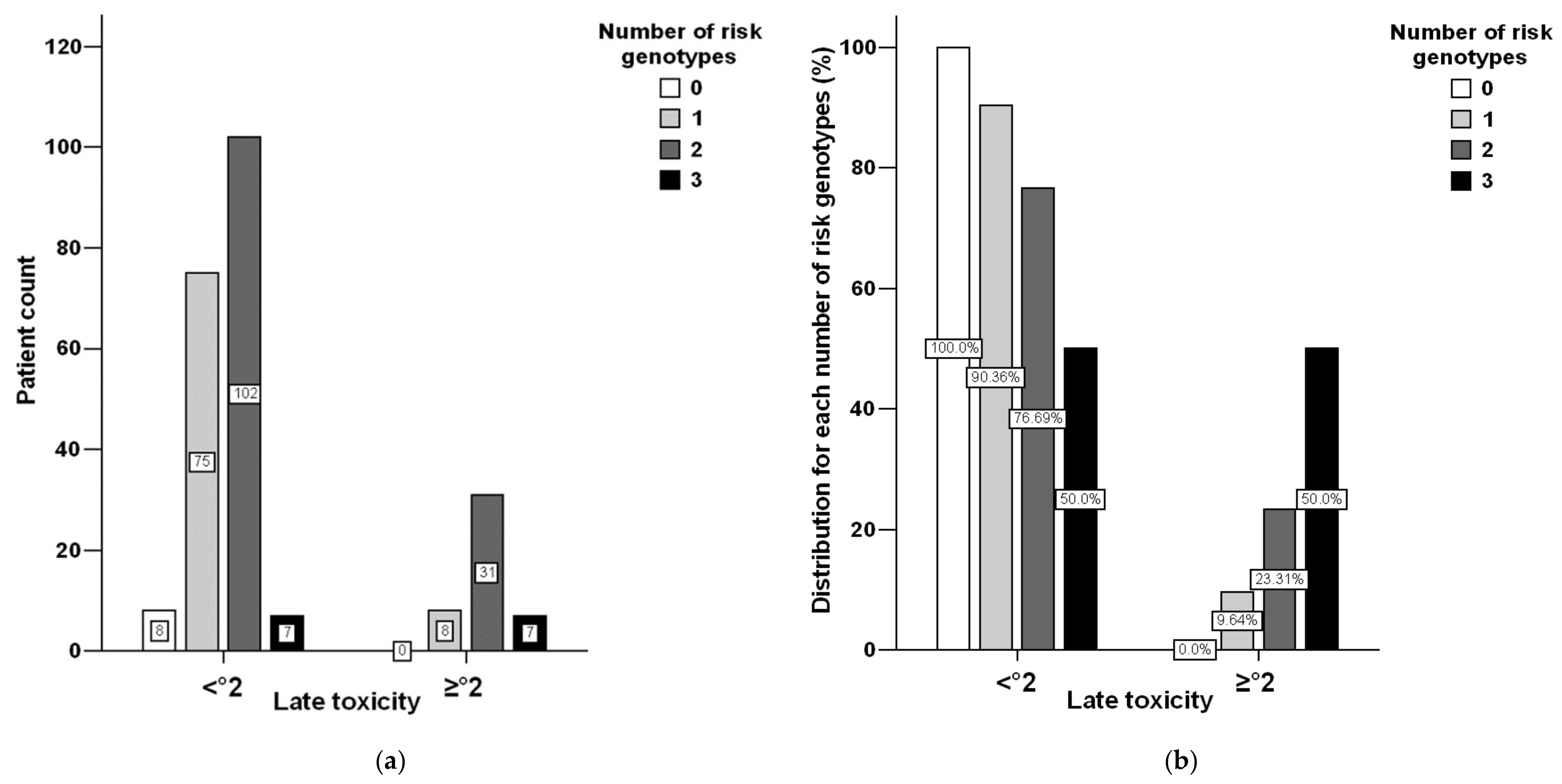
3.1. Toxicity Distribution
3.2. Genotyping Performance
3.3. Genetic Polymorphisms and Radiation Toxicity
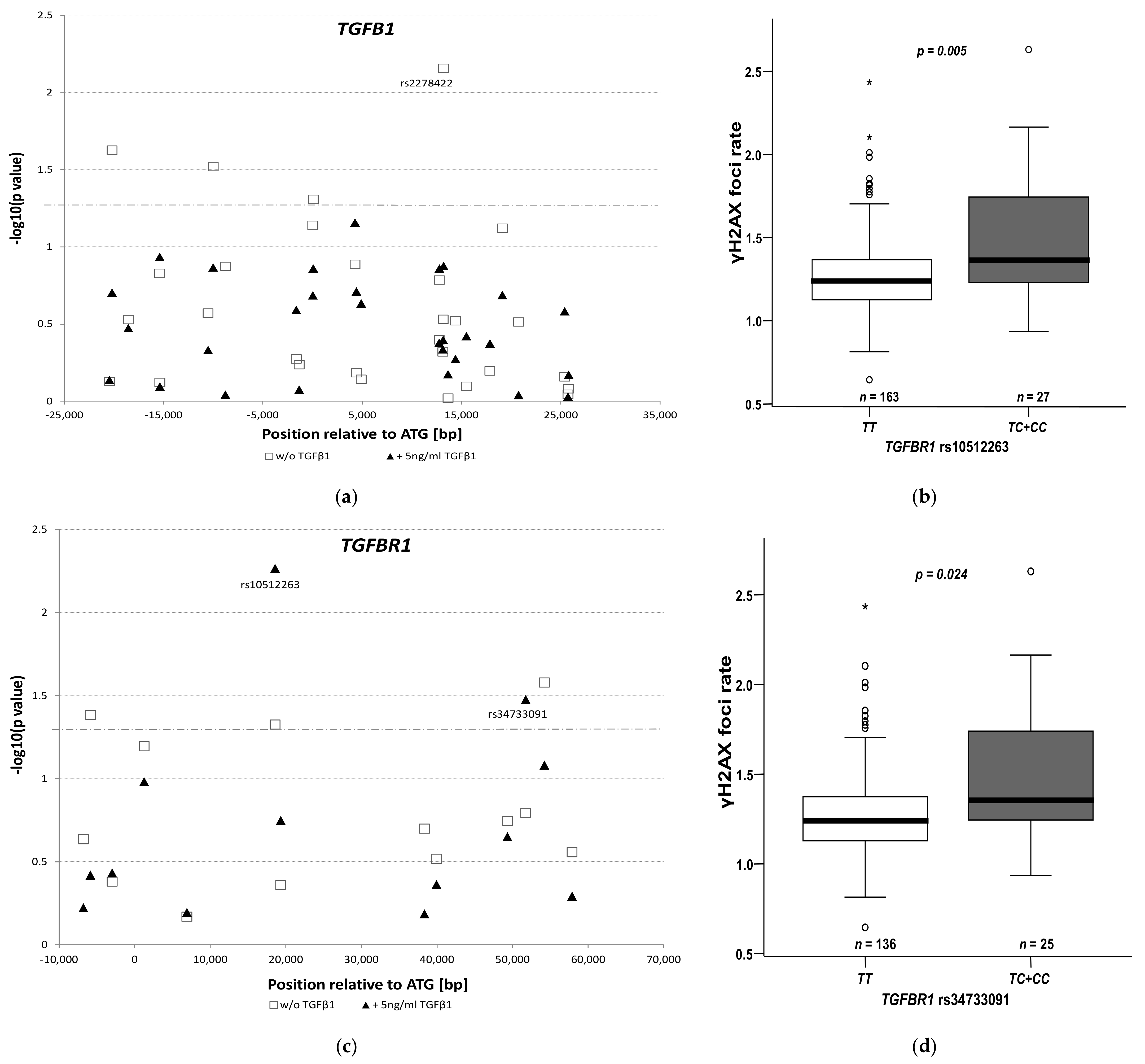
3.4. Impact of rs10512263 on DNA Repair
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Mph, K.D.M.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Mohler, J.L.; Antonarakis, E.S.; Armstrong, A.J.; D’Amico, A.V.; Davis, B.J.; Dorff, T.; Eastham, J.A.; Enke, C.A.; Farrington, T.A.; Higano, C.S.; et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 479–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, D.; Garmo, H.; Lissbrant, I.F.; Widmark, A.; Pettersson, A.; Gunnlaugsson, A.; Adolfsson, J.; Bratt, O.; Nilsson, P.; Stattin, P. Prostate Cancer Death After Radiotherapy or Radical Prostatectomy: A Nationwide Population-based Observational Study. Eur. Urol. 2017, 73, 502–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lardas, M.; Liew, M.; Bergh, R.C.V.D.; De Santis, M.; Bellmunt, J.; Broeck, T.V.D.; Cornford, P.; Cumberbatch, M.G.; Fossati, N.; Gross, T.; et al. Quality of Life Outcomes after Primary Treatment for Clinically Localised Prostate Cancer: A Systematic Review. Eur. Urol. 2017, 72, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Donovan, J.L.; Hamdy, F.C.; Lane, J.A.; Mason, M.; Metcalfe, C.; Walsh, E.; Blazeby, J.; Peters, T.; Holding, P.; Bonnington, S.; et al. Patient-Reported Outcomes after Monitoring, Surgery, or Radiotherapy for Prostate Cancer. N. Engl. J. Med. 2016, 375, 1425–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelefsky, M.J.; Levin, E.J.; Hunt, M.; Yamada, Y.; Shippy, A.M.; Jackson, A.; Amols, H.I. Incidence of Late Rectal and Urinary Toxicities After Three-Dimensional Conformal Radiotherapy and Intensity-Modulated Radiotherapy for Localized Prostate Cancer. Int. J. Radiat. Oncol. 2008, 70, 1124–1129. [Google Scholar] [CrossRef]
- Uhl, M.; Herfarth, K.; Eble, M.J.; Pinkawa, M.; Van Triest, B.; Kalisvaart, R.; Weber, D.C.; Miralbell, R.; Song, D.Y.; Deweese, T.L. Absorbable hydrogel spacer use in men undergoing prostate cancer radiotherapy: 12 month toxicity and proctoscopy results of a prospective multicenter phase II trial. Radiat. Oncol. 2014, 9, 96. [Google Scholar] [CrossRef] [Green Version]
- Andreassen, C.N.; Alsner, J.; Overgaard, J. Does variability in normal tissue reactions after radiotherapy have a genetic basis – where and how to look for it? Radiother. Oncol. 2002, 64, 131–140. [Google Scholar] [CrossRef]
- Azria, D.; Lapierre, A.; Gourgou, S.; De Ruysscher, D.; Colinge, J.; Lambin, P.; Brengues, M.; Ward, T.; Bentzen, S.M.; Thierens, H.; et al. Data-Based Radiation Oncology: Design of Clinical Trials in the Toxicity Biomarkers Era. Front. Oncol. 2017, 7, 83. [Google Scholar] [CrossRef] [Green Version]
- De Langhe, S.; De Meerleer, G.; De Ruyck, K.; Ost, P.; Fonteyne, V.; De Neve, W.; Thierens, H. Integrated models for the prediction of late genitourinary complaints after high-dose intensity modulated radiotherapy for prostate cancer: Making informed decisions. Radiother. Oncol. 2014, 112, 95–99. [Google Scholar] [CrossRef]
- Rosenstein, B.S. Identification of SNPs associated with susceptibility for development of adverse reactions to radiotherapy. Pharmacogenomics 2011, 12, 267–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farhood, B.; Khodamoradi, E.; Hoseini-Ghahfarokhi, M.; Motevaseli, E.; Mahyari, H.M.; Musa, A.E.; Najafi, M. TGF-β in radiotherapy: Mechanisms of tumor resistance and normal tissues injury. Pharmacol. Res. 2020, 155, 104745. [Google Scholar] [CrossRef] [PubMed]
- Barcellos-Hoff, M.H.; Derynck, R.; Tsang, M.L.; A Weatherbee, J. Transforming growth factor-beta activation in irradiated murine mammary gland. J. Clin. Investig. 1994, 93, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Anscher, M.; Crocker, I.R.; Jirtle, R.L. Transforming growth factor-beta 1 expression in irradiated liver. Radiat. Res. 1990, 122, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Aula, H.; Skyttä, T.; Tuohinen, S.; Luukkaala, T.; Hämäläinen, M.; Virtanen, V.; Raatikainen, P.; Moilanen, E.; Kellokumpu-Lehtinen, P.-L. Transforming growth factor beta 1 levels predict echocardiographic changes at three years after adjuvant radiotherapy for breast cancer. Radiat. Oncol. 2019, 14, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grainger, D.J. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum. Mol. Genet. 1999, 8, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Liao, Z.; Liu, Z.; Wang, L.-E.; Tucker, S.L.; Mao, L.; Wang, X.S.; Martel, M.; Komaki, R.; Cox, J.D.; et al. Single Nucleotide Polymorphism at rs1982073:T869C of the TGFβ1 Gene Is Associated With the Risk of Radiation Pneumonitis in Patients With Non–Small-Cell Lung Cancer Treated With Definitive Radiotherapy. J. Clin. Oncol. 2009, 27, 3370–3378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossberg, A.; Lei, X.; Xu, T.; Shaitelman, S.F.; Hoffman, K.E.; Bloom, E.S.; Stauder, M.C.; Tereffe, W.; Schlembach, P.J.; Woodward, W.; et al. Association of Transforming Growth Factor β Polymorphism C−509T With Radiation-Induced Fibrosis Among Patients With Early-Stage Breast Cancer. JAMA Oncol. 2018, 4, 1751–1757. [Google Scholar] [CrossRef]
- Damaraju, S.; Murray, D.; Dufour, J.; Carandang, D.; Myrehaug, S.; Fallone, B.G.; Field, C.; Greiner, R.; Hanson, J.; Cass, C.E.; et al. Association of DNA Repair and Steroid Metabolism Gene Polymorphisms with Clinical Late Toxicity in Patients Treated with Conformal Radiotherapy for Prostate Cancer. Clin. Cancer Res. 2006, 12, 2545–2554. [Google Scholar] [CrossRef] [Green Version]
- De Langhe, S.; De Ruyck, K.; Ost, P.; Fonteyne, V.; Werbrouck, J.; De Meerleer, G.; De Neve, W.; Thierens, H. Acute Radiation-Induced Nocturia in Prostate Cancer Patients Is Associated With Pretreatment Symptoms, Radical Prostatectomy, and Genetic Markers in the TGFβ1 Gene. Int. J. Radiat. Oncol. 2013, 85, 393–399. [Google Scholar] [CrossRef]
- Peters, C.A.; Stock, R.G.; Cesaretti, J.A.; Atencio, D.P.; Peters, S.; Burri, R.J.; Stone, N.N.; Ostrer, H.; Rosenstein, B.S. TGFB1 Single Nucleotide Polymorphisms Are Associated With Adverse Quality of Life in Prostate Cancer Patients Treated With Radiotherapy. Int. J. Radiat. Oncol. 2008, 70, 752–759. [Google Scholar] [CrossRef] [PubMed]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef] [Green Version]
- The 1000 Genomes Project Consortium; Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hille, A.; Schmidberger, H.; Hermann, R.M.; Christiansen, H.; Saile, B.; Pradier, O.; Hess, C.F. A phase III randomized, placebo-controlled, double-blind study of misoprostol rectal suppositories to prevent acute radiation proctitis in patients with prostate cancer. Int. J. Radiat. Oncol. 2005, 63, 1488–1493. [Google Scholar] [CrossRef] [PubMed]
- Hille, A.; Schmidberger, H.; Töws, N.; Weiss, E.; Vorwerk, H.; Hess, C.F. The Impact of Varying Volumes in Rectal Balloons on Rectal Dose Sparing in Conformal Radiation Therapy of Prostate Cancer. Strahlentherapie und Onkologie 2005, 181, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2004, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muslimović, A.; Ismail, I.H.; Gao, Y.; Hammarsten, O. An optimized method for measurement of gamma-H2AX in blood mononuclear and cultured cells. Nat. Protoc. 2008, 3, 1187–1193. [Google Scholar] [CrossRef]
- I Epstein, J.; Allsbrook, W.C.; Amin, M.B.; Egevad, L.L. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2005, 29, 1228–1242. [Google Scholar] [CrossRef] [Green Version]
- Bouquet, F.; Pal, A.; Pilones, K.A.; Demaria, S.; Hann, B.; Akhurst, R.; Babb, J.; Lonning, S.M.; Dewyngaert, J.K.; Formenti, S.C.; et al. TGFβ1 Inhibition Increases the Radiosensitivity of Breast Cancer Cells In Vitro and Promotes Tumor Control by Radiation In Vivo. Clin. Cancer Res. 2011, 17, 6754–6765. [Google Scholar] [CrossRef] [Green Version]
- Scollen, S.; Luccarini, C.; Baynes, C.; Driver, K.; Humphreys, M.K.; Garcia-Closas, M.; Figueroa, J.; Lissowska, J.; Pharoah, P.D.; Easton, D.F.; et al. TGF-β Signaling Pathway and Breast Cancer Susceptibility. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1112–1119. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Miao, L.; Jin, G.; Ren, C.; Ke, Q.; Qian, Y.; Dong, M.; Li, H.; Zhang, Q.; Ding, Y.; et al. TGFBR1tagging SNPs and gastric cancer susceptibility: A two-stage case-control study in chinese population. Mol. Carcinog. 2012, 53, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Y.-J.; Zheng, L.-Y.; Jia, Y.-M.; Chen, Y.-L.; Chen, L.; Liu, D.-G.; Li, X.-H.; Guo, H.-Y.; Sun, Y.-L.; et al. Genetic Polymorphisms of TGFB1, TGFBR1, SNAI1 and TWIST1 Are Associated with Endometrial Cancer Susceptibility in Chinese Han Women. PLoS ONE 2016, 11, e0155270. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Lopez, K.; Murnane, J.; Humphrey, T.; Barcellos-Hoff, M.H. Misrepair in Context: TGFβ Regulation of DNA Repair. Front. Oncol. 2019, 9, 799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.-R.; Lee, J.; An, Y.S.; Jin, Y.B.; Park, I.-C.; Chung, E.; Shin, I.; Barcellos-Hoff, M.H.; Yi, J.Y. TGFβ1 Protects Cells from γ-IR by Enhancing the Activity of the NHEJ Repair Pathway. Mol. Cancer Res. 2014, 13, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Palomero, L.; Moore, J.; Guix, I.; Espín, R.; Aytés, A.; Mao, J.-H.; Paulovich, A.G.; Whiteaker, J.R.; Ivey, R.G.; et al. Loss of TGFβ signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Sci. Transl. Med. 2021, 13, 580. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Sedlarikova, L.; Hajek, R.; Sevcikova, S. TGF-β—An excellent servant but a bad master. J. Transl. Med. 2012, 10, 183. [Google Scholar] [CrossRef] [Green Version]
- Roedel, F.; Kley, N.; Beuscher, H.U.; Hildebrandt, G.; Keilholz, L.; Kern, P.; Voll, R.; Herrmann, M.; Sauer, R. Anti-inflammatory effect of low-dose X-irradiation and the involvement of a TGF-β 1 -induced down-regulation of leukocyte/endothelial cell adhesion. Int. J. Radiat. Biol. 2002, 78, 711–719. [Google Scholar] [CrossRef]
- Barnett, G.C.; Coles, C.E.; Elliott, R.M.; Baynes, C.; Luccarini, C.; Conroy, D.; Wilkinson, J.S.; Tyrer, J.; Misra, V.; Platte, R.; et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: A prospective analysis study. Lancet Oncol. 2012, 13, 65–77. [Google Scholar] [CrossRef]
- Dunning, A.M.; Ellis, P.D.; McBride, S.; Kirschenlohr, H.L.; Healey, C.S.; Kemp, P.R.; Luben, R.N.; Chang-Claude, J.; Mannermaa, A.; Kataja, V.; et al. A transforming growth factorbeta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003, 63, 2610–2615. [Google Scholar]
- Tao, Y.; Sturgis, E.M.; Huang, Z.; Wang, Y.; Wei, P.; Wang, J.R.; Wei, Q.; Li, G. TGFβ1 Genetic Variants Predict Clinical Outcomes of HPV-Positive Oropharyngeal Cancer Patients after Definitive Radiotherapy. Clin. Cancer Res. 2018, 24, 2225–2233. [Google Scholar] [CrossRef] [Green Version]
- Dai, L.; Gast, A.; Horska, A.; Schrappe, M.; Bartram, C.R.; Hemminki, K.; Kumar, R.; Bermejo, J.L. A case-control study of childhood acute lymphoblastic leukaemia and polymorphisms in the TGF-β and receptor genes. Pediatr. Blood Cancer 2009, 52, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.-L.; Wang, M.; Shi, T.-Y.; Li, Q.-X.; Xi, P.; Xia, K.-Q.; Zheng, L.; Wei, Q.-Y. No Association between TGFB1 Polymorphisms and Late Radiotherapy Toxicity: A Meta-Analysis. PLoS ONE 2013, 8, e76964. [Google Scholar] [CrossRef]
- Delobel, J.-B.; Gnep, K.; Ospina, J.D.; Beckendorf, V.; Chira, C.; Zhu, J.; Bossi, A.; Messai, T.; Acosta, O.; Castelli, J.; et al. Nomogram to predict rectal toxicity following prostate cancer radiotherapy. PLoS ONE 2017, 12, e0179845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Oorschot, B.; Hovingh, S.; Dekker, A.; Stalpers, L.J.; Franken, N.A.P. Predicting Radiosensitivity with Gamma-H2AX Foci Assay after Single High-Dose-Rate and Pulsed Dose-Rate Ionizing Irradiation. Radiat. Res. 2016, 185, 190–198. [Google Scholar] [CrossRef]
| Parameter | All | Acute Toxicity ≥ °2 n = 71 | Late Toxicity ≥ °2 n = 46 |
|---|---|---|---|
| Age (years), median (IQR, min-max) | 70 (67–73, 53–83) | 69 (66–74, 57–79) | 70 (67–74, 62–79) |
| p = 0.688 | p = 0.178 | ||
| BMI (kg/m²), median (IQR, min-max) | 27 (25–30, 19–44) | 27 (25–30, 21–41) | 27 (25–28, 22–41) |
| p = 0.708 | p = 0.329 | ||
| T stage, No. (%) | |||
| 1 | 60 (25.0) | 17 (23.9) | 14 (30.4) |
| 2 | 155 (64.5) | 45 (63.4) | 26 (56.5) |
| 3 | 21 (8.8) | 8 (11.3) | 5 (10.9) |
| 4 | 4 (1.7) | 1 (1.4) | 1 (2.2) |
| p = 0.622 | p = 0.829 | ||
| N stage, No. (%) | |||
| cN0 | 234 (97.5) | 67 (94.4) | 45 (97.8) |
| cN1 $ | 6 (2.5) | 4 (5.6) | 1 (2.2) |
| f.s. | f.s. | ||
| Gleason score #, No. (%) | |||
| ≤5 & | 46 | 10 | 9 |
| 6 | 104 | 34 | 18 |
| 7 | 68 | 22 | 14 |
| ≥8 | 18 | 4 | 5 |
| p = 0.538 | p = 0.511 | ||
| PSA at diagnosis (ng/mL), median (IQR, min-max) | 9.4 (6.2–15.6, 1.1–186) | 10.0 (7.1–19.9, 2.1–179) | 8.6 (6.2–13.9, 1.7–77) |
| p = 0.174 | p = 0.510 | ||
| PLDA, No. (%) | |||
| No | 190 (79.2) | 51 (71.8) | 39 (84.8) |
| Yes | 50 (20.8) | 20 (28.2) | 7 (15.2) |
| p = 0.063 | p = 0.280 | ||
| HDR, No. (%) | |||
| No | 176 (73.3) | 55 (77.5) | 37 (80.4) |
| Yes | 64 (26.7) | 16 (22.5) | 9 (19.6) |
| p = 0.600 | p = 0.319 | ||
| RT dose, median | 72 | 72 | 72 |
| (IQR, min-max) | (68.4–72, 64–77) | (71–72, 64–72) | (71–72, 66–72) |
| p = 0.526 | p = 0.223 | ||
| ADT, ever, No. (%) | |||
| No | 77 (32.1) | 19 (26.8) | 14 (30.4) |
| Yes | 163 (67.9) | 52 (73.2) | 32 (69.6) |
| p = 0.253 | p = 0.861 | ||
| ADT, concomitant to RT §, No. (%) | |||
| No | 84 (35.0) | 20 (28.2) | 14 (30.4) |
| Yes | 156 (65.0) | 51 (71.8) | 32 (69.6) |
| p = 0.152 | p = 0.524 | ||
| Follow-up ‡, median (IQR, min-max) | 59.5 (49.8–62.5, 0.0–150.6) | 55.7 (47.5–61.8, 0.6–130.5) | 54.0 (40.5–62.2, 12.7–109.3) |
| Parameter | RR (95%-CI), Puni, Raw * | |
|---|---|---|
| Acute | Late | |
| Non-genetic | ||
| Radiation field: with PLDA vs. without | 1.51 (1.00–2.29), 0.080 | |
| Acute toxicity ≥ °2 | - | 2.00 (1.37–2.92), 0.002 |
| Genetic | ||
| TGFB1 rs1800470 (L10P): TC + CC vs. TT | 0.71 (0.48–1.05), 0.087 | |
| TGFB1 rs2241713: GC + CC vs. GG | 0.70 (0.47–1.03), 0.084 | |
| TGFBR1 rs10512263: TC + CC vs. TT | 2.14 (1.46–3.14), 0.001 | |
| TGFBR1 rs34733091: TC + CC vs. TT | 1.61 (1.07–2.42), 0.034 | |
| TGFB1 rs10417924: GA + AA vs. GG | 0.52 (0.28–0.97), 0.041 | |
| TGFB1 rs1800470 (L10P): CC vs. TT + TC | 1.95 (1.11–3.45), 0.035 | |
| TGFB1 rs2241713: CC vs. GG + GC | 1.95 (1.11–3.45), 0.035 | |
| TGFB1 rs75041078: TT vs. CC + CT | 2.17 (1.14–4.14), 0.056 | |
| TGFB1 rs8108357: GG vs. AA + AG | 2.02 (1.15–3.56), 0.029 | |
| TGFBR1 rs78471739: TC + CC vs TT | 0.17 (0.02–1.16), 0.022 | |
| Parameter | OR (95%-CI), Puni, Raw * | |
|---|---|---|
| Acute toxicity | ||
| Radiation field: with PLDA vs. without | 1.87 (0.95–3.69), 0.072 | |
| TGFB1 rs1800470 (L10P) #: TC + CC vs. TT | 0.53 (0.29–0.95), 0.034 | |
| TGFBR1 rs10512263: TC + CC vs. TT | 3.80 (1.78–8.12), 0.0006 | |
| Late toxicity | ||
| Acute toxicity ≥ °2 | 2.62 (1.31–5.21), 0.006 | |
| TGFB1 rs10417924: GA + AA vs. GG | 0.39 (0.18–0.86), 0.019 | |
| TGFB1 rs1800470 (L10P) #: CC vs. TT + TC | 2.70 (1.11–6.53), 0.028 | |
| TGFBR1 rs78471739: TC + CC vs. TT | 0.16 (0.02–1.20), 0.075 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guhlich, M.; Hubert, L.; Mergler, C.P.N.; Rave-Fraenk, M.; Dröge, L.H.; Leu, M.; Schmidberger, H.; Rieken, S.; Hille, A.; Schirmer, M.A. Identification of Risk Loci for Radiotoxicity in Prostate Cancer by Comprehensive Genotyping of TGFB1 and TGFBR1. Cancers 2021, 13, 5585. https://doi.org/10.3390/cancers13215585
Guhlich M, Hubert L, Mergler CPN, Rave-Fraenk M, Dröge LH, Leu M, Schmidberger H, Rieken S, Hille A, Schirmer MA. Identification of Risk Loci for Radiotoxicity in Prostate Cancer by Comprehensive Genotyping of TGFB1 and TGFBR1. Cancers. 2021; 13(21):5585. https://doi.org/10.3390/cancers13215585
Chicago/Turabian StyleGuhlich, Manuel, Laura Hubert, Caroline Patricia Nadine Mergler, Margret Rave-Fraenk, Leif Hendrik Dröge, Martin Leu, Heinz Schmidberger, Stefan Rieken, Andrea Hille, and Markus Anton Schirmer. 2021. "Identification of Risk Loci for Radiotoxicity in Prostate Cancer by Comprehensive Genotyping of TGFB1 and TGFBR1" Cancers 13, no. 21: 5585. https://doi.org/10.3390/cancers13215585
APA StyleGuhlich, M., Hubert, L., Mergler, C. P. N., Rave-Fraenk, M., Dröge, L. H., Leu, M., Schmidberger, H., Rieken, S., Hille, A., & Schirmer, M. A. (2021). Identification of Risk Loci for Radiotoxicity in Prostate Cancer by Comprehensive Genotyping of TGFB1 and TGFBR1. Cancers, 13(21), 5585. https://doi.org/10.3390/cancers13215585








