Role and Function of O-GlcNAcylation in Cancer
Abstract
Simple Summary
Abstract
1. Introduction
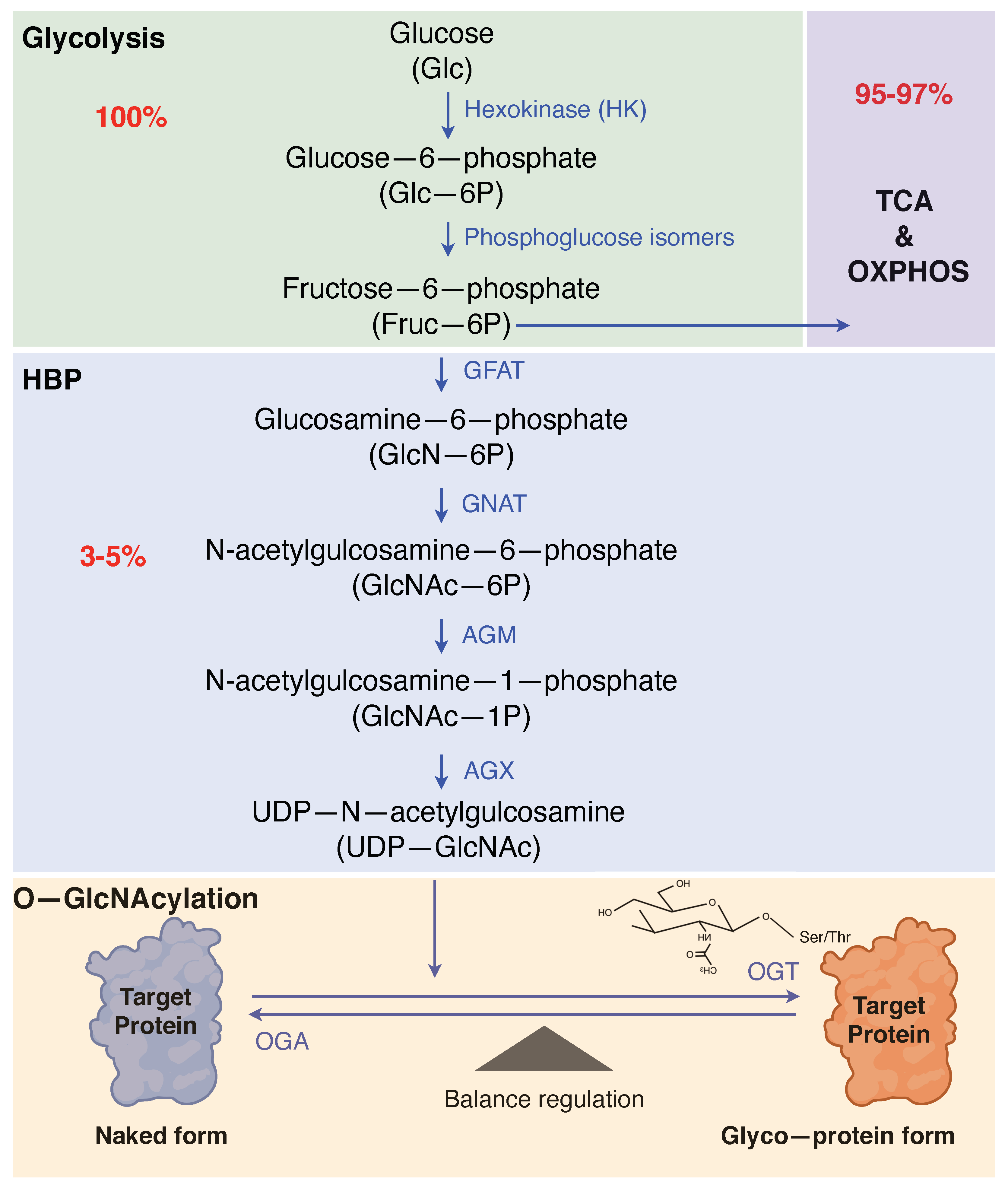
2. O-GlcNAcylation
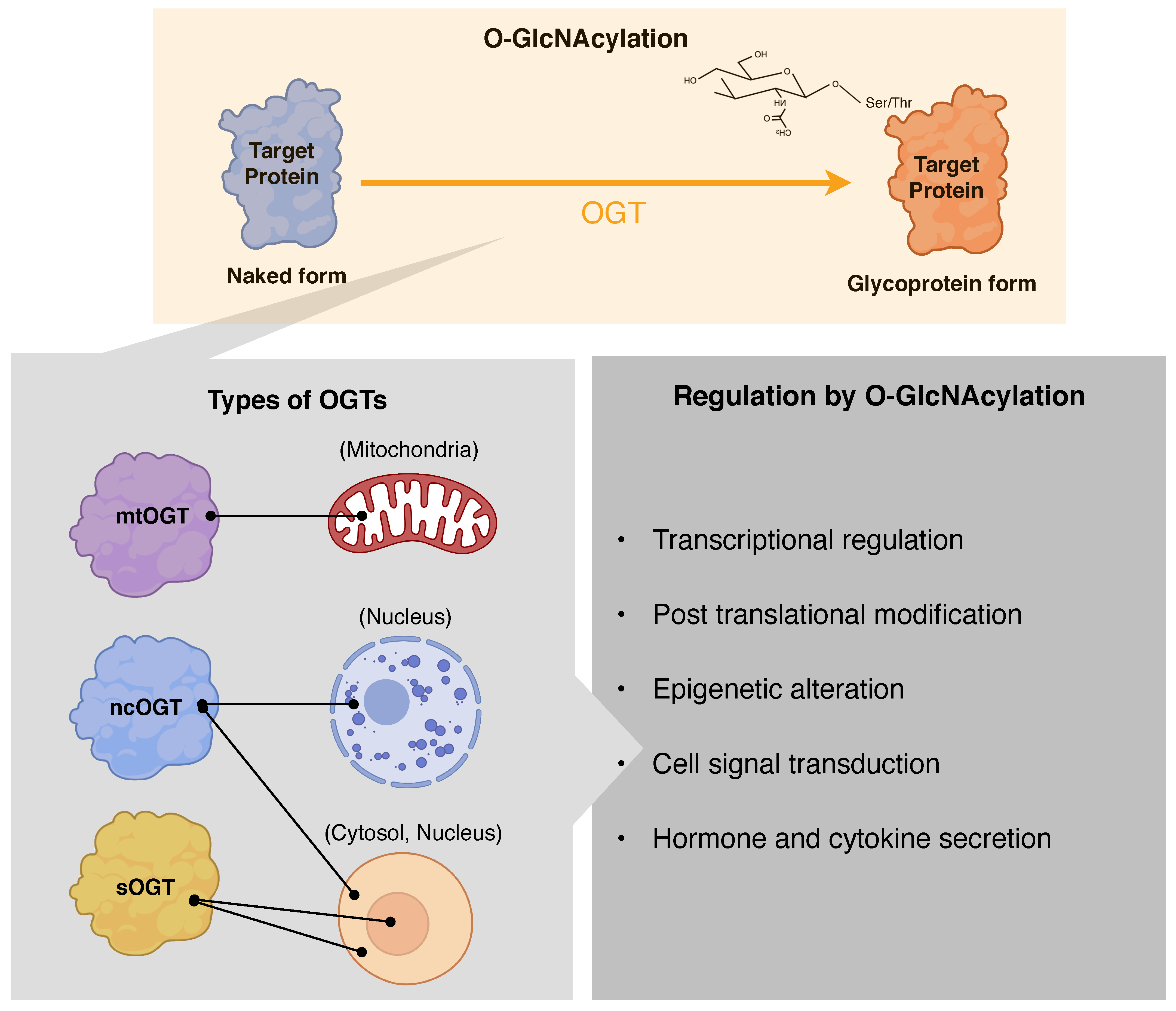
2.1. Substrate Recognition by OGT and OGA
2.2. Functions of O-GlcNAcylation
3. O-GlcNAcylation and Cancer
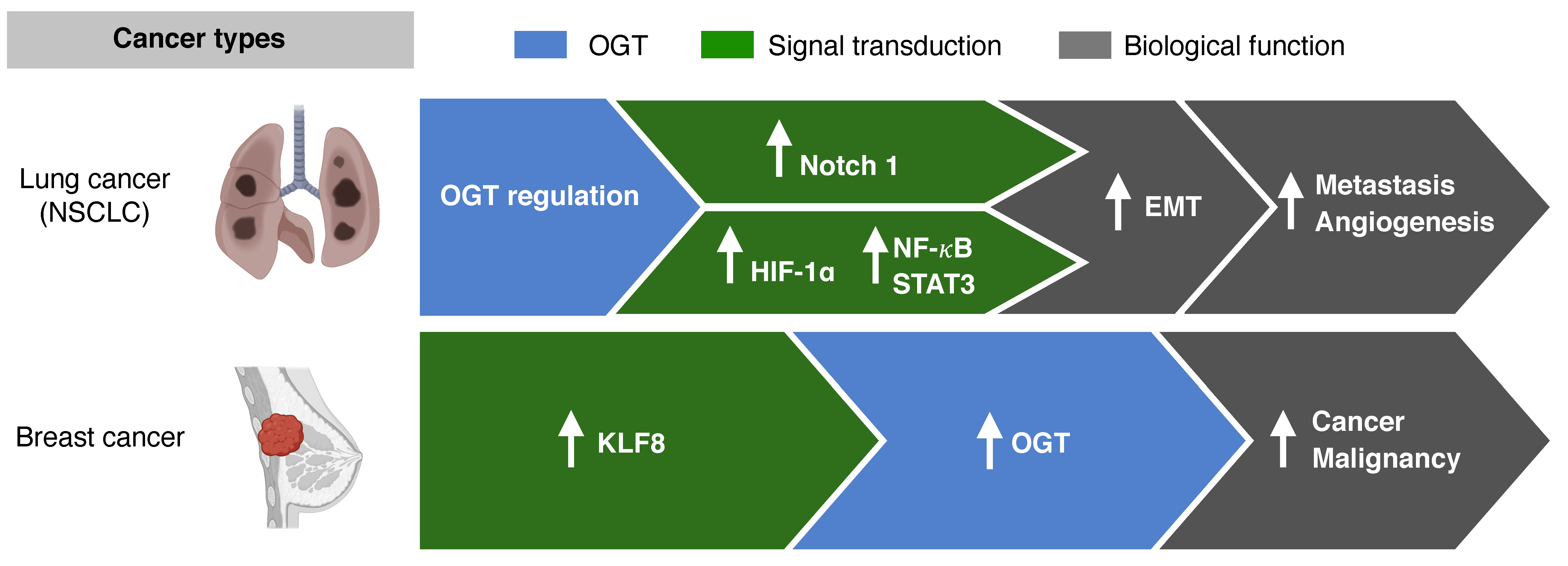
3.1. O-GlcNAcylation and Metastasis
3.2. O-GlcNAcylation and Receptor Tyrosine Kinase
3.3. O-GlcNAcylation and Resistance to Chemotherapy
3.4. O-GlcNAcylation as Prognostic Marker
4. O-GlcNAcylation and Immune Responses in Cancer
4.1. Overview of Immune System and O-GlcNAcylation
4.2. T Cell Activation and Differentiation Regulated by O-GlcNAcylation
4.3. Macrophage Differentiation and Activation by O-GlcNAcylation
4.4. O-GlcNAcylation and Tumor Microenvironment
5. Cancer Therapeutics Targeting O-GlcNAcylation
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Fardini, Y.; Dehennaut, V.; Lefebvre, T.; Issad, T. O-GlcNAcylation: A New Cancer Hallmark? Front. Endocrinol. 2013, 4, 99. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.; Bacote, V.; Traxinger, R.R. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 1991, 266, 4706–4712. [Google Scholar] [CrossRef]
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 2015, 208, 869–880. [Google Scholar] [CrossRef]
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.R.; Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984, 259, 3308–3317. [Google Scholar] [CrossRef]
- Hart, G.W.; Housley, M.P.; Slawson, C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007, 446, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Shimoji, S.; Hart, G.W. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010, 584, 2526–2538. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Clark, P.M.; Mason, D.E.; Keenan, M.C.; Hill, C.; Goddard, W.A., 3rd; Peters, E.C.; Driggers, E.M.; Hsieh-Wilson, L.C. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.; Remigante, A.; Civello, D.A.; Bernardinelli, E.; Szabó, Z.; Morabito, R.; Marino, A.; Sarikas, A.; Patsch, W.; Paulmichl, M.; et al. O-GlcNAcylation Suppresses the Ion Current IClswell by Preventing the Binding of the Protein ICln to α-Integrin. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Ruan, H.-B.; Singh, J.P.; Li, M.-D.; Wu, J.; Yang, X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. TEM 2013, 24, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.R.; Hanover, J.A. O-GlcNAc cycling: A link between metabolism and chronic disease. Annu. Rev. Nutr. 2013, 33, 205–229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shan, X.; Yuzwa, S.A.; Vocadlo, D.J. The Emerging Link between O-GlcNAc and Alzheimer Disease. J. Biol. Chem. 2014, 289, 34472–34481. [Google Scholar] [CrossRef]
- Ferrer, C.M.; Sodi, V.L.; Reginato, M.J. O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J. Mol. Biol. 2016, 428, 3282–3294. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Jin, J.; Qiu, Z.; Liu, D.; Luo, H. Functional Analysis of O-GlcNAcylation in Cancer Metastasis. Front. Oncol. 2020, 10, 585288. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.-W.; Fei, G.; Paulo, J.A.; Bellaousov, S.; Martin, S.E.S.; Duveau, D.Y.; Thomas, C.J.; Gygi, S.P.; Boutz, P.L.; Walker, S. O-GlcNAc regulates gene expression by controlling detained intron splicing. Nucleic Acids Res. 2020, 48, 5656–5669. [Google Scholar] [CrossRef]
- Love, D.C.; Kochran, J.; Cathey, R.L.; Shin, S.-H.; Hanover, J.A. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 2003, 116, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, B.D.; Love, D.C.; Hanover, J.A. Recombinant O-GlcNAc transferase isoforms: Identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 2006, 16, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Trapannone, R.; Mariappa, D.; Ferenbach, A.T.; van Aalten, D.M.F. Nucleocytoplasmic human O-GlcNAc transferase is sufficient for O-GlcNAcylation of mitochondrial proteins. Biochem. J. 2016, 473, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Sacoman, J.L.; Dagda, R.Y.; Burnham-Marusich, A.R.; Dagda, R.K.; Berninsone, P.M. Mitochondrial O-GlcNAc Transferase (mOGT) Regulates Mitochondrial Structure, Function, and Survival in HeLa Cells. J. Biol. Chem. 2017, 292, 4499–4518. [Google Scholar] [CrossRef]
- Jóźwiak, P.; Ciesielski, P.; Zakrzewski, P.K.; Kozal, K.; Oracz, J.; Budryn, G.; Żyżelewicz, D.; Flament, S.; Vercoutter-Edouart, A.S.; Bray, F.; et al. Mitochondrial O-GlcNAc Transferase Interacts with and Modifies Many Proteins and Its Up-Regulation Affects Mitochondrial Function and Cellular Energy Homeostasis. Cancers 2021, 13, 2956. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.D.; Hart, G.W. AMP-activated Protein Kinase and p38 MAPK Activate O-GlcNAcylation of Neuronal Proteins during Glucose Deprivation. J. Biol. Chem. 2008, 283, 13009–13020. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.-B.; Han, X.; Li, M.-D.; Singh, J.P.; Qian, K.; Azarhoush, S.; Zhao, L.; Bennett, A.M.; Samuel, V.T.; Wu, J.; et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 2012, 16, 226–237. [Google Scholar] [CrossRef]
- Whisenhunt, T.R.; Yang, X.; Bowe, D.B.; Paterson, A.J.; Van Tine, B.A.; Kudlow, J.E. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology 2006, 16, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, M.B.; Nam, Y.; Jiang, J.; Sliz, P.; Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 2011, 469, 564–567. [Google Scholar] [CrossRef]
- Vucic, D.; Dixit, V.M.; Wertz, I.E. Ubiquitylation in apoptosis: A post-translational modification at the edge of life and death. Nat. Rev. Mol. Cell Biol. 2011, 12, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Zachara, N.E.; O’Donnell, N.; Cheung, W.D.; Mercer, J.J.; Marth, J.D.; Hart, G.W. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 2004, 279, 30133–30142. [Google Scholar] [CrossRef]
- Ruan, H.B.; Nie, Y.; Yang, X. Regulation of protein degradation by O-GlcNAcylation: Crosstalk with ubiquitination. Mol. Cell Proteom. 2013, 12, 3489–3497. [Google Scholar] [CrossRef]
- Guinez, C.; Losfeld, M.-E.; Cacan, R.; Michalski, J.-C.; Lefebvre, T. Modulation of HSP70 GlcNAc-directed lectin activity by glucose availability and utilization. Glycobiology 2005, 16, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Guinez, C.; Mir, A.-M.; Leroy, Y.; Cacan, R.; Michalski, J.-C.; Lefebvre, T. Hsp70-GlcNAc-binding activity is released by stress, proteasome inhibition, and protein misfolding. Biochem. Biophys. Res. Commun. 2007, 361, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, T.-W.; Cecioni, S.; Eskandari, R.; Zandberg, W.F.; Vocadlo, D.J. O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol. 2015, 11, 319–325. [Google Scholar] [CrossRef]
- Hanover, J.A.; Krause, M.W.; Love, D.C. Bittersweet memories: Linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 2012, 13, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, H.; Lu, L.; Jiang, J. Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode. Nat. Struct. Mol. Biol. 2017, 24, 362–369. [Google Scholar] [CrossRef]
- Elsen, N.L.; Patel, S.B.; Ford, R.E.; Hall, D.L.; Hess, F.; Kandula, H.; Kornienko, M.; Reid, J.; Selnick, H.; Shipman, J.M.; et al. Insights into activity and inhibition from the crystal structure of human O-GlcNAcase. Nat. Chem. Biol. 2017, 13, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Chan, S.; Offen, W.A.; Hemsworth, G.R.; Willems, L.I.; King, D.T.; Varghese, V.; Britton, R.; Vocadlo, D.J.; Davies, G.J. Structural and functional insight into human O-GlcNAcase. Nat. Chem. Biol. 2017, 13, 610–612. [Google Scholar] [CrossRef] [PubMed]
- Dennis, R.J.; Taylor, E.J.; Macauley, M.S.; Stubbs, K.A.; Turkenburg, J.P.; Hart, S.J.; Black, G.N.; Vocadlo, D.J.; Davies, G.J. Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 2006, 13, 365–371. [Google Scholar] [CrossRef]
- Rao, F.V.; Dorfmueller, H.C.; Villa, F.; Allwood, M.; Eggleston, I.M.; van Aalten, D.M.F. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006, 25, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Schimpl, M.; Borodkin, V.S.; Gray, L.J.; van Aalten, D.M. Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chem. Biol. 2012, 19, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Su, K.; Roos, M.D.; Chang, Q.; Paterson, A.J.; Kudlow, J.E. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. USA 2001, 98, 6611–6616. [Google Scholar] [CrossRef]
- Housley, M.P.; Rodgers, J.T.; Udeshi, N.D.; Kelly, T.J.; Shabanowitz, J.; Hunt, D.F.; Puigserver, P.; Hart, G.W. O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 2008, 283, 16283–16292. [Google Scholar] [CrossRef]
- Lewis, B.A.; Hanover, J.A. O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 2014, 289, 34440–34448. [Google Scholar] [CrossRef]
- Singh, J.P.; Zhang, K.; Wu, J.; Yang, X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 2015, 356, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yin, R.; Yang, X. O-GlcNAc: A Bittersweet Switch in Liver. Front. Endocrinol. 2014, 5, 221. [Google Scholar] [CrossRef]
- Taylor, R.P.; Parker, G.J.; Hazel, M.W.; Soesanto, Y.; Fuller, W.; Yazzie, M.J.; McClain, D.A. Glucose Deprivation Stimulates GlcNAc Modification of Proteins through Up-regulation of O-Linked-N-Acetylglucosaminyltransferase. J. Biol. Chem. 2008, 283, 6050–6057. [Google Scholar] [CrossRef]
- Taylor, R.P.; Geisler, T.S.; Chambers, J.H.; McClain, D.A. Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. J. Biol. Chem. 2009, 284, 3425–3432. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008, 451, 964–969. [Google Scholar] [CrossRef]
- Kreppel, L.K.; Blomberg, M.A.; Hart, G.W. Dynamic Glycosylation of Nuclear and Cytosolic Proteins: Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9308–9315. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Park, S.Y.; Nam, H.W.; Kim, D.H.; Kang, J.G.; Kang, E.S.; Kim, Y.S.; Lee, H.C.; Kim, K.S.; Cho, J.W. NFκB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. USA 2008, 105, 17345–17350. [Google Scholar] [CrossRef] [PubMed]
- Dehennaut, V.; Leprince, D.; Lefebvre, T. O-GlcNAcylation, an Epigenetic Mark. Focus on the Histone Code, TET Family Proteins, and Polycomb Group Proteins. Front. Endocrinol. 2014, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.A.; Dias, W.B.; Thiruneelakantapillai, L.; Lane, M.D.; Hart, G.W. Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes. J. Biol. Chem. 2010, 285, 5204–5211. [Google Scholar] [CrossRef]
- Barkovskaya, A.; Prasmickaite, L.; Duveau, D.Y.; Mills, I.G.; Mælandsmo, G.M.; Moestue, S.A.; Itkonen, H.M. Abstract 1131: O-GlcNAc transferase inhibition in breast cancer cells. Cancer Res. 2017, 77, 1131. [Google Scholar] [CrossRef]
- Akella, N.M.; Le Minh, G.; Ciraku, L.; Mukherjee, A.; Bacigalupa, Z.A.; Mukhopadhyay, D.; Sodi, V.L.; Reginato, M.J. O-GlcNAc Transferase Regulates Cancer Stem–like Potential of Breast Cancer Cells. Mol. Cancer Res. 2020, 18, 585–598. [Google Scholar] [CrossRef]
- Ma, Z.; Vosseller, K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 2014, 289, 34457–34465. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Wieland, E.; Rodriguez-Vita, J.; Liebler, S.S.; Mogler, C.; Moll, I.; Herberich, S.E.; Espinet, E.; Herpel, E.; Menuchin, A.; Chang-Claude, J.; et al. Endothelial Notch1 Activity Facilitates Metastasis. Cancer Cell 2017, 31, 355–367. [Google Scholar] [CrossRef]
- Ruland, J. Colon Cancer: Epithelial Notch Signaling Recruits Neutrophils to Drive Metastasis. Cancer Cell 2019, 36, 213–214. [Google Scholar] [CrossRef] [PubMed]
- Lignitto, L.; LeBoeuf, S.E.; Homer, H.; Jiang, S.; Askenazi, M.; Karakousi, T.R.; Pass, H.I.; Bhutkar, A.J.; Tsirigos, A.; Ueberheide, B.; et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell 2019, 178, 316–329.e318. [Google Scholar] [CrossRef]
- Szymura, S.J.; Zaemes, J.P.; Allison, D.F.; Clift, S.H.; D’Innocenzi, J.M.; Gray, L.G.; McKenna, B.D.; Morris, B.B.; Bekiranov, S.; LeGallo, R.D.; et al. NF-κB upregulates glutamine-fructose-6-phosphate transaminase 2 to promote migration in non-small cell lung cancer. Cell Commun. Signal. 2019, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-cruz, P.; Alisson-Silva, F.; Todeschini, A.R.; Dias, W.B. Cellular glycosylation senses metabolic changes and modulates cell plasticity during epithelial to mesenchymal transition. Dev. Dyn. 2018, 247, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sai, B.; Wang, F.; Wang, L.; Wang, Y.; Zheng, L.; Li, G.; Tang, J.; Xiang, J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 2019, 18, 40. [Google Scholar] [CrossRef]
- Diepenbruck, M.; Christofori, G. Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr. Opin. Cell Biol. 2016, 43, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Kim, S.H.; Kim, M.J.; Choi, M.Y.; Kang, S.S.; Cho, G.J.; Kim, Y.S.; Choi, J.-Y.; Choi, W.S. O-GlcNAcylation of NF-κB Promotes Lung Metastasis of Cervical Cancer Cells via Upregulation of CXCR4 Expression. Mol. Cells 2017, 40, 476–484. [Google Scholar] [CrossRef]
- Yan, M.; Xu, Q.; Zhang, P.; Zhou, X.J.; Zhang, Z.Y.; Chen, W.T. Correlation of NF-kappaB signal pathway with tumor metastasis of human head and neck squamous cell carcinoma. BMC Cancer 2010, 10, 437. [Google Scholar] [CrossRef]
- Hu, C.-M.; Tien, S.-C.; Hsieh, P.-K.; Jeng, Y.-M.; Chang, M.-C.; Chang, Y.-T.; Chen, Y.-J.; Chen, Y.-J.; Lee, E.Y.H.P.; Lee, W.-H. High Glucose Triggers Nucleotide Imbalance through O-GlcNAcylation of Key Enzymes and Induces KRAS Mutation in Pancreatic Cells. Cell Metab. 2019, 29, 1334–1349.e1310. [Google Scholar] [CrossRef] [PubMed]
- Kaleem, A.; Ahmad, I.; Hoessli, D.C.; Walker-Nasir, E.; Saleem, M.; Shakoori, A.R.; Nasir ud, D. Epidermal growth factor receptors: Function modulation by phosphorylation and glycosylation interplay. Mol. Biol. Rep. 2009, 36, 631–639. [Google Scholar] [CrossRef][Green Version]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect Med. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- da Cunha Santos, G.; Shepherd, F.A.; Tsao, M.S. EGFR mutations and lung cancer. Annu. Rev. Pathol. 2011, 6, 49–69. [Google Scholar] [CrossRef]
- Hsu, W.H.; Yang, J.C.; Mok, T.S.; Loong, H.H. Overview of current systemic management of EGFR-mutant NSCLC. Ann. Oncol. 2018, 29, i3–i9. [Google Scholar] [CrossRef] [PubMed]
- Stateva, S.R.; Villalobo, A. O-GlcNAcylation of the human epidermal growth factor receptor. Org. Biomol. Chem. 2015, 13, 8196–8204. [Google Scholar] [CrossRef]
- Luanpitpong, S.; Angsutararux, P.; Samart, P.; Chanthra, N.; Chanvorachote, P.; Issaragrisil, S. Hyper-O-GlcNAcylation induces cisplatin resistance via regulation of p53 and c-Myc in human lung carcinoma. Sci. Rep. 2017, 7, 10607. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wu, J.; Wang, D.; Huang, X.; Zhang, N.; Shi, Y. Cisplatin enhances protein O-GlcNAcylation by altering the activity of OGT, OGA and AMPK in human non-small cell lung cancer cells. Int. J. Oncol. 2021, 58, 27. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Maio, M.D.; Chiodini, P.; Rudd, R.M.; Okamoto, H.; Skarlos, D.V.; Früh, M.; Qian, W.; Tamura, T.; Samantas, E.; et al. Carboplatin- or Cisplatin-Based Chemotherapy in First-Line Treatment of Small-Cell Lung Cancer: The COCIS Meta-Analysis of Individual Patient Data. J. Clin. Oncol. 2012, 30, 1692–1698. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Lee, S.J.; Kwon, O.-S. O-GlcNAc Transferase Inhibitor Synergistically Enhances Doxorubicin-Induced Apoptosis in HepG2 Cells. Cancers 2020, 12, 3154. [Google Scholar] [CrossRef]
- Itkonen, H.M.; Minner, S.; Guldvik, I.J.; Sandmann, M.J.; Tsourlakis, M.C.; Berge, V.; Svindland, A.; Schlomm, T.; Mills, I.G. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 2013, 73, 5277–5287. [Google Scholar] [CrossRef] [PubMed]
- Mi, W.; Gu, Y.; Han, C.; Liu, H.; Fan, Q.; Zhang, X.; Cong, Q.; Yu, W. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim. Biophys. Acta 2011, 1812, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Lin, C.H.; Yeh, Y.C.; Ho, H.L.; Wu, Y.C.; Chen, M.Y.; Chou, T.Y. High O-linked N-acetylglucosamine transferase expression predicts poor survival in patients with early stage lung adenocarcinoma. Oncotarget 2018, 9, 31032–31044. [Google Scholar] [CrossRef] [PubMed]
- Chiaradonna, F.; Ricciardiello, F.; Palorini, R. The Nutrient-Sensing Hexosamine Biosynthetic Pathway as the Hub of Cancer Metabolic Rewiring. Cells 2018, 7, 53. [Google Scholar] [CrossRef]
- Qiang, A.; Slawson, C.; Fields, P.E. The Role of O-GlcNAcylation in Immune Cell Activation. Front. Endocrinol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- McClain, D.A.; Lubas, W.A.; Cooksey, R.C.; Hazel, M.; Parker, G.J.; Love, D.C.; Hanover, J.A. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc. Natl. Acad. Sci. USA 2002, 99, 10695–10699. [Google Scholar] [CrossRef] [PubMed]
- Machacek, M.; Saunders, H.; Zhang, Z.; Tan, E.P.; Li, J.; Li, T.; Villar, M.T.; Artigues, A.; Lydic, T.; Cork, G.; et al. Elevated O-GlcNAcylation enhances pro-inflammatory Th17 function by altering the intracellular lipid microenvironment. J. Biol. Chem. 2019, 294, 8973–8990. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Salgado, O.C.; Singh, S.; Hippen, K.L.; Maynard, J.C.; Burlingame, A.L.; Ball, L.E.; Blazar, B.R.; Farrar, M.A.; Hogquist, K.A.; et al. The lineage stability and suppressive program of regulatory T cells require protein O-GlcNAcylation. Nat. Commun. 2019, 10, 354. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Weng, C.L.; Lin, K.I. O-GlcNAcylation and its role in the immune system. J. Biomed. Sci. 2020, 27, 57. [Google Scholar] [CrossRef]
- Golks, A.; Tran, T.T.; Goetschy, J.F.; Guerini, D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 2007, 26, 4368–4379. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.; Pathak, S.; Grzes, K.M.; Damerow, S.; Sinclair, L.V.; van Aalten, D.M.; Cantrell, D.A. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 2016, 17, 712–720. [Google Scholar] [CrossRef]
- Hwang, J.S.; Kim, K.H.; Park, J.; Kim, S.M.; Cho, H.; Lee, Y.; Han, I.O. Glucosamine improves survival in a mouse model of sepsis and attenuates sepsis-induced lung injury and inflammation. J. Biol Chem 2019, 294, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- He, Y.; Ma, X.; Li, D.; Hao, J. Thiamet G mediates neuroprotection in experimental stroke by modulating microglia/macrophage polarization and inhibiting NF-κB p65 signaling. J. Cereb. Blood Flow Metab. 2017, 37, 2938–2951. [Google Scholar] [CrossRef]
- Li, T.; Li, X.; Attri, K.S.; Liu, C.; Li, L.; Herring, L.E.; Asara, J.M.; Lei, Y.L.; Singh, P.K.; Gao, C.; et al. O-GlcNAc Transferase Links Glucose Metabolism to MAVS-Mediated Antiviral Innate Immunity. Cell Host Microbe 2018, 24, 791–803.e796. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Qi, Q.; Cao, R.; Qin, B.; Wang, B.; Wang, Y.; Zhao, L.; Li, W.; Du, X.; Liu, F.; et al. MAVS O-GlcNAcylation Is Essential for Host Antiviral Immunity against Lethal RNA Viruses. Cell Rep. 2019, 28, 2386–2396.e2385. [Google Scholar] [CrossRef] [PubMed]
- Allison, D.F.; Wamsley, J.J.; Kumar, M.; Li, D.; Gray, L.G.; Hart, G.W.; Jones, D.R.; Mayo, M.W. Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-κB acetylation and transcription. Proc. Natl. Acad. Sci. USA 2012, 109, 16888–16893. [Google Scholar] [CrossRef]
- Li, X.; Gong, W.; Wang, H.; Li, T.; Attri, K.S.; Lewis, R.E.; Kalil, A.C.; Bhinderwala, F.; Powers, R.; Yin, G.; et al. O-GlcNAc Transferase Suppresses Inflammation and Necroptosis by Targeting Receptor-Interacting Serine/Threonine-Protein Kinase 3. Immunity 2019, 50, 576–590.e576. [Google Scholar] [CrossRef]
- Hou, C.-W.; Mohanan, V.; Zachara, N.E.; Grimes, C.L. Identification and biological consequences of the O-GlcNAc modification of the human innate immune receptor, Nod2. Glycobiology 2015, 26, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Andrejeva, G.; Rathmell, J.C. Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors. Cell Metab. 2017, 26, 49–70. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers 2014, 6, 1670. [Google Scholar] [CrossRef]
- Zheng, X.; Mansouri, S.; Krager, A.; Grimminger, F.; Seeger, W.; Pullamsetti, S.S.; Wheelock, C.E.; Savai, R. Metabolism in tumour-associated macrophages: A quid pro quo with the tumour microenvironment. Eur. Respir. Rev. 2020, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Geng, X.; Hou, J.; Wu, G. New insights into M1/M2 macrophages: Key modulators in cancer progression. Cancer Cell Int. 2021, 21, 389. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Prenen, H.; Mazzone, M. Tumor-associated macrophages: A short compendium. Cell Mol. Life Sci. 2019, 76, 1447–1458. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, P.; Clark, P.M.; Mason, D.E.; Peters, E.C.; Hsieh-Wilson, L.C.; Baltimore, D. Activation of the transcriptional function of the NF-κB protein c-Rel by O-GlcNAc glycosylation. Sci. Signal. 2013, 6, ra75. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Xu, B.; Li, X.; Shang, Y.; Chu, Y.; Wang, W.; Chen, D.; Wu, N.; Hu, S.; Zhang, S.; et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene 2019, 38, 301–316. [Google Scholar] [CrossRef]
- Itkonen, H.M.; Poulose, N.; Steele, R.E.; Martin, S.E.S.; Levine, Z.G.; Duveau, D.Y.; Carelli, R.; Singh, R.; Urbanucci, A.; Loda, M.; et al. Inhibition of O-GlcNAc transferase renders prostate cancer cells dependent on CDK9. Mol. Cancer Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, H.; Cao, Y.; Wu, Q.; Li, W.; Zhang, J. Suppression of OGT by microRNA24 reduces FOXA1 stability and prevents breast cancer cells invasion. Biochem. Biophys. Res. Commun. 2017, 487, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Itkonen, H.M.; Gorad, S.S.; Duveau, D.Y.; Martin, S.E.; Barkovskaya, A.; Bathen, T.F.; Moestue, S.A.; Mills, I.G. Inhibition of O-GlcNAc transferase activity reprograms prostate cancer cell metabolism. Oncotarget 2016, 7, 12464–12476. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, X.; Wu, J.L.; Fu, L.; Liu, K.; Liu, D.; Chen, G.G.; Lai, P.B.; Wong, N.; Yu, J. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J. Hepatol. 2017, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Trapannone, R.; Rafie, K.; van Aalten, D.M. O-GlcNAc transferase inhibitors: Current tools and future challenges. Biochem. Soc. Trans. 2016, 44, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Borodkin, V.S.; Schimpl, M.; Gundogdu, M.; Rafie, K.; Dorfmueller, H.C.; Robinson, D.A.; van Aalten, D.M.F. Bisubstrate UDP-peptide conjugates as human O-GlcNAc transferase inhibitors. Biochem. J. 2014, 457, 497–502. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.B.; Pyo, K.-H.; Kim, H.R. Role and Function of O-GlcNAcylation in Cancer. Cancers 2021, 13, 5365. https://doi.org/10.3390/cancers13215365
Lee JB, Pyo K-H, Kim HR. Role and Function of O-GlcNAcylation in Cancer. Cancers. 2021; 13(21):5365. https://doi.org/10.3390/cancers13215365
Chicago/Turabian StyleLee, Jii Bum, Kyoung-Ho Pyo, and Hye Ryun Kim. 2021. "Role and Function of O-GlcNAcylation in Cancer" Cancers 13, no. 21: 5365. https://doi.org/10.3390/cancers13215365
APA StyleLee, J. B., Pyo, K.-H., & Kim, H. R. (2021). Role and Function of O-GlcNAcylation in Cancer. Cancers, 13(21), 5365. https://doi.org/10.3390/cancers13215365






