Simple Summary
The mutational load of BRAFV600E in melanomas has been described as a possible prognostic biomarker but there is no information about the mutant allele frequency (MAF) variability of BRAFV600E within cutaneous melanomas and its potential prognostic implications. Our study suggests that the variation degree of BRAFV600E MAF within primary cutaneous melanoma could act as a determinant in the location of primary melanomas as well as their first metastases and could influence prognostic indicators such as Breslow and mitotic indexes. BRAFV600E MAF variation is also related to the neoplastic cell phenotype and tumor lymphocytic infiltrate of primary tumors. For all these reasons, detection of BRAFV600E MAF variation could be a useful prognostic biomarker. It is worth exploring the role of BRAFV600E MAF variation with regards to the response of melanoma to targeted therapies.
Abstract
Background: Cutaneous melanoma shows high variability regarding clinicopathological presentation, evolution and prognosis. Methods: Next generation sequencing was performed to analyze hotspot mutations in different areas of primary melanomas (MMp) and their paired metastases. Clinicopathological features were evaluated depending on the degree of variation of the BRAFV600E mutant allele frequency (MAF) in MMp. Results: In our cohort of 14 superficial spreading, 10 nodular melanomas and 52 metastases, 17/24 (71%) melanomas had a BRAFV600E mutation and 5/24 (21%) had a NRASQ61 mutation. We observed a high variation of BRAFV600E MAF (H-BRAFV600E) in 7/17 (41%) MMp. The H-BRAFV600E MMp were all located on the trunk, had lower Breslow and mitotic indexes and predominantly, a first nodal metastasis. Regions with spindled tumor cells (Spin) and high lymphocytic infiltrate (HInf) were more frequent in the H-BRAFV600E patients (4/7; 57%), whereas regions with epithelial tumor cells (Epit) and low lymphocytic infiltrate (LInf) were predominant (6/10; 60%) and exclusive in the low BRAFV600E MAF variation tumors (L-BRAFV600E). The H-BRAFV600E/Spin/HInf MMp patients had better prognostic features and nodal first metastasis. Conclusions: The H-BRAFV600E MMp were located on the trunk, had better prognostic characteristics, such as lower Breslow and mitotic indexes as well as high lymphocytic infiltrate.
1. Introduction
Cutaneous melanoma (CMM) is the most aggressive skin malignancy. Although CMM can be cured with wide local excision when it is diagnosed in the early stages, advanced melanoma is associated with poor outcomes [1]. Nevertheless, knowledge advances in melanoma pathogenesis and the subsequent development of targeted and immunobased therapies have improved metastatic melanoma treatment [2].
The MAPK–ERK pathway plays a major role in the development and progression of melanoma [3]. Overall, about 35–50% of melanomas of all clinical types have mutations in the V600 codon of BRAF [4]. Approximately 90% of BRAF mutations result in the substitution of valine (V) for glutamic acid (E) at position 600 (BRAFV600E) [5,6,7,8,9]. The BRAFV600E mutation confers to this kinase the ability to activate MEK [4,10]. This ligand-independent pathway activation is essential in melanoma development and progression [11,12]. Combinations of BRAF and MEK inhibitors have emerged as new treatment options for patients with BRAF-mutated melanoma with a good response rate [13,14]. Unfortunately, most patients that initially show an extremely good response to BRAF inhibitors develop resistance to those drugs within a relatively short period of time (<1 year) [10,14].
Besides the usefulness of the BRAFV600E driver mutation as a therapeutic target, a recent study has suggested that the mutational load of BRAFV600E is a possible prognostic biomarker [15], expanding the classical histopathological prognostic criteria such as ulceration, Breslow, Clark or mitotic indexes [1,16,17,18,19,20,21,22]. Moreover, some studies in recent years have attempted to relate the mutational load to the response in order to target therapies, but controversial results have been obtained [23,24,25,26].
Next-generation sequencing (NGS) technologies have played an essential role in understanding the altered genetic pathways involved in human cancer. It is a high-throughput method which allows for the sequencing of multiple samples and targeted genes in the same run. NGS can quantitate the proportion of reads for a given mutation, also known as mutant allele frequency (MAF), which represents the percentage of tumor cells that harbor a specific mutation in a neoplastic tissue. The sensitivity of NGS is higher than that observed with Sanger sequencing (2–10% versus 15–25% allele frequency, respectively) [27,28].
It is widely accepted that melanomas are compound by tumor cells, with different histophenotypic characteristics (morphology or pigmentation), and different non-tumor cells as inflammatory cells. These characteristics can also be different between the primary tumor and its metastases [29]. Therefore, understanding MAF using NGS technology could provide invaluable information regarding melanomagenesis and dissemination.
In this work, we used NGS to study the variation of BRAFV600E MAF between different regions of 24 primary melanomas and their paired metastases as well as its relation to their histophenotypic and clinical characteristics, in an attempt to identify the predictors of disease outcome.
2. Materials and Methods
2.1. Patient Selection and Data Collection
The Pathology Department archive was retrospectively reviewed from 1 January 2000 to 30 June 2020. Patients who had paired primary cutaneous melanoma (MMp) and metastases (MMx) were selected. In order to avoid excessive genetic diversity, the study was restricted to superficial spreading melanoma (SSM) and nodular melanoma (NM), according to the WHO classifications [8].
Gender, age, MMp subtype, Breslow and Clark indexes, presence of ulceration, number of mitoses, MMp location, number and location of the MMx, disease free survival (DFS) and overall survival (OS) were recorded.
2.2. Diagnostic Criteria of Histological Features
A detailed microscopic analysis of routine H&E sections for the identification of areas of different cell morphology (spindled, epithelioid and mixed) and different degree of pigmentation (absent/low, moderate and high) was performed. A selection of 3 and 2 different areas of the MMp and the MMx, respectively, was done (Figure 1a).
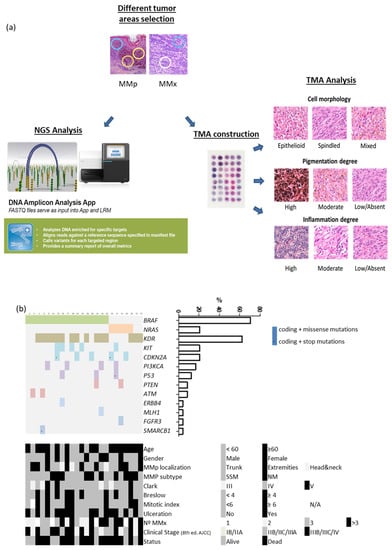
Figure 1.
(a) Study workflow design. (b) Heatmap showing mutated genes, demographic, clinical and tumor characteristics of our patient cohort.
2.3. DNA Extraction and NGS Analysis
In total, 191 areas selected were microdissected from the formalin-fixed paraffin-embedded blocs. DNA extracted by Maxwell® FFPE Plus DNA Kit (Promega, Madison, WI, USA) were quantified by Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
We performed NGS with the Ampliseq Cancer HotSpot panel v2 (Illumina, San Diego, CA, USA) to detect genetic mutations from 50 oncogenes and tumor suppressor genes. Genomic DNA (10 ng) was used to construct the amplicon libraries according to the manufacturer’s protocol and quality control of the libraries was done with Agilent 2100 Bioanalyzer. Resulting data files were analyzed by DNA amplicon analysis workflow to perform alignment and variant calling. We excluded non-coding variants and synonymous changes of each polymorphism. Single nucleotide variants with a variant allele frequency of at least 10% and a minimum depth coverage of 150 reads were used as cut-off values for variant identification. (Figure 1a).
2.4. TMA Elaboration and Analysis
Four tissue microarrays (TMAs) containing the 191 areas, were constructed using a tissue arrayer device (Beecher Instrument, Sun Prairie, WI, USA). Histopathological tumor characteristics (cell morphology, degree of pigmentation and lymphocytic infiltrate) were qualitatively analyzed within each TMA spot. Cell shape was visually graded as epithelioid or spindled when the majority of the tumor cells expressed the same phenotype (>80) and mixed when both phenotypes were observed and none of them were >80%.
Pigmentation was scored on a three-point scale (high, moderate and low/absent). Melanophages were not considered. Lymphocytic infiltrate was scored on a three-point scale (high, moderate and low/absent). The dissected areas contained at least 50% of tumor nuclei. (Figure 1a).
Immunohistochemical Analysis of BRAFV600E Specific Target Protein
TMA BRAFV600E immunostaining was performed with the VE1 anti-BRAFV600E primary antibody (Ventana Medical Systems, Inc., Roche, Basel, Switzerland) as described [30]. Staining quantification was performed with QuPath 0.2.3 software (Centre for Cancer Research & Cell Biology, Queen’s University Belfast, UK). Values ranged from 0 to 300 [31].
2.5. Statistical Analysis
The Fisher exact test and T-test were used to examine associations between high or low intratumoral BRAFV600E MAF variation (H-BRAFV600E or L-BRAFV600E) and demographic, clinical and MMp histopathological variables. Overall survival (OS) and disease-free survival (DFS) curves were calculated using the Kaplan–Meier method with log-rank test. The Mann–Whitney test was used to assess immunohistochemical characteristics. The p-values are indicated by asterisks * p < 0.05; ** p < 0.01; *** p < 0.001. All statistical analyses and figures were performed using SPSS Statistics software and Graphpad Prism software.
3. Results
3.1. Clinicopathological Characteristics
Our series comprised 29 patients with 29 MMp and 52 paired MMx, which supposed a total number of 87 MMp and 104 MMx spots. Due to low quality samples, 5 patients were rejected and 24 patients were elected for further analysis (Figure S1). This cohort was composed of 16 (67%) men and 8 (33%) women, with a median age of 63 years (24–87). The MM subtype was SSM in 14 (58%) and NM in 10 (42%) biopsies. In total, 12 (50%) MMp were located on the trunk, 6 (25%) on the lower extremities, 3 (12.5%) on the upper extremities and 3 (12.5%) on the head and neck. The mean MMp Breslow and Clark indexes were 4.22 mm (1.04–13.50) and 4 (3–5), respectively. Ulceration was present in 14 (58%) cases and the mean mitotic index was 5/cm2 (1–11). Eight patients (33.33%) were alive at the end of the study with a mean survival time of 60 months (median 40.5 (9–177)) (Table 1 and Table S1).

Table 1.
Clinicopathological Characteristics.
3.2. Genetic Analysis: There Is Variation in BRAFV600E Mutant Allele Frequency in Primary Melanomas
NSG of 66 MMp and 28 MMx regions identified mutations in seven oncogenes (BRAF, NRAS, KIT, KDR, FGFR3, ERBB4 and PI3KCA) and six tumor suppressor genes (CDKN2A, TP53, ATM, PTEN, MLH1 and SMARCB1) in the MMp (Figure 1b). The BRAFV600E mutation was present in 17/24 (71%) patients, NRASQ61 in 5/24 (21%) and 2/24 (8%) MMp were of the wild type for both genes. We compared the clinicopathological characteristics of BRAFV600E and NRASQ61 patients and no statistical differences were detected. The BRAFV600E and NRASQ61 mutations where highly preserved in the MMp and their paired MMx. Only 2/17 (11.8%) BRAFV600E mutated patients presented an absence of BRAFV600E mutation in one of the three MMp spots (intratumor heterogeneity). Additionally, in 2/17 (11.8%) patients whose three MMp spots were BRAFV600E mutated, both spots of their paired MMx were the BRAF wild type (intertumor heterogeneity) [32,33]. Across the cohort, 11.8% intratumoral and intertumoral heterogeneity was detected in the BRAFV600E mutation.
Striking differences in the MAF of BRAFV600E between spots of the same MMp were detected. Therefore, we decided to quantify such variations. First, to obtain the real BRAFV600E MAF we took into account the percentage (%) of tumoral cells present in each spot by three investigator-blinded assessments on H&E staining. Discrepant cases were discussed until an agreement was reached. Only spots composed of at least 50% tumor cells were considered. In all cases, the mean BRAFV600E MAF obtained by NGS was recalculated depending on the percentage of tumor cells observed in each spot from the TMA created. We defined H-BRAFV600E primary tumors as those that contained one spot with a two-fold increase MAF value regarding at least one of the other MMp spots. Samples that lacked this diversity were considered L-BRAFV600E. In our cohort of 17 MMp with the BRAFV600E mutation, 7 (41.18%) showed H-BRAFV600E while the remaining 10 (58.82%) had L-BRAFV600E (Figure 2a).
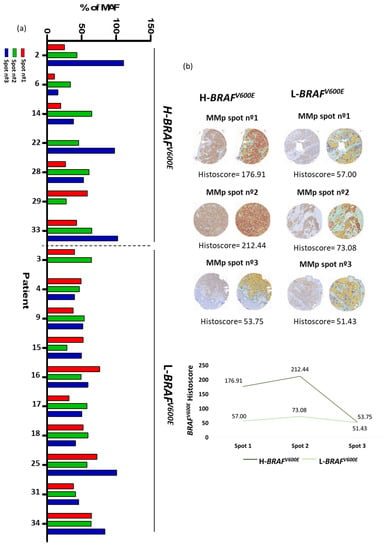
Figure 2.
MAF of BRAFV600E (a) NGS analysis of BRAFV600E MAF corrected by % of tumor cells. Patients grouped in H-BRAFV600E and L-BRAFV600E. (b) Immunohistochemical examples of primary melanoma tumor with high immunohistochemical histoscore variation of BRAFV600E between its spots and primary melanoma tumor with low immunohistochemical histoscore variation of BRAFV600E between its spots.
3.3. Immunohistochemical Analysis: There Is Variation in BRAFV600E Immunoexpression in Primary Melanomas
High and low variation in BRAFV600E MAF was validated by an immunohistochemical study.
First, histoscores of BRAFV600E were obtained by Qupath analysis from all of the TMA spots. As expected, NRAS mutated MMp spots presented a low histoscore (<20). Then, we considered a histoscore of ≥20 as the cut-off for the immunohistochemical histoscore scale for BRAFV600E positivity with a sensitivity of 87.5% and a specificity of 100% [34,35,36].
Then, we defined tumors with high immunohistochemical variation of BRAFV600E as those primary tumors that contained one spot with a two-fold increase histoscore value regarding at least one of the other MMp spots. Samples that lacked this diversity were considered to show a low immunohistochemical variation of BRAFV600E (Figure 2b). We observed that immunohistochemical evaluation of BRAFV600E showed significant consistency with the NGS analysis, as it was able to identify patients with H-BRAFV600E with a sensitivity of 80% and a specificity of 77.78%.
3.4. Correlation between BRAFV600E Allelic Frequency Variation and Histomorphological Features
3.4.1. H-BRAFV600E Correlation with Demographic, Clinical and Tumor Characteristics
First, we analyzed all clinicopathological characteristics of H-BRAFV600E patients compared with those of the L-BRAFV600E group (Figure 3A).
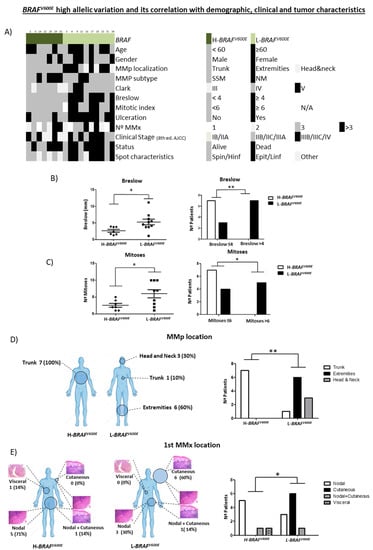
Figure 3.
Clinicopathological characteristics of MMp depending on BRAFV600E MAF variation (high vs. low). (A) Heatmap showing demographic, clinical and tumor characteristics of MMp patients depending on BRAFV600E MAF variation. (B) Breslow thickness depending on BRAFV600E MAF variation and number of patients according to Breslow thickness (<4 mm vs. ≥4 mm) in primary melanoma tumors. (C) Number of mitoses and number of patients according to mitotic index (<6 vs. ≥6) depending on BRAFV600E MAF variation. (D) MMp and (E) first MMx location depending on H-BRAFV600E vs. L-BRAFV600E patients. Statistical analysis was performed using t-test, Fischer exact tests (* p < 0.05; ** p < 0.01).
H-BRAFV600E patients had statistically lower Breslow and mitotic indexes than L-BRAFV600E (Mean 2.617 ± 0.4426 vs. 5.280 ± 0.8373 and Mean 2.571 ± 0.5714 vs. 6.000 ± 1.202). These results indicated that the H-BRAFV600E MMp had better prognostic features than the L-BRAFV600E since their Breslow tended to be <4 mm and they had a lower mitotic index [1,20,22] (Figure 3B,C). No statistical differences were observed regarding age and gender, MMp subtype, Clark or ulceration.
In addition, we analyzed the MMp and MMx locations in both groups. H-BRAFV600E patients presented all their MMp on the trunk (7/7 100%), while L-BRAFV600E patients presented most of their MMp in other locations, mainly on the extremities (6/10; 60%) but also on the head and neck (3/10; 30%) and trunk (1/10; 10%) (Figure 3D).
Moreover, we observed important differences in the first MMx location. Most H-BRAFV600E patients presented a nodal first MMx (6/7; 86%), while the first MMx in L-BRAFV600E patients was mostly cutaneous/subcutaneous (7/10; 70%) (Figure 3E). Interestingly, in H-BRAFV600E patients the nodal MMx was detected in the sentinel lymph node biopsy in 4/6 (67%) cases, whereas in 4/7 (57%), skin MMx of the L-BRAFV600E patients were already present at diagnosis of the primary tumor as a satellitoses or in-transit metastases. No statistical differences were observed in OS and DFS between the groups (Figure S2a).
All these results indicate that H-BRAFV600E seems to correlate with the development of the MMp on the trunk, better prognostic features such as lower Breslow and mitotic indexes, and nodal first MMx.
3.4.2. Demographic, Clinical and Tumor Characteristics of H-BRAFV600E/Spindled/ High Lymphocytic Infiltrate Group vs. L-BRAFV600E/Epithelioid/Low Lymphocytic Infiltrate Group
We wanted to assess if the variation in BRAFV600E MAF was correlated with a certain cytological morphology, pigmentation or lymphocytic infiltrate degree. We identified that 7/7 (100%) H-BRAFV600E patients had at least one spot with high-moderated lymphocytic infiltrate (HInf) while 7/10 (70%) L-BRAFV600E patients had low lymphocytic infiltrate (LInf) in all their spots (Figure 4a). Moreover, the melanoma cells in 4/7 (57%) H-BRAFV600E patients with the HInf spot had a spindled phenotype while L-BRAFV600E patients with the HInf spot and spindled phenotype were observed in 3/10 (30%) patients. On the other hand, 6/10 (60%) L-BRAFV600E patients had all LInf spots and their melanoma cells were epithelioid, while none of the H-BRAFV600E patients had this late characteristic (0%) (Figure 4b). Therefore, we divided the patients into two groups: patients who presented H-BRAFV600E and had at least one spindled and high-moderated lymphocytic infiltrate spot (H-BRAFV600E/Spin/HInf) vs. patients with L-BRAFV600E and epithelioid and low lymphocytic infiltrate (L-BRAFV600E/Epit/LInf) spots.
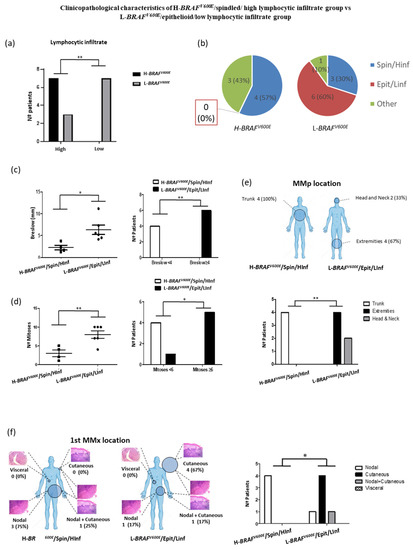
Figure 4.
(a) Patient distribution according to variation of BRAFV600E MAF (high vs. low) and lymphocytic infiltrate degree of their primary melanoma spots. (b) Patient distribution according to variation of BRAFV600E MAF and the combination of their histophenotypic characteristics and lymphocytic infiltrate degree of the primary melanoma spots. (c) Breslow thickness depending on variation of BRAFV600E MAF, cytological characteristics and lymphocytic infiltration degree and number of patients according to Breslow thickness (<4 mm vs. ≥4 mm) in primary melanoma tumors. (d) Number of mitoses and number of patients according to mitotic index (<6 vs. ≥6) depending on variation of BRAFV600E MAF, cytological characteristics and lymphocytic infiltration degree. (e) MMp and (f) first MMx location depending on H-BRAFV600E/Spin/HInf vs. L-BRAFV600E/Epit/LInf patients. Statistical analysis was performed using t-test and Fischer exact tests (* p < 0.05; ** p < 0.01).
The H-BRAFV600E/Spin/HInf group showed a significantly lower Breslow and mitotic index than the L-BRAFV600E/Epit/Linf group (Mean 2.270 ± 0.5533 vs. 6.308 ± 1.067 and Mean 3.000 ± 0.9129 vs. 8.000 ± 1.000) (Figure 4c,d). H-BRAFV600E/Spin/HInf patients presented all their MMp on the trunk (4/4; 100%) while L-BRAFV600E/Epit/LInf patients presented all their MMp on the extremities (4/6; 67%) and head and neck (2/6; 33%) (Figure 4e). There were no statistical differences in age, gender, MMp subtype, Clark index, presence of ulceration or number of MMx. Nevertheless, we observed differences in the first MMx location. All H-BRAFV600E/Spin/HInf patients had a nodal MMx (4/4; 100%) whereas 5/6 (83.33%) L-BRAFV600E/Epit/Linf patients had a cutaneous first MMx (Figure 4f). Interestingly, in H-BRAFV600E patients the nodal MMx was detected by sentinel lymph node biopsy in 3/4 (75%) cases, whereas 5/5 (100%) skin MMx of the L-BRAFV600E patients were present at MMp diagnosis as a satellitoses or in-transit metastases. No statistical differences were observed in OS and DFS between the groups (Figure S2b).
All these results suggest that the spindle phenotype and high/moderate lymphocytic infiltrate correlate with the H-BRAFV600E samples. This confers better clinicopathological prognostic characteristics for these MM patients, such as lower Breslow and mitotic indexes [1,20,22].
4. Discussion
Melanoma can show different morphological characteristics such as different cytological subtypes or degrees of pigmentation either between different MMp or even within the same MMp (intratumoral heterogeneity) and between the MMp and paired MMx (intertumoral heterogeneity).
This study attempts to investigate the relationship between the diverse histophenotypic features present in different regions of MMp compared with the genetic characteristics found by NGS assay in a series of 24 patients with primary SSM or NM.
We detected mutations in seven oncogenes (BRAF, NRAS, KIT, KDR, FGFR3, ERBB4 and PI3KCA) and six tumor suppressor genes (CDKN2A, TP53, ATM, PTEN, MLH1 and SMARCB1) in our MMp series. Other studies using NGS and cancer hotspot panels in paired melanoma samples found the same main driver mutations (BRAF, NRAS, KIT) [29,37]. However, none of these studies analyzed the BRAFV600E MAF variation.
BRAF and NRAS mutation frequencies differ between cutaneous melanoma subtypes. BRAF mutation occurs more frequently in SSM (20–65%) and in second place in NM (20–43%) [38,39,40,41,42,43], while NM is the most frequent MM subtype with NRAS mutations (21–27%) [41,44]. Our series only included SSM and NM, which harbored the BRAFV600E mutation in 71% and NRASQ61 in 21% of MMp. These findings were similar to those previously described, although the presence of BRAFV600E was slightly higher. As the number of SSM and NM in our series are quite balanced (14 and 10 cases, respectively) differences in sequencing methodology could perhaps explain this fact, since a greater sensibility of NGS has been described in the detection of BRAF mutations compared to allele specific PCR [37,45].
We found that the BRAFV600E and NRASQ61 mutations where highly preserved in MMp and their paired MMx. Only 2/17 (11.8%) cases presented intratumor heterogeneity and 2/17 (11.6%) cases presented intertumor heterogeneity regarding BRAFV600E mutation. These findings support the idea that driver mutations occur early during the development of malignant melanoma [23,38,39,46]. Then, melanoma can show intratumoral and intertumoral BRAFV600E mutation heterogeneity, although its frequency is low.
In recent years, controversial results have been obtained by studies which related the BRAFV600E MAF of melanoma tumors with clinicopathological and prognostic characteristics as well as the melanoma response, in order to target therapies [15,23,24,25,26]. All these studies analyzed the MAF value of one specific region for each tumor, so we wanted to know whether the variation degree of BRAFV600E MAF within different regions of the same tumor could influence these characteristics.
Interestingly, our study suggests that variation of BRAFV600E MAF within MMp is related to the clinical features of the MMp, histopathological prognostic factors and progression behavior, although all the patients in our series developed metastasis at some point (since it was one of the inclusion criteria). Thus, H-BRAFV600E MMp would settle more frequently on the trunk and their first MMx would more frequently be nodal, while the L-BRAFV600E melanomas would be located more frequently on the extremities and develop most often cutaneous/subcutaneous, regional first MMx. The preferential location of the first MMx on the skin in L-BRAFV600E MMp cases could be explained because all were satellite or in-transit MMx, and this type of MMx are more frequently associated with MMp located on the extremities [47,48]. In addition, the H-BRAFV600E patients seem to have better histopathological prognostic indicators such as lower Breslow and mitotic indexes [1]. These associations were even more evident when we analyzed the H-BRAFV600E/Spin/Hinf and L-BRAFV600E/Epit/Linf subgroups. We identified an even stronger predilection of the MMp and first MMx for the trunk and nodal regions, respectively, with lower Breslow and mitotic indexes in the H-BRAFV600E/Spin/Hinf patients, while the L-BRAFV600E/Epit/Linf MMp were preferentially located on the extremities and their first MMx was significantly more frequently cutaneous, showing higher Breslow and mitotic indexes. Our findings could be explained in part by the suggestion of some authors that the presence of tumor-infiltrating lymphocytes (TILs) in melanoma is associated with a lower Breslow and mitotic rate [49,50,51]. Moreover, with regards to cytological features, a recent study showed that melanoma cells with rounded behavior had a higher capacity for tumor formation, tumor progression and metastasis than elongated cells [52], indicating that an increase in epithelioid cells in MMp could mean a higher proliferative and invasive behavior.
Even though we are aware of the modest sample size, our results suggest that variation of BRAFV600E MAF can constitute a potential biomarker that is worth exploring further. Moreover, these findings could serve as initial step for the future development of therapies focused on the presence of worse genetic and histopathological characteristics, such as L-BRAFV600E, epithelioid cell compound and low lymphocytic infiltrate in MMp.
5. Conclusions
The attained results suggest that MAF variation of BRAFV600E within primary cutaneous melanomas (MMp) could influence their histopathological characteristics, as well as their anatomic location and that of their first metastases (MMx). Here we described for the first time in the biomedical literature, that H-BRAFV600E could correlate with a spindle phenotype, high/moderate lymphocytic infiltrate and the development of the MMp on the trunk and nodal first MMx. On the contrary, L-BRAFV600E seems to correlate with an epithelioid phenotype, low lymphocytic infiltrate, the development of the MMp predominantly on the extremities and a cutaneous first MMx.
In addition, we suggest a possible new link between H-BRAFV600E and better classical prognostic indicators such as lower Breslow and mitotic indexes.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13205073/s1, Figure S1: Patient selection; Figure S2: (a) Overall survival and disease free survival of patients depending on BRAFV600E MAF variation (high vs. low); (b) Overall survival and disease free survival of H-BRAFV600E/Spin/HInf vs. L-BRAFV600E/Epit/Linf patients.; Table S1: Demographic, clinical and tumor characteristics of our series.; Table S2: Histophenotypic sample characteristics.
Author Contributions
Conceptualization, X.M.-G., R.M.M. and A.M.; methodology, X.S., A.V., R.M.M. and A.M.; validation, F.V., S.G. and M.S.; formal analysis, X.S., C.B., Ó.M. and A.M.; investigation, X.S., F.V. Ó.M., C.B., P.S., I.d.l.R., A.R. and A.M.; resources, X.S., D.C., M.S., A.M. and R.M.M.; writing—original draft preparation, X.S.; writing—review and editing, R.M.M., Ó.M. and A.M.; visualization, X.S., R.M.M. and A.M.; supervision, R.M.M. and A.M.; project administration, R.M.M. and A.M.; funding acquisition, R.M.M., A.M. and X.M.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from ISCIII and FEDER (“una manera de hacer Europa”) PI15/00711 to RMM and PI18/00573 to RMM & AM. CIBERONC (CB16/12/00231) to XMG. Postdoctoral AECC (POSTD004MACI - POSTD16) to AM. Predoctoral UdL to PS and predoctoral AECC fellowship to IR. The work was also supported by the Xarxa de Bancs de Tumors de Catalunya sponsored by “Pla Director d’Oncologia de Catalunya (XBTC)”and IRBLleida Biobank (B.0000682) and PLATAFORMA BIOBANCOS (PT17/0015/0027; PT20/00021).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Hospital Universitary Arnau de Vilanova Ethics Committee (CEIm (ref 1465)). Samples were obtained with the support of the Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d’Oncología de Catalunya (XBTC)”, IRBLleida Biobank (B.0000682) and PLATAFORMA BIOBANCOS (PT17/0015/0027 and PT20/00021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available in this article (and Supplementary material).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma Staging: Evidence-Based Changes in the American Joint Committee on Cancer Eighth Edition Cancer Staging Manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flaherty, K.T.; Hodi, F.S.; Fisher, D.E. From Genes to Drugs: Targeted Strategies for Melanoma. Nat. Rev. Cancer 2012, 12, 349–361. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF Gene in Human Cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Akbani, R.; Akdemir, K.C.; Aksoy, B.A.; Albert, M.; Ally, A.; Amin, S.B.; Arachchi, H.; Arora, A.; Auman, J.T.; Ayala, B.; et al. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menzies, A.M.; Haydu, L.E.; Visintin, L.; Carlino, M.S.; Howle, J.R.; Thompson, J.F.; Kefford, R.F.; Scolyer, R.A.; Long, G.V. Distinguishing Clinicopathologic Features of Patients with V600E and V600K BRAF -Mutant Metastatic Melanoma. Clin. Cancer Res. 2012, 18, 3242–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siroy, A.E.; Boland, G.M.; Milton, D.R.; Roszik, J.; Frankian, S.; Malke, J.; Haydu, L.; Prieto, V.G.; Tetzlaff, M.; Ivan, D.; et al. Beyond BRAF V600: Clinical Mutation Panel Testing by Next-Generation Sequencing in Advanced Melanoma. J. Investig. Dermatol. 2015, 135, 508–515. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Kuo, H.; Li, W.-Q.; Canales, A.L.; Han, J.; Qureshi, A.A. Association between BRAF V600E and NRAS Q61R Mutations and Clinicopathologic Characteristics, Risk Factors and Clinical Outcome of Primary Invasive Cutaneous Melanoma. Cancer Causes Control 2014, 25, 1379–1386. [Google Scholar] [CrossRef] [Green Version]
- Elder, D.E.; Bastian, B.C.; Cree, I.A.; Massi, D.; Scolyer, R.A. The 2018 World Health Organization Classification of Cutaneous, Mucosal, and Uveal Melanoma: Detailed Analysis of 9 Distinct Subtypes Defined by Their Evolutionary Pathway. Arch. Pathol. Lab. Med. 2020, 144, 500–522. [Google Scholar] [CrossRef] [Green Version]
- Chiappetta, C.; Proietti, I.; Soccodato, V.; Puggioni, C.; Zaralli, R.; Pacini, L.; Porta, N.; Skroza, N.; Petrozza, V.; Potenza, C.; et al. BRAF and NRAS Mutations Are Heterogeneous and Not Mutually Exclusive in Nodular Melanoma. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Shtivelman, E.; Davies, M.A.; Hwu, P.; Yang, J.; Lotem, M.; Oren, M.; Flaherty, K.T.; Fisher, D.E. Pathways and Therapeutic Targets in Melanoma. Oncotarget 2014, 5, 1701–1752. [Google Scholar] [CrossRef] [Green Version]
- Savoia, P.; Fava, P.; Casoni, F.; Cremona, O. Targeting the ERK Signaling Pathway in Melanoma. Int. J. Mol. Sci. 2019, 20, 1483. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Liu, W.-Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.-F. Signaling Pathway of MAPK/ERK in Cell Proliferation, Differentiation, Migration, Senescence and Apoptosis. J. Recept. Signal Transduct. Res. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Tétu, P.; Baroudjian, B.; Lebbe, C. Targeting BRAF and MEK Inhibitors in Melanoma in the Metastatic, Neoadjuvant and Adjuvant Setting. Curr. Opin. Oncol. 2020, 32, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, A.M.; Bartnik, E.; Fiedorowicz, M.; Rutkowski, P. Targeted Therapy in Melanoma and Mechanisms of Resistance. Int. J. Mol. Sci. 2020, 21, 4576. [Google Scholar] [CrossRef]
- Sevilla, A.; Morales, M.C.; Ezkurra, P.A.; Rasero, J.; Velasco, V.; Cancho-Galan, G.; Sánchez-Diez, A.; Mujika, K.; Penas, C.; Smith, I.; et al. BRAF V600E Mutational Load as a Prognosis Biomarker in Malignant Melanoma. PLoS ONE 2020, 15, e0230136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breslow, A. Thickness, Cross-Sectional Areas and Depth of Invasion in the Prognosis of Cutaneous Melanoma. Ann. Surg. 1970, 172, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Gimotty, P.A.; Elder, D.E.; Fraker, D.L.; Botbyl, J.; Sellers, K.; Elenitsas, R.; Ming, M.E.; Schuchter, L.; Spitz, F.R.; Czerniecki, B.J.; et al. Identification of High-Risk Patients Among Those Diagnosed With Thin Cutaneous Melanomas. JCO 2007, 25, 1129–1134. [Google Scholar] [CrossRef]
- Balch, C.M.; Murad, T.M.; Soong, S.J.; Ingalls, A.L.; Halpern, N.B.; Maddox, W.A. A Multifactorial Analysis of Melanoma: Prognostic histopathological features comparing Clark’s and Breslow’s staging methods. Ann. Surg. 1978, 188, 732–742. [Google Scholar] [CrossRef]
- Green, A.C.; Baade, P.; Coory, M.; Aitken, J.F.; Smithers, M. Population-Based 20-Year Survival Among People Diagnosed With Thin Melanomas in Queensland, Australia. JCO 2012, 30, 1462–1467. [Google Scholar] [CrossRef] [Green Version]
- Piñero-Madrona, A.; Ruiz-Merino, G.; Cerezuela Fuentes, P.; Martínez-Barba, E.; Rodríguez-López, J.N.; Cabezas-Herrera, J. Mitotic Rate as an Important Prognostic Factor in Cutaneous Malignant Melanoma. Clin. Transl. Oncol. 2019, 21, 1348–1356. [Google Scholar] [CrossRef]
- Mandalà, M.; Galli, F.; Cattaneo, L.; Merelli, B.; Rulli, E.; Ribero, S.; Quaglino, P.; De Giorgi, V.; Pigozzo, J.; Sileni, V.C.; et al. Mitotic Rate Correlates with Sentinel Lymph Node Status and Outcome in Cutaneous Melanoma Greater than 1 Millimeter in Thickness: A Multi-Institutional Study of 1524 Cases. J. Am. Acad. Dermatol. 2017, 76, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.L.; Vidri, R.J.; MacGillivray, D.C.; Fitzgerald, T.L. Tumor Mitotic Rate Is an Independent Predictor of Survival for Nonmetastatic Melanoma. Surgery 2018, 164, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Satzger, I.; Marks, L.; Kerick, M.; Klages, S.; Berking, C.; Herbst, R.; Völker, B.; Schacht, V.; Timmermann, B.; Gutzmer, R. Allele Frequencies of BRAF V600 Mutations in Primary Melanomas and Matched Metastases and Their Relevance for BRAF Inhibitor Therapy in Metastatic Melanoma. Oncotarget 2015, 6, 37895–37905. [Google Scholar] [CrossRef] [Green Version]
- Lebbé, C.; How-Kit, A.; Battistella, M.; Sadoux, A.; Podgorniak, M.-P.; Sidina, I.; Pages, C.; Roux, J.; Porcher, R.; Tost, J.; et al. BRAF V600 Mutation Levels Predict Response to Vemurafenib in Metastatic Melanoma. Melanoma Res. 2014, 24, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Stagni, C.; Zamuner, C.; Elefanti, L.; Zanin, T.; Bianco, P.D.; Sommariva, A.; Fabozzi, A.; Pigozzo, J.; Mocellin, S.; Montesco, M.C.; et al. BRAF Gene Copy Number and Mutant Allele Frequency Correlate with Time to Progression in Metastatic Melanoma Patients Treated with MAPK Inhibitors. Mol. Cancer Ther. 2018, 17, 1332–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrino, E.; Balsamo, A.; Pisacane, A.; Gallo, S.; Becco, P.; Miglio, U.; Caravelli, D.; Poletto, S.; Paruzzo, L.; Debernardi, C.; et al. High BRAF Variant Allele Frequencies Are Associated with Distinct Pathological Features and Responsiveness to Target Therapy in Melanoma Patients. ESMO Open 2021, 6, 100133. [Google Scholar] [CrossRef] [PubMed]
- Serratì, S.; De Summa, S.; Pilato, B.; Petriella, D.; Lacalamita, R.; Tommasi, S.; Pinto, R. Next-Generation Sequencing: Advances and Applications in Cancer Diagnosis. OTT 2016, 9, 7355–7365. [Google Scholar] [CrossRef] [Green Version]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef] [PubMed]
- Mejbel, H.A.; Arudra, S.K.C.; Pradhan, D.; Torres-Cabala, C.A.; Nagarajan, P.; Tetzlaff, M.T.; Curry, J.L.; Ivan, D.; Duose, D.Y.; Luthra, R.; et al. Immunohistochemical and Molecular Features of Melanomas Exhibiting Intratumor and Intertumor Histomorphologic Heterogeneity. Cancers 2019, 11, 1714. [Google Scholar] [CrossRef] [Green Version]
- Maiques, O.; Macià, A.; Moreno, S.; Barceló, C.; Santacana, M.; Vea, A.; Herreros, J.; Gatius, S.; Ortega, E.; Valls, J.; et al. Immunohistochemical Analysis of T-Type Calcium Channels in Acquired Melanocytic Naevi and Melanoma. Br. J. Dermatol. 2017, 176, 1247–1258. [Google Scholar] [CrossRef] [Green Version]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open Source Software for Digital Pathology Image Analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Grzywa, T.M.; Paskal, W.; Włodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, H.; Zebary, A.; Vassilaki, I.; Omholt, K.; Ghaderi, M.; Hansson, J. BRAF V600E Protein Expression in Primary Cutaneous Malignant Melanomas and Paired Metastases. JAMA Dermatol. 2015, 151, 410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verlinden, I.; van den Hurk, K.; Clarijs, R.; Willig, A.P.; Stallinga, C.M.H.A.; Roemen, G.M.J.M.; van den Oord, J.J.; zur Hausen, A.; Speel, E.-J.M.; Winnepenninckx, V.J.L. BRAFV600E Immunopositive Melanomas Show Low Frequency of Heterogeneity and Association With Epithelioid Tumor Cells: A STROBE-Compliant Article. Medicine 2014, 93, e285. [Google Scholar] [CrossRef]
- Tetzlaff, M.T.; Pattanaprichakul, P.; Wargo, J.; Fox, P.S.; Patel, K.P.; Estrella, J.S.; Broaddus, R.R.; Williams, M.D.; Davies, M.A.; Routbort, M.J.; et al. Utility of BRAF V600E Immunohistochemistry Expression Pattern as a Surrogate of BRAF Mutation Status in 154 Patients with Advanced Melanoma. Hum. Pathol. 2015, 46, 1101–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, I.P.; Salhi, A.; Giles, K.M.; Vogelsang, M.; Han, S.W.; Ismaili, N.; Lui, K.P.; Robinson, E.M.; Wilson, M.A.; Shapiro, R.L.; et al. Identification of a Novel Pathogenic Germline KDR Variant in Melanoma. Clin. Cancer Res. 2016, 22, 2377–2385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omholt, K.; Platz, A.; Kanter, L.; Ringborg, U.; Hansson, J. NRAS and BRAF Mutations Arise Early during Melanoma Pathogenesis and Are Preserved throughout Tumor Progression. Clin. Cancer Res. 2003, 9, 6483–6488. [Google Scholar]
- Edlundh-Rose, E.; Egyha, S.; Omholt, K.; Mansson-Brahme, E.; Platz, A.; Hansson, J.; Lundeberg, J. NRAS and BRAF Mutations in Melanoma Tumours in Relation to Clinical Characteristics: A Study Based on Mutation Screening by Pyrosequencing. Melanoma Res. 2006, 16, 471–478. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, S.N.; Hahn, H.J.; Lee, Y.W.; Choe, Y.B.; Ahn, K.J. Metaanalysis of BRAF Mutations and Clinicopathologic Characteristics in Primary Melanoma. J. Am. Acad. Dermatol. 2015, 72, 1036–1046. [Google Scholar] [CrossRef]
- Lopez-Martin, J.A.; Arance Fernández, A.; Ríos-Martín, J.J.; Hernández-Losa, J.; Alós Hernández, L.; Cerezuela Fuentes, P.; Ortiz Reina, S.; Ortega Izquierdo, E.; Martí, R.M.; Soberino García, J.; et al. Frequency and Clinicopathological Profile Associated with Braf Mutations in Patients with Advanced Melanoma in Spain. Transl. Oncol. 2020, 13, 100750. [Google Scholar] [CrossRef]
- Lee, J.-H.; Choi, J.-W.; Kim, Y.-S. Frequencies of BRAF and NRAS Mutations Are Different in Histological Types and Sites of Origin of Cutaneous Melanoma: A Meta-Analysis: BRAF and NRAS Mutations in Melanoma. Br. J. Dermatol. 2011, 164, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, G.; Potter, L.; DaForno, P.; Pringle, J.H. Cutaneous Melanoma Subtypes Show Different BRAF and NRAS Mutation Frequencies. Clin. Cancer Res. 2006, 12, 4499–4505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez-Castañeda, L.D.; Nova, J.A.; Tovar-Parra, J.D. Frequency of Mutations in BRAF, NRAS, and KIT in Different Populations and Histological Subtypes of Melanoma: A Systemic Review. Melanoma Res. 2020, 30, 62–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.-L.; Zhou, L.; Sadri, N. Comparison of Targeted next Generation Sequencing (NGS) versus Isolated BRAF V600E Analysis in Patients with Metastatic Melanoma. Virchows Arch. 2018, 473, 371–377. [Google Scholar] [CrossRef]
- Menzies, A.M.; Lum, T.; Wilmott, J.S.; Hyman, J.; Kefford, R.F.; Thompson, J.F.; O’Toole, S.; Long, G.V.; Scolyer, R.A. Intrapatient Homogeneity of BRAFV600E Expression in Melanoma. Am. J. Surg. Pathol. 2014, 38, 377–382. [Google Scholar] [CrossRef]
- Bann, D.V.; Chaikhoutdinov, I.; Zhu, J.; Andrews, G. Satellite and In-Transit Metastatic Disease in Melanoma Skin Cancer: A Retrospective Review of Disease Presentation, Treatment, and Outcomes. Dermatol. Surg. 2019, 45, 371–380. [Google Scholar] [CrossRef]
- Testori, A.; Ribero, S.; Bataille, V. Diagnosis and Treatment of In-Transit Melanoma Metastases. Eur. J. Surg. Oncol. (EJSO) 2017, 43, 544–560. [Google Scholar] [CrossRef]
- Azimi, F.; Scolyer, R.A.; Rumcheva, P.; Moncrieff, M.; Murali, R.; McCarthy, S.W.; Saw, R.P.; Thompson, J.F. Tumor-Infiltrating Lymphocyte Grade Is an Independent Predictor of Sentinel Lymph Node Status and Survival in Patients With Cutaneous Melanoma. JCO 2012, 30, 2678–2683. [Google Scholar] [CrossRef]
- Thomas, N.E.; Busam, K.J.; From, L.; Kricker, A.; Armstrong, B.K.; Anton-Culver, H.; Gruber, S.B.; Gallagher, R.P.; Zanetti, R.; Rosso, S.; et al. Tumor-Infiltrating Lymphocyte Grade in Primary Melanomas Is Independently Associated With Melanoma-Specific Survival in the Population-Based Genes, Environment and Melanoma Study. JCO 2013, 31, 4252–4259. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Chen, N.; Ge, C.; Li, R.; Li, Z.; Zeng, B.; Li, C.; Wang, Y.; Xue, Y.; Song, X.; et al. Prognostic Value of Tumor-Infiltrating Lymphocytes in Melanoma: A Systematic Review and Meta-Analysis. OncoImmunology 2019, 8, e1593806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Hernandez, I.; Maiques, O.; Kohlhammer, L.; Cantelli, G.; Perdrix-Rosell, A.; Monger, J.; Fanshawe, B.; Bridgeman, V.L.; Karagiannis, S.N.; Penin, R.M.; et al. WNT11-FZD7-DAAM1 Signalling Supports Tumour Initiating Abilities and Melanoma Amoeboid Invasion. Nat. Commun. 2020, 11, 5315. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).