Immune Functions of Signaling Lymphocytic Activation Molecule Family Molecules in Multiple Myeloma
Abstract
Simple Summary
Abstract
1. Introduction
2. Overview of SLAM Family Receptors
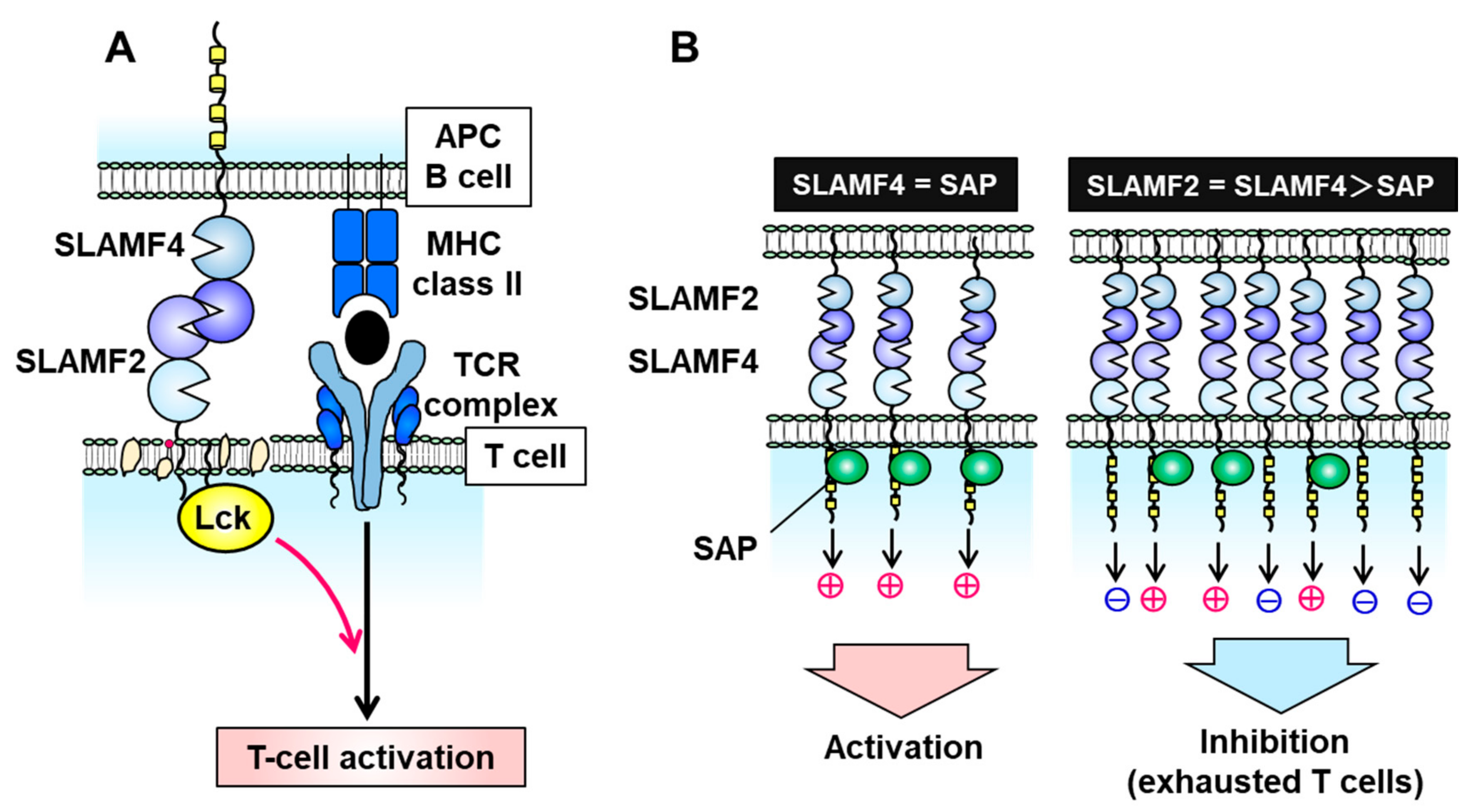
3. SLAMF2 (CD48)
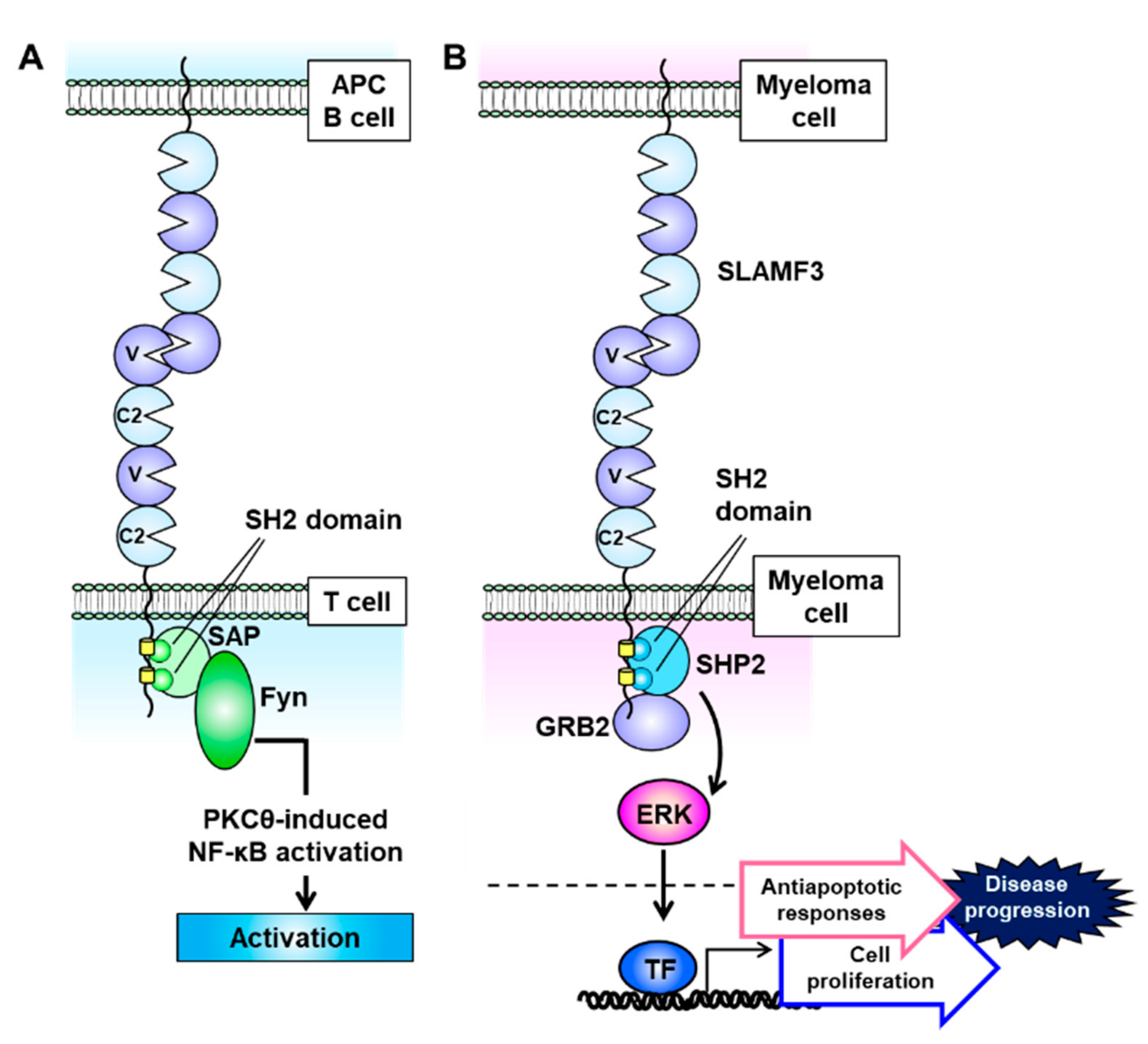
4. SLAMF3 (CD229, Ly9)
5. SLAMF6 (CD352, NTB-A)
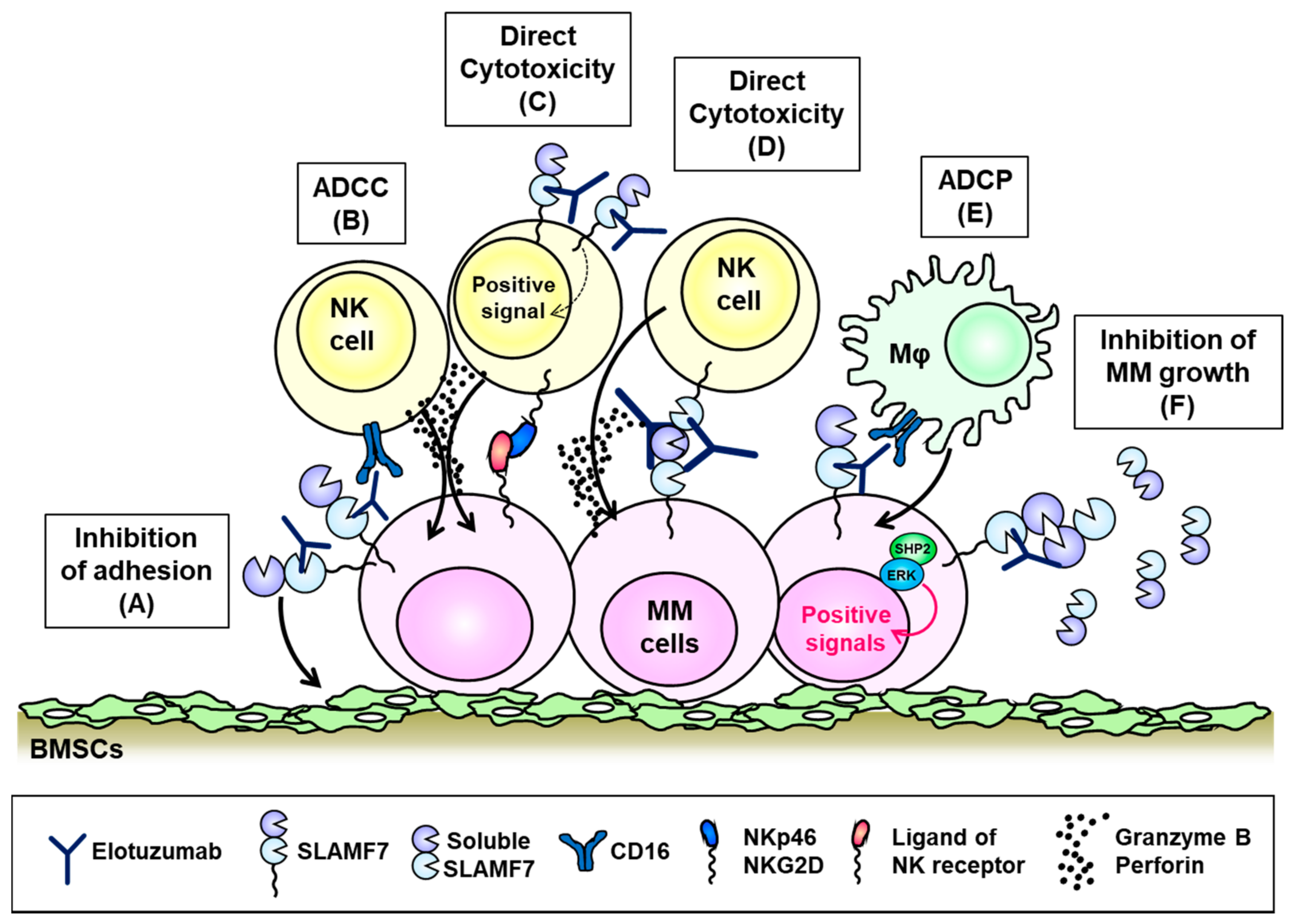
6. SLAMF7 (CD319, CS1)
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rajkumar, S.V. Multiple myeloma: 2018 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2018, 93. [Google Scholar] [CrossRef]
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
- Prideaux, S.M.; Conway-O’Brien, E.; Chevassut, T.J. The genetic architecture of multiple myeloma. Adv. Hematol. 2014, 2014, 864058. [Google Scholar] [CrossRef]
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.-V.; Gay, F.; Anderson, K.C. Multiple myeloma. Nat. Rev. Dis. Primers 2017, 3, 17046. [Google Scholar] [CrossRef]
- Gooding, S.; Edwards, C.M. New approaches to targeting the bone marrow microenvironment in multiple myeloma. Curr. Opin. Pharmacol. 2016, 28, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Tamura, H.; Ishibashi, M.; Sunakawa, M.; Inokuchi, K. Immunotherapy for Multiple Myeloma. Cancers 2019, 11, 9. [Google Scholar] [CrossRef]
- Swan, D.; Lynch, K.; Gurney, M.; O’Dwyer, M. Current and emerging immunotherapeutic approaches to the treatment of multiple myeloma. Ther. Adv. Hematol. 2019, 10. [Google Scholar] [CrossRef]
- Moreau, P.; Zamagni, E.; Mateos, M.V. Treatment of patients with multiple myeloma progressing on frontline-therapy with lenalidomide. Blood Cancer J. 2019, 9, 38. [Google Scholar] [CrossRef]
- Neri, P.; Bahlis, N.J.; Lonial, S. New Strategies in Multiple Myeloma: Immunotherapy as a Novel Approach to Treat Patients with Multiple Myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5959–5965. [Google Scholar] [CrossRef]
- Anderson, K.C. Progress and Paradigms in Multiple Myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5419–5427. [Google Scholar] [CrossRef]
- Ma, J.; Li, Q.; Yu, Z.; Cao, Z.; Liu, S.; Chen, L.; Li, H.; Gao, S.; Yan, T.; Wang, Y.; et al. Immunotherapy Strategies Against Multiple Myeloma. Technol. Cancer Res. Treat. 2017, 16, 717–726. [Google Scholar] [CrossRef]
- Hosen, N.; Matsunaga, Y.; Hasegawa, K.; Matsuno, H.; Nakamura, Y.; Makita, M.; Watanabe, K.; Yoshida, M.; Satoh, K.; Morimoto, S.; et al. The activated conformation of integrin β(7) is a novel multiple myeloma-specific target for CAR T cell therapy. Nat. Med. 2017, 23, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Chari, A.; Scott, E.; Mezzi, K.; Usmani, S.Z. B-cell maturation antigen (BCMA) in multiple myeloma: Rationale for targeting and current therapeutic approaches. Leukemia 2020, 34, 985–1005. [Google Scholar] [CrossRef] [PubMed]
- Veillette, A.; Latour, S. The SLAM family of immune-cell receptors. Curr. Opin. Immunol. 2003, 15, 277–285. [Google Scholar] [CrossRef]
- Veillette, A. SLAM-family receptors: Immune regulators with or without SAP-family adaptors. Cold Spring Harb. Perspect. Biol. 2010, 2, a002469. [Google Scholar] [CrossRef]
- Schwartzberg, P.L.; Mueller, K.L.; Qi, H.; Cannons, J.L. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat. Rev. Immunol. 2009, 9, 39–46. [Google Scholar] [CrossRef]
- Cannons, J.L.; Tangye, S.G.; Schwartzberg, P.L. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 2011, 29, 665–705. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Deenick, E.K. The role of SAP and SLAM family molecules in the humoral immune response. Ann. N. Y. Acad. Sci. 2011, 1217, 32–44. [Google Scholar] [CrossRef]
- Fouquet, G.; Marcq, I.; Debuysscher, V.; Bayry, J.; Rabbind-Singh, A.; Bengrine, A.; Nguyen-Khac, E.; Naassila, M.; Bouhlal, H. Signaling lymphocytic activation molecules Slam and cancers: Friends or foes? Oncotarget 2018, 9, 16248–16262. [Google Scholar] [CrossRef] [PubMed]
- Detre, C.; Keszei, M.; Romero, X.; Tsokos, G.C.; Terhorst, C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin. Immunopathol. 2010, 32, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Veillette, A. SLAM family receptors in normal immunity and immune pathologies. Curr. Opin. Immunol. 2016, 38, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Morra, M.; Lu, J.; Poy, F.; Martin, M.; Sayos, J.; Calpe, S.; Gullo, C.; Howie, D.; Rietdijk, S.; Thompson, A.; et al. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. EMBO J. 2001, 20, 5840–5852. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Quintero, L.A.; Roncagalli, R.; Guo, H.; Latour, S.; Davidson, D.; Veillette, A. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cγ, Ca++, and Erk, leading to granule polarization. J. Exp. Med. 2014, 211, 727–742. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Veillette, A. How do SAP family deficiencies compromise immunity? Trends Immunol. 2010, 31, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Le Borgne, M.; Shaw, A.S. SAP signaling: A dual mechanism of action. Immunity 2012, 36, 899–901. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, S.; Dong, Z. NK cell recognition of hematopoietic cells by SLAM-SAP families. Cell. Mol. Immunol. 2019, 16, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Atanackovic, D.; Panse, J.; Hildebrandt, Y.; Jadczak, A.; Kobold, S.; Cao, Y.; Templin, J.; Meyer, S.; Reinhard, H.; Bartels, K.; et al. Surface molecule CD229 as a novel target for the diagnosis and treatment of multiple myeloma. Haematologica 2011, 96, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Gordiienko, I.M.; Shlapatska, L.M.; Kovalevska, L.M.; Sidorenko, S.P. Differential expression of CD150/SLAMF1 in normal and malignant B cells on the different stages of maturation. Exp. Oncol. 2016, 38, 101–107. [Google Scholar] [CrossRef]
- Hsi, E.D.; Steinle, R.; Balasa, B.; Szmania, S.; Draksharapu, A.; Shum, B.P.; Huseni, M.; Powers, D.; Nanisetti, A.; Zhang, Y.; et al. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 2775–2784. [Google Scholar] [CrossRef]
- Radhakrishnan, S.V.; Bhardwaj, N.; Luetkens, T.; Atanackovic, D. Novel anti-myeloma immunotherapies targeting the SLAM family of receptors. Oncoimmunology 2017, 6, e1308618. [Google Scholar] [CrossRef]
- Muccio, V.E.; Saraci, E.; Gilestro, M.; Gattei, V.; Zucchetto, A.; Astolfi, M.; Ruggeri, M.; Marzanati, E.; Passera, R.; Palumbo, A.; et al. Multiple myeloma: New surface antigens for the characterization of plasma cells in the era of novel agents. Cytom. Part B Clin. Cytom. 2016, 90, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Eck, M.J.; Terhorst, C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat. Rev. Immunol. 2003, 3, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Sayos, J.; Wu, C.; Morra, M.; Wang, N.; Zhang, X.; Allen, D.; van Schaik, S.; Notarangelo, L.; Geha, R.; Roncarolo, M.G.; et al. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature 1998, 395, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Leone, P.E.; Chiecchio, L.; Dickens, N.J.; Jenner, M.W.; Boyd, K.D.; Johnson, D.C.; Gonzalez, D.; Dagrada, G.P.; Protheroe, R.K.; et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 2010, 116, e56–e65. [Google Scholar] [CrossRef] [PubMed]
- Smetana, J.; Oppelt, J.; Štork, M.; Pour, L.; Kuglík, P. Chromothripsis 18 in multiple myeloma patient with rapid extramedullary relapse. Mol. Cytogenet. 2018, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Sklavenitis-Pistofidis, R.; Reidy, M.; Huynh, D.; Salem, K.Z.; Park, J.; Glavey, S.; Leleu, X.; Roo, D.; Ghobrial, I.M.; Manier, S. Founding Precision Therapy in 1q-Amplified Multiple Myeloma. Blood 2018, 132 (Suppl. 1), 1007. [Google Scholar] [CrossRef]
- McArdel, S.L.; Terhorst, C.; Sharpe, A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016, 164, 10–20. [Google Scholar] [CrossRef]
- Brown, M.H.; Boles, K.; van der Merwe, P.A.; Kumar, V.; Mathew, P.A.; Barclay, A.N. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 1998, 188, 2083–2090. [Google Scholar] [CrossRef]
- Arulanandam, A.R.; Moingeon, P.; Concino, M.F.; Recny, M.A.; Kato, K.; Yagita, H.; Koyasu, S.; Reinherz, E.L. A soluble multimeric recombinant CD2 protein identifies CD48 as a low affinity ligand for human CD2: Divergence of CD2 ligands during the evolution of humans and mice. J. Exp. Med. 1993, 177, 1439–1450. [Google Scholar] [CrossRef]
- Moran, M.; Miceli, M.C. Engagement of GPI-linked CD48 contributes to TCR signals and cytoskeletal reorganization: A role for lipid rafts in T cell activation. Immunity 1998, 9, 787–796. [Google Scholar] [CrossRef]
- Kis-Toth, K.; Tsokos, G.C. Engagement of SLAMF2/CD48 prolongs the time frame of effective T cell activation by supporting mature dendritic cell survival. J. Immunol. 2014, 192, 4436–4442. [Google Scholar] [CrossRef] [PubMed]
- Meinke, S.; Watzl, C. NK cell cytotoxicity mediated by 2B4 and NTB-A is dependent on SAP acting downstream of receptor phosphorylation. Front. Immunol. 2013, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Morandi, B.; Costa, R.; Falco, M.; Parolini, S.; de Maria, A.; Ratto, G.; Mingari, M.C.; Melioli, G.; Moretta, A.; Ferlazzo, G. Distinctive lack of CD48 expression in subsets of human dendritic cells tunes NK cell activation. J. Immunol. 2005, 175, 3690–3697. [Google Scholar] [CrossRef] [PubMed]
- Agresta, L.; Hoebe, K.H.N.; Janssen, E.M. The Emerging Role of CD244 Signaling in Immune Cells of the Tumor Microenvironment. Front. Immunol. 2018, 9, 2809. [Google Scholar] [CrossRef] [PubMed]
- Zelle-Rieser, C.; Thangavadivel, S.; Biedermann, R.; Brunner, A.; Stoitzner, P.; Willenbacher, E.; Greil, R.; Jöhrer, K. T cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J. Hematol. Oncol. 2016, 9, 116. [Google Scholar] [CrossRef] [PubMed]
- Baitsch, L.; Baumgaertner, P.; Devêvre, E.; Raghav, S.K.; Legat, A.; Barba, L.; Wieckowski, S.; Bouzourene, H.; Deplancke, B.; Romero, P.; et al. Exhaustion of tumor-specific CD8⁺ T cells in metastases from melanoma patients. J. Clin. Investig. 2011, 121, 2350–2360. [Google Scholar] [CrossRef]
- Wu, Y.; Kuang, D.M.; Pan, W.D.; Wan, Y.L.; Lao, X.M.; Wang, D.; Li, X.F.; Zheng, L. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology 2013, 57, 1107–1116. [Google Scholar] [CrossRef]
- Tan, J.; Chen, S.; Lu, Y.; Yao, D.; Xu, L.; Zhang, Y.; Yang, L.; Chen, J.; Lai, J.; Yu, Z.; et al. Higher PD-1 expression concurrent with exhausted CD8+ T cells in patients with de novo acute myeloid leukemia. Chin. J. Cancer Res. = Chung-Kuo Yen Cheng Yen Chiu 2017, 29, 463–470. [Google Scholar] [CrossRef]
- Hosen, N.; Ichihara, H.; Mugitani, A.; Aoyama, Y.; Fukuda, Y.; Kishida, S.; Matsuoka, Y.; Nakajima, H.; Kawakami, M.; Yamagami, T.; et al. CD48 as a novel molecular target for antibody therapy in multiple myeloma. Br. J. Haematol. 2012, 156, 213–224. [Google Scholar] [CrossRef]
- Ashour, R.; Ri, M.; Aly, S.S.; Yoshida, T.; Tachita, T.; Kanamori, T.; Aoki, S.; Kinoshita, S.; Narita, T.; Totani, H.; et al. Expression analysis of two SLAM family receptors, SLAMF2 and SLAMF7, in patients with multiple myeloma. Int. J. Hematol. 2019, 110, 69–76. [Google Scholar] [CrossRef]
- Lewis, T.S.; Olson, D.; Gordon, K.; Sandall, S.; Quick, M.; Finn, M.; Westendorf, L.; Linares, G.; Leiske, C.; Nesterova, A.; et al. SGN-CD48A: A Novel Humanized Anti-CD48 Antibody-Drug Conjugate for the Treatment of Multiple Myeloma. Blood 2016, 76, 4470. [Google Scholar] [CrossRef]
- Olson, D.J.; Liu, B.A.; Zaval, M.; Cao, A.; Gurgel, J.; Cochran, J.; Stevens, N.; Anderson, M.; Lewis, T.S. Additional mechanisms of action of SGN-CD48A in multiple myeloma and improved antitumor activity in combination with daratumumab. Cancer Res. 2018, 78, 5619. [Google Scholar] [CrossRef]
- Sintes, J.; Vidal-Laliena, M.; Romero, X.; Tovar, V.; Engel, P. Characterization of mouse CD229 (Ly9), a leukocyte cell surface molecule of the CD150 (SLAM) family. Tissue Antigens 2007, 70, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Sandrin, M.S.; Henning, M.M.; Lo, M.F.; Baker, E.; Sutherland, G.R.; McKenzie, I.F. Isolation and characterization of cDNA clones for Humly9: The human homologue of mouse Ly9. Immunogenetics 1996, 43, 13–19. [Google Scholar] [CrossRef]
- de la Fuente, M.A.; Tovar, V.; Villamor, N.; Zapater, N.; Pizcueta, P.; Campo, E.; Bosch, J.; Engel, P. Molecular characterization and expression of a novel human leukocyte cell-surface marker homologous to mouse Ly-9. Blood 2001, 97, 3513–3520. [Google Scholar] [CrossRef]
- Romero, X.; Zapater, N.; Calvo, M.; Kalko, S.G.; de la Fuente, M.A.; Tovar, V.; Ockeloen, C.; Pizcueta, P.; Engel, P. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. J. Immunol. 2005, 174, 7033–7042. [Google Scholar] [CrossRef]
- Cannons, J.L.; Yu, L.J.; Hill, B.; Mijares, L.A.; Dombroski, D.; Nichols, K.E.; Antonellis, A.; Koretzky, G.A.; Gardner, K.; Schwartzberg, P.L. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity 2004, 21, 693–706. [Google Scholar] [CrossRef]
- Sayós, J.; Martín, M.; Chen, A.; Simarro, M.; Howie, D.; Morra, M.; Engel, P.; Terhorst, C. Cell surface receptors Ly-9 and CD84 recruit the X-linked lymphoproliferative disease gene product SAP. Blood 2001, 97, 3867–3874. [Google Scholar] [CrossRef]
- Simarro, M.; Lanyi, A.; Howie, D.; Poy, F.; Bruggeman, J.; Choi, M.; Sumegi, J.; Eck, M.J.; Terhorst, C. SAP increases FynT kinase activity and is required for phosphorylation of SLAM and Ly9. Int. Immunol. 2004, 16, 727–736. [Google Scholar] [CrossRef]
- Graham, D.B.; Bell, M.P.; McCausland, M.M.; Huntoon, C.J.; van Deursen, J.; Faubion, W.A.; Crotty, S.; McKean, D.J. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J. Immunol. 2006, 176, 291–300. [Google Scholar] [CrossRef]
- Tembhare, P.R.; Ghogale, S.; Tauro, W.; Badrinath, Y.; Deshpande, N.; Kedia, S.; Cherian, K.; Patkar, N.V.; Chatterjee, G.; Gujral, S.; et al. Evaluation of CD229 as a new alternative plasma cell gating marker in the flow cytometric immunophenotyping of monoclonal gammopathies. Cytom. Part B Clin. Cytom. 2018, 94, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Yousef, S.; Kovacsovics-Bankowski, M.; Salama, M.E.; Bhardwaj, N.; Steinbach, M.; Langemo, A.; Kovacsovics, T.; Marvin, J.; Binder, M.; Panse, J.; et al. CD229 is expressed on the surface of plasma cells carrying an aberrant phenotype and chemotherapy-resistant precursor cells in multiple myeloma. Hum. Vaccines Immunother. 2015, 11, 1606–1611. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Takahashi, R.; Tsubota, A.; Sasaki, M.; Handa, H.; Imai, Y.; Tanaka, N.; Tsukune, Y.; Tanosaki, S.; Ito, S.; et al. SLAMF3-Mediated Signaling via ERK Pathway Activation Promotes Aggressive Phenotypic Behaviors in Multiple Myeloma. Mol. Cancer Res. MCR 2020, 18, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Pojero, F.; Flores-Montero, J.; Sanoja, L.; Pérez, J.J.; Puig, N.; Paiva, B.; Bottcher, S.; van Dongen, J.J.; Orfao, A. Utility of CD54, CD229, and CD319 for the identification of plasma cells in patients with clonal plasma cell diseases. Cytom. Part B Clin. Cytom. 2016, 90, 91–100. [Google Scholar] [CrossRef]
- Ijaz, M.; Wang, F.; Shahbaz, M.; Jiang, W.; Fathy, A.H.; Nesa, E.U. The Role of Grb2 in Cancer and Peptides as Grb2 Antagonists. Protein Pept. Lett. 2018, 24, 1084–1095. [Google Scholar] [CrossRef]
- Lin, Q.; Zhao, J.; Song, Y.; Liu, D. Recent updates on CAR T clinical trials for multiple myeloma. Mol. Cancer 2019, 18, 154. [Google Scholar] [CrossRef]
- Cunninghame-Graham, D.S.; Vyse, T.J.; Fortin, P.R.; Montpetit, A.; Cai, Y.C.; Lim, S.; McKenzie, T.; Farwell, L.; Rhodes, B.; Chad, L.; et al. Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008, 9, 93–102. [Google Scholar] [CrossRef]
- Margraf, S.; Garner, L.I.; Wilson, T.J.; Brown, M.H. A polymorphism in a phosphotyrosine signalling motif of CD229 (Ly9, SLAMF3) alters SH2 domain binding and T-cell activation. Immunology 2015, 146, 392–400. [Google Scholar] [CrossRef]
- Ishibashi, M.; Sunakawa-Kii, M.; Kaito, Y.; Kinoshita, R.; Asayama, T.; Kuribayashi, Y.; Inokuchi, K.; Morita, R.; Tamura, H. The SLAMF3 rs509749 polymorphism correlates with malignant potential in multiple myeloma. Exp. Hematol. 2020. [Google Scholar] [CrossRef]
- Radhakrishnan, S.V.; Luetkens, T.; Scherer, S.D.; Davis, P.; Vander-Mause, E.R.; Olson, M.L.; Yousef, S.; Panse, J.; Abdiche, Y.; Li, K.D.; et al. CD229 CAR T cells eliminate multiple myeloma and tumor propagating cells without fratricide. Nat. Commun. 2020, 11, 798. [Google Scholar] [CrossRef]
- Cao, E.; Ramagopal, U.A.; Fedorov, A.; Fedorov, E.; Yan, Q.; Lary, J.W.; Cole, J.L.; Nathenson, S.G.; Almo, S.C. NTB-A receptor crystal structure: Insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family. Immunity 2006, 25, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Dragovich, M.A.; Adam, K.; Strazza, M.; Tocheva, A.S.; Peled, M.; Mor, A. SLAMF6 clustering is required to augment T cell activation. PLoS ONE 2019, 14, e0218109. [Google Scholar] [CrossRef] [PubMed]
- Valdez, P.A.; Wang, H.; Seshasayee, D.; van Lookeren-Campagne, M.; Gurney, A.; Lee, W.P.; Grewal, I.S. NTB-A, a new activating receptor in T cells that regulates autoimmune disease. J. Biol. Chem. 2004, 279, 18662–18669. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Kis-Toth, K.; Thai, T.H.; Terhorst, C.; Tsokos, G.C. SLAMF6-driven co-stimulation of human peripheral T cells is defective in SLE T cells. Autoimmunity 2011, 44, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Radomir, L.; Cohen, S.; Kramer, M.P.; Bakos, E.; Lewinsky, H.; Barak, A.; Porat, Z.; Bucala, R.; Stepensky, P.; Becker-Herman, S.; et al. T Cells Regulate Peripheral Naive Mature B Cell Survival by Cell-Cell Contact Mediated through SLAMF6 and SAP. J. Immunol. 2017, 199, 2745–2757. [Google Scholar] [CrossRef]
- Lewis, T.; Olson, D.J.; Gordon, K.A.; Sandall, S.L.; Miyamoto, J.; Westendorf, L.; Linares, G.; Leiske, C.; Kostner, H.; Stone, I.; et al. SGN-CD352A: A novel humanized anti-CD352 antibody-drug conjugate for the treatment of multiple myeloma. Cancer Res. 2016, 76, 1195. [Google Scholar] [CrossRef]
- Veillette, A.; Guo, H. CS1, a SLAM family receptor involved in immune regulation, is a therapeutic target in multiple myeloma. Crit. Rev. Oncol. Hematol. 2013, 88, 168–177. [Google Scholar] [CrossRef]
- Cruz-Munoz, M.E.; Dong, Z.; Shi, X.; Zhang, S.; Veillette, A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat. Immunol. 2009, 10, 297–305. [Google Scholar] [CrossRef]
- Tassi, I.; Colonna, M. The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase Cgamma signaling pathways in human NK cells. J. Immunol. 2005, 175, 7996–8002. [Google Scholar] [CrossRef]
- Tai, Y.T.; Dillon, M.; Song, W.; Leiba, M.; Li, X.F.; Burger, P.; Lee, A.I.; Podar, K.; Hideshima, T.; Rice, A.G.; et al. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood 2008, 112, 1329–1337. [Google Scholar] [CrossRef]
- Ritchie, D.; Colonna, M. Mechanisms of Action and Clinical Development of Elotuzumab. Clin. Transl. Sci. 2018, 11, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Szmania, S.; van Rhee, F. Profile of elotuzumab and its potential in the treatment of multiple myeloma. Blood Lymphat. Cancer Targets Ther. 2014, 2014, 15–27. [Google Scholar] [CrossRef]
- Weisel, K. Spotlight on elotuzumab in the treatment of multiple myeloma: The evidence to date. OncoTargets Ther. 2016, 9, 6037–6048. [Google Scholar] [CrossRef]
- Collins, S.M.; Bakan, C.E.; Swartzel, G.D.; Hofmeister, C.C.; Efebera, Y.A.; Kwon, H.; Starling, G.C.; Ciarlariello, D.; Bhaskar, S.; Briercheck, E.L.; et al. Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: Evidence for augmented NK cell function complementing ADCC. Cancer Immunol. Immunother. CII 2013, 62, 1841–1849. [Google Scholar] [CrossRef] [PubMed]
- Pazina, T.; James, A.M.; MacFarlane, A.W.T.; Bezman, N.A.; Henning, K.A.; Bee, C.; Graziano, R.F.; Robbins, M.D.; Cohen, A.D.; Campbell, K.S. The anti-SLAMF7 antibody elotuzumab mediates NK cell activation through both CD16-dependent and -independent mechanisms. Oncoimmunology 2017, 6, e1339853. [Google Scholar] [CrossRef] [PubMed]
- Pazina, T.; James, A.M.; Colby, K.B.; Yang, Y.; Gale, A.; Jhatakia, A.; Kearney, A.Y.; Graziano, R.F.; Bezman, N.A.; Robbins, M.D.; et al. Enhanced SLAMF7 Homotypic Interactions by Elotuzumab Improves NK Cell Killing of Multiple Myeloma. Cancer Immunol. Res. 2019, 7, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Kurdi, A.T.; Glavey, S.V.; Bezman, N.A.; Jhatakia, A.; Guerriero, J.L.; Manier, S.; Moschetta, M.; Mishima, Y.; Roccaro, A.; Detappe, A.; et al. Antibody-Dependent Cellular Phagocytosis by Macrophages is a Novel Mechanism of Action of Elotuzumab. Mol. Cancer Ther. 2018, 17, 1454–1463. [Google Scholar] [CrossRef]
- Zonder, J.A.; Mohrbacher, A.F.; Singhal, S.; van Rhee, F.; Bensinger, W.I.; Ding, H.; Fry, J.; Afar, D.E.; Singhal, A.K. A phase 1, multicenter, open-label, dose escalation study of elotuzumab in patients with advanced multiple myeloma. Blood 2012, 120, 552–559. [Google Scholar] [CrossRef]
- van Rhee, F.; Szmania, S.M.; Dillon, M.; van Abbema, A.M.; Li, X.; Stone, M.K.; Garg, T.K.; Shi, J.; Moreno-Bost, A.M.; Yun, R.; et al. Combinatorial efficacy of anti-CS1 monoclonal antibody elotuzumab (HuLuc63) and bortezomib against multiple myeloma. Mol. Cancer Ther. 2009, 8, 2616–2624. [Google Scholar] [CrossRef]
- Lonial, S.; Dimopoulos, M.; Palumbo, A.; White, D.; Grosicki, S.; Spicka, I.; Walter-Croneck, A.; Moreau, P.; Mateos, M.V.; Magen, H.; et al. Elotuzumab Therapy for Relapsed or Refractory Multiple Myeloma. N. Engl. J. Med. 2015, 373, 621–631. [Google Scholar] [CrossRef]
- Dimopoulos, M.A.; Lonial, S.; Betts, K.A.; Chen, C.; Zichlin, M.L.; Brun, A.; Signorovitch, J.E.; Makenbaeva, D.; Mekan, S.; Sy, O.; et al. Elotuzumab plus lenalidomide and dexamethasone in relapsed/refractory multiple myeloma: Extended 4-year follow-up and analysis of relative progression-free survival from the randomized ELOQUENT-2 trial. Cancer 2018, 124, 4032–4043. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, M.A.; Dytfeld, D.; Grosicki, S.; Moreau, P.; Takezako, N.; Hori, M.; Leleu, X.; LeBlanc, R.; Suzuki, K.; Raab, M.S.; et al. Elotuzumab plus Pomalidomide and Dexamethasone for Multiple Myeloma. N. Engl. J. Med. 2018, 379, 1811–1822. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Cruz-Munoz, M.E.; Wu, N.; Robbins, M.; Veillette, A. Immune cell inhibition by SLAMF7 is mediated by a mechanism requiring src kinases, CD45, and SHIP-1 that is defective in multiple myeloma cells. Mol. Cell. Biol. 2015, 35, 41–51. [Google Scholar] [CrossRef]
- Kikuchi, J.; Hori, M.; Iha, H.; Toyama-Sorimachi, N.; Hagiwara, S.; Kuroda, Y.; Koyama, D.; Izumi, T.; Yasui, H.; Suzuki, A.; et al. Soluble SLAMF7 promotes the growth of myeloma cells via homophilic interaction with surface SLAMF7. Leukemia 2020, 34, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Soeda, S.; Sasaki, M.; Handa, H.; Imai, Y.; Tanaka, N.; Tanosaki, S.; Ito, S.; Odajima, T.; Sugimori, H.; et al. Clinical impact of serum soluble SLAMF7 in multiple myeloma. Oncotarget 2018, 9, 34784–34793. [Google Scholar] [CrossRef]
- Xie, Z.; Gunaratne, J.; Cheong, L.L.; Liu, S.C.; Koh, T.L.; Huang, G.; Blackstock, W.P.; Chng, W.J. Plasma membrane proteomics identifies biomarkers associated with MMSET overexpression in T(4;14) multiple myeloma. Oncotarget 2013, 4, 1008–1018. [Google Scholar] [CrossRef]
- Dimopoulos, M.A.; Lonial, S.; White, D.; Moreau, P.; Palumbo, A.; San-Miguel, J.; Shpilberg, O.; Anderson, K.; Grosicki, S.; Spicka, I.; et al. Elotuzumab plus lenalidomide/dexamethasone for relapsed or refractory multiple myeloma: ELOQUENT-2 follow-up and post-hoc analyses on progression-free survival and tumour growth. Br. J. Haematol. 2017, 178, 896–905. [Google Scholar] [CrossRef]
- Chu, J.; He, S.; Deng, Y.; Zhang, J.; Peng, Y.; Hughes, T.; Yi, L.; Kwon, C.H.; Wang, Q.E.; Devine, S.M.; et al. Genetic modification of T cells redirected toward CS1 enhances eradication of myeloma cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 3989–4000. [Google Scholar] [CrossRef]
- Gogishvili, T.; Danhof, S.; Prommersberger, S.; Rydzek, J.; Schreder, M.; Brede, C.; Einsele, H.; Hudecek, M. SLAMF7-CAR T cells eliminate myeloma and confer selective fratricide of SLAMF7(+) normal lymphocytes. Blood 2017, 130, 2838–2847. [Google Scholar] [CrossRef]
- Zhao, J.; Lin, Q.; Song, Y.; Liu, D. Universal CARs, universal T cells, and universal CAR T cells. J. Hematol. Oncol. 2018, 11, 132. [Google Scholar] [CrossRef]
- Mathur, R.; Zhang, Z.; He, J.; Galetto, R.; Gouble, A.; Chion-Sotinel, I.; Filipe, S.; Gariboldi, A.; Veeramachaneni, T.; Manasanch, E.E.; et al. Universal SLAMF7-Specific CAR T-Cells As Treatment for Multiple Myeloma. Blood 2017, 130 (Suppl. 1), 502. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishibashi, M.; Morita, R.; Tamura, H. Immune Functions of Signaling Lymphocytic Activation Molecule Family Molecules in Multiple Myeloma. Cancers 2021, 13, 279. https://doi.org/10.3390/cancers13020279
Ishibashi M, Morita R, Tamura H. Immune Functions of Signaling Lymphocytic Activation Molecule Family Molecules in Multiple Myeloma. Cancers. 2021; 13(2):279. https://doi.org/10.3390/cancers13020279
Chicago/Turabian StyleIshibashi, Mariko, Rimpei Morita, and Hideto Tamura. 2021. "Immune Functions of Signaling Lymphocytic Activation Molecule Family Molecules in Multiple Myeloma" Cancers 13, no. 2: 279. https://doi.org/10.3390/cancers13020279
APA StyleIshibashi, M., Morita, R., & Tamura, H. (2021). Immune Functions of Signaling Lymphocytic Activation Molecule Family Molecules in Multiple Myeloma. Cancers, 13(2), 279. https://doi.org/10.3390/cancers13020279





