Characterization of SOX2, OCT4 and NANOG in Ovarian Cancer Tumor-Initiating Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Spheroid Formation Assay
2.3. siRNA Transfections and Western Blots
2.4. RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
2.5. Cell Viability Assay
2.6. Flow Cytometry
2.7. Animal Experiments
2.8. Immunohistochemistry (IHC)
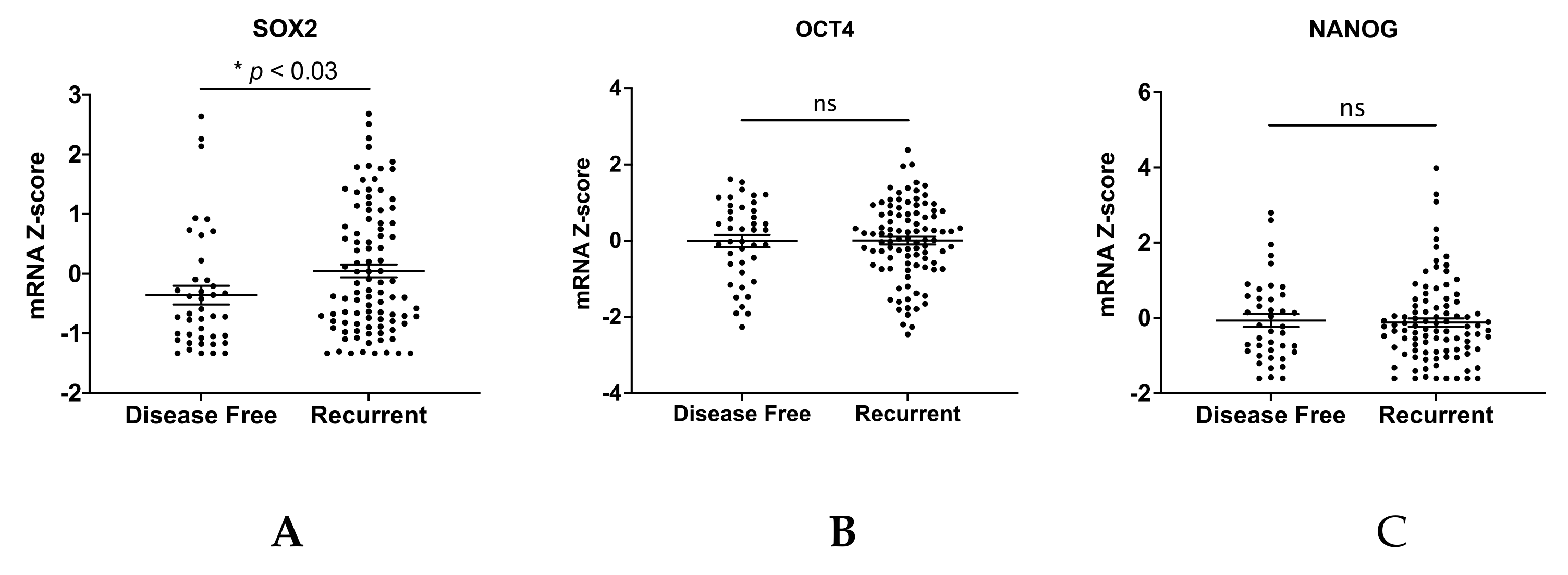
2.9. Public Database Analysis
2.10. Statistical Analysis
2.11. Data Availability
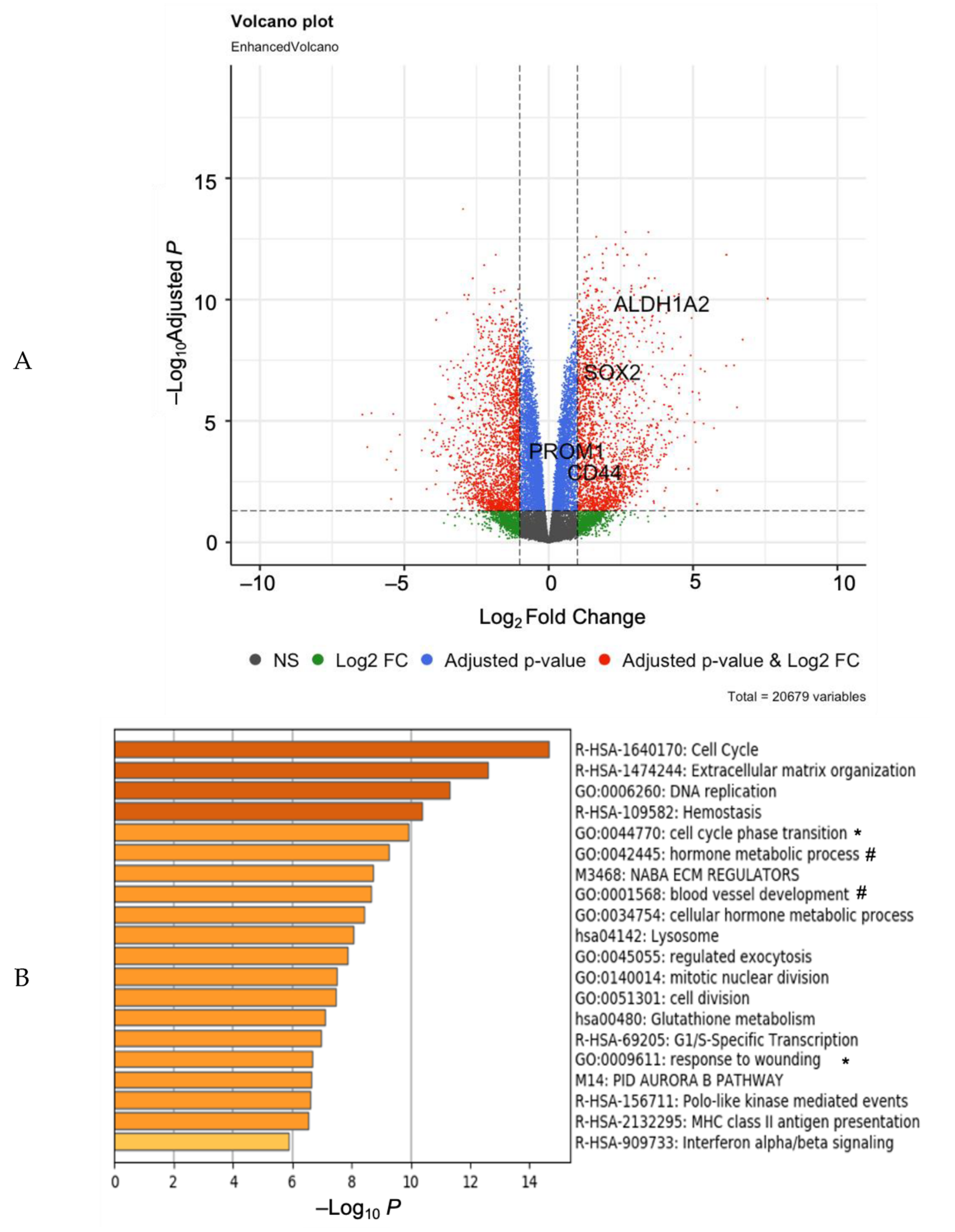
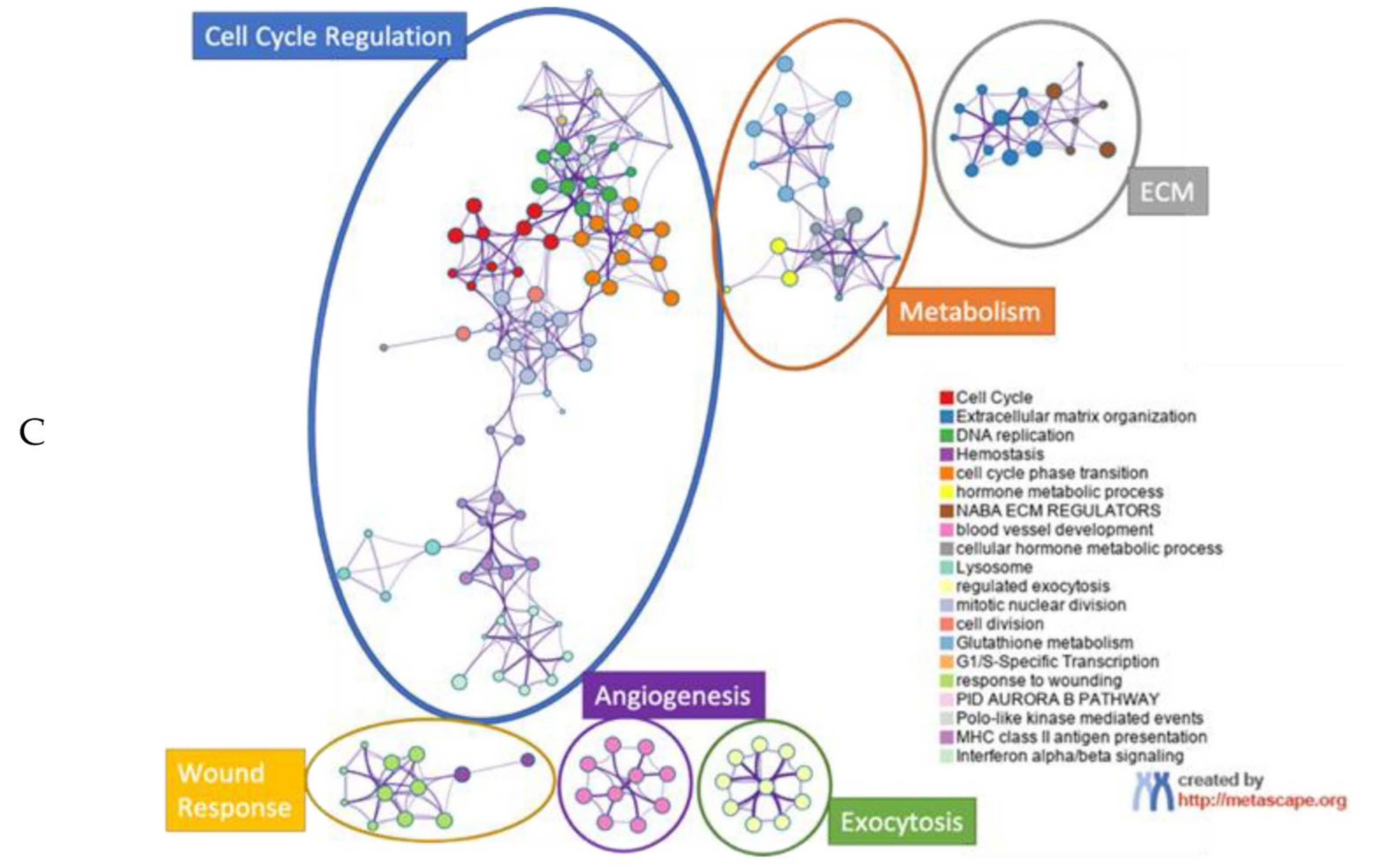
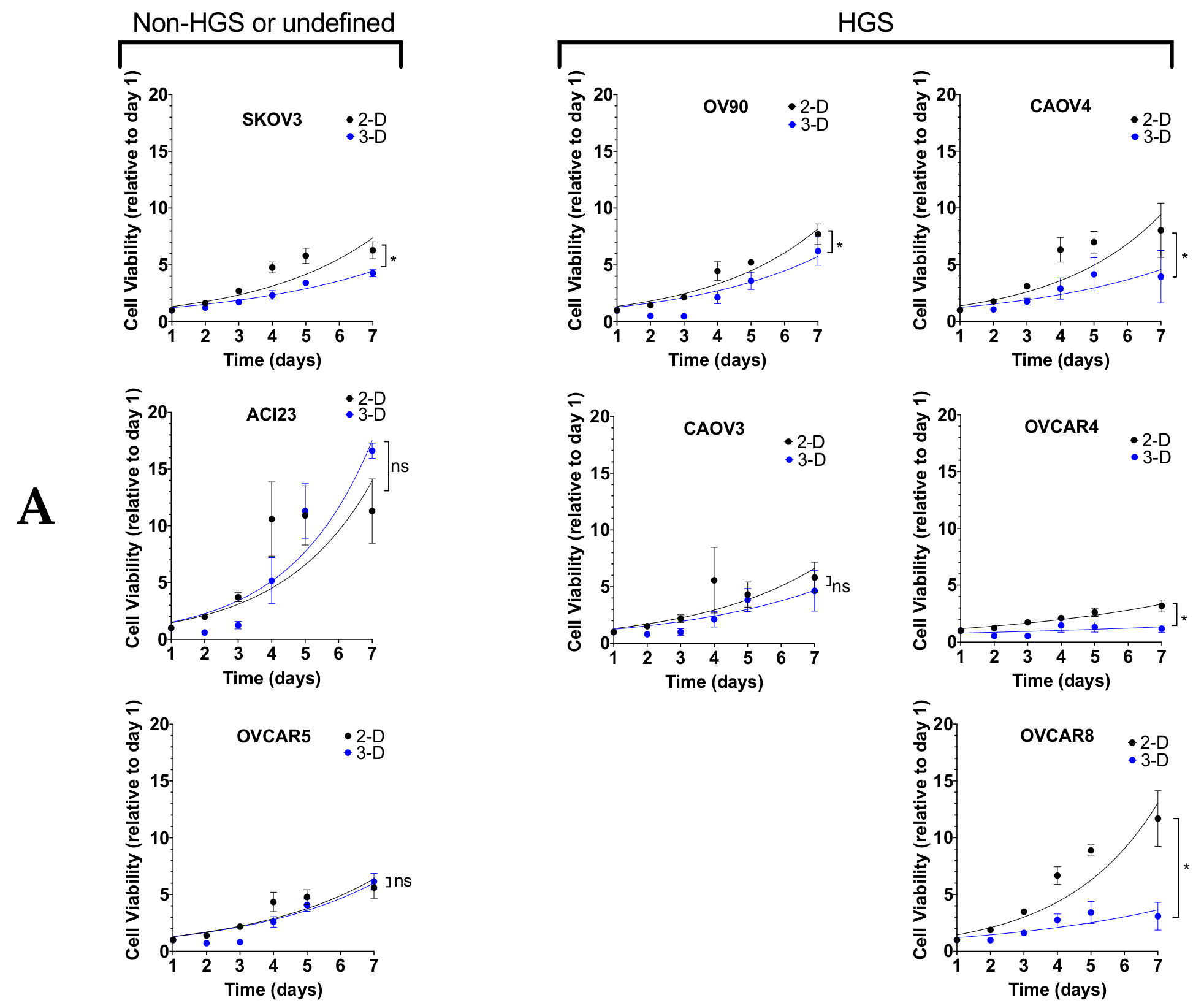
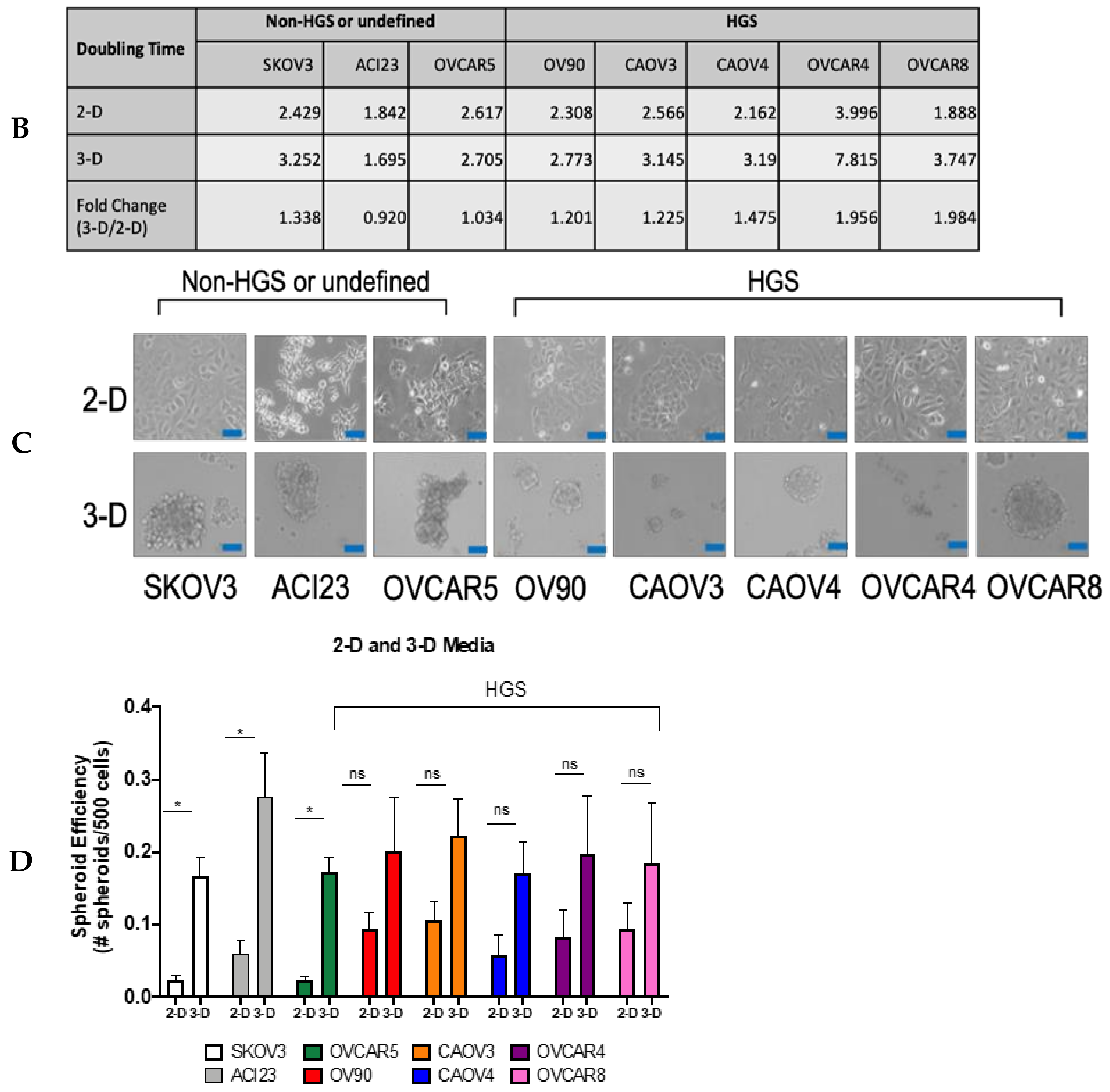
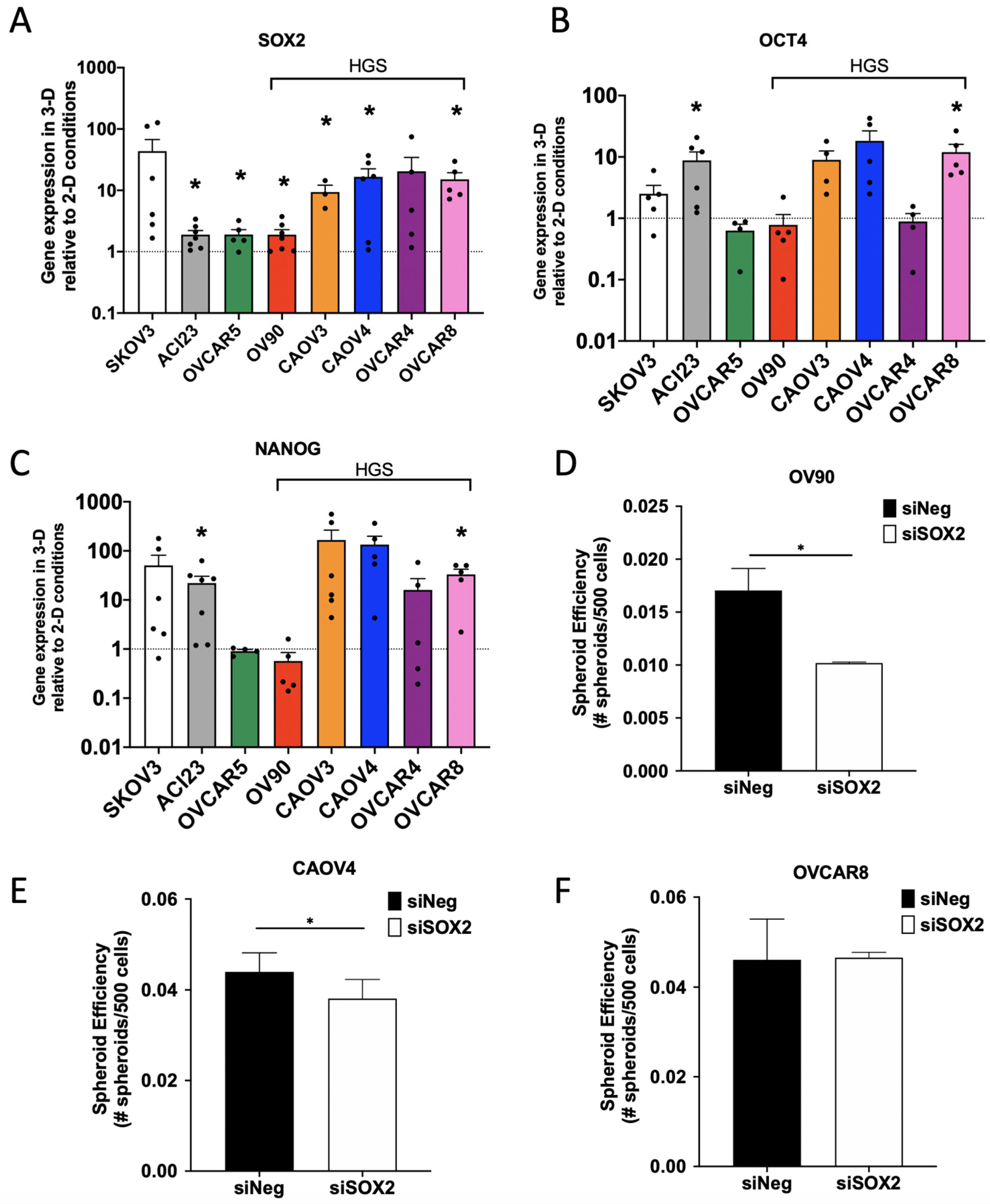
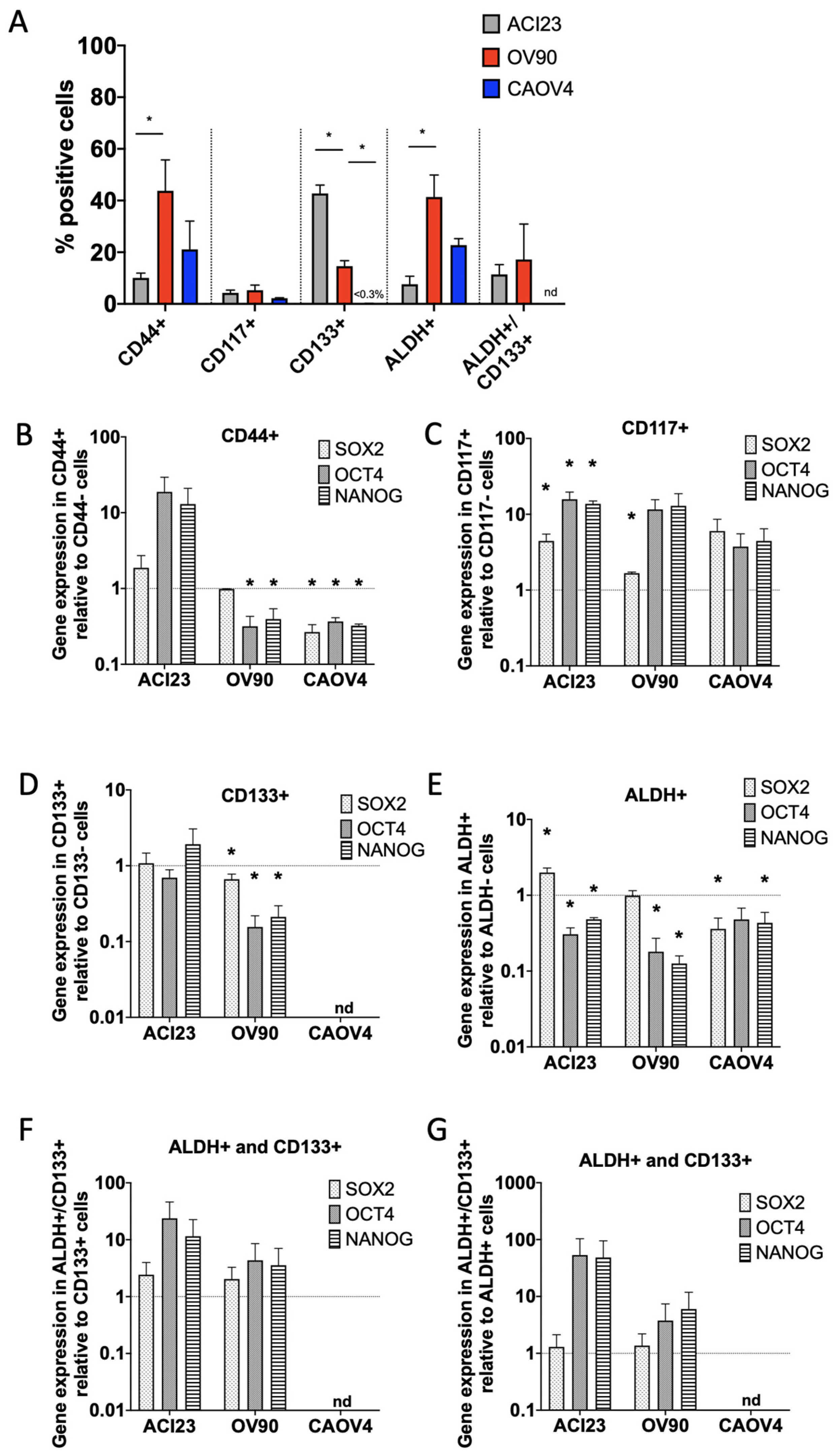
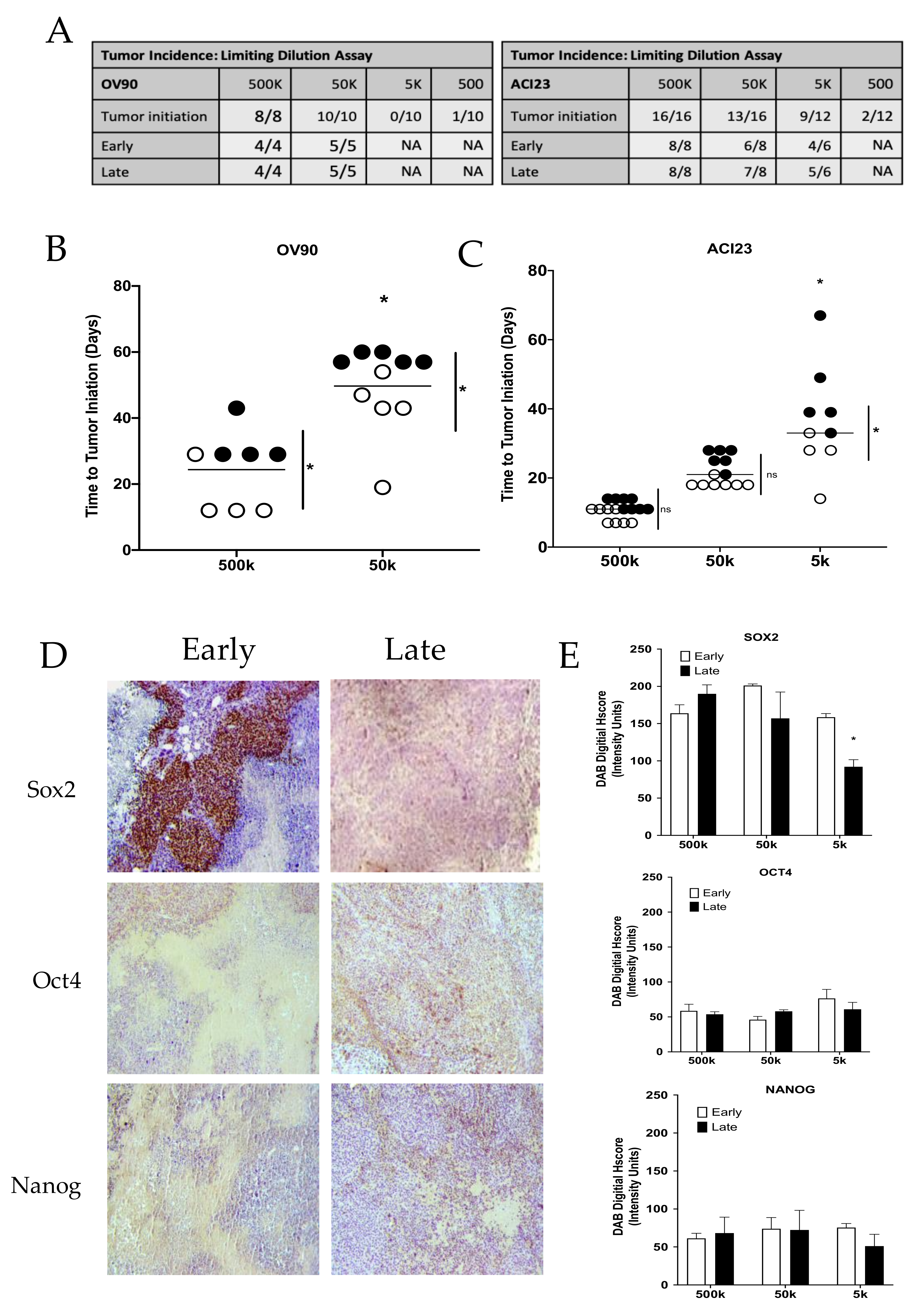
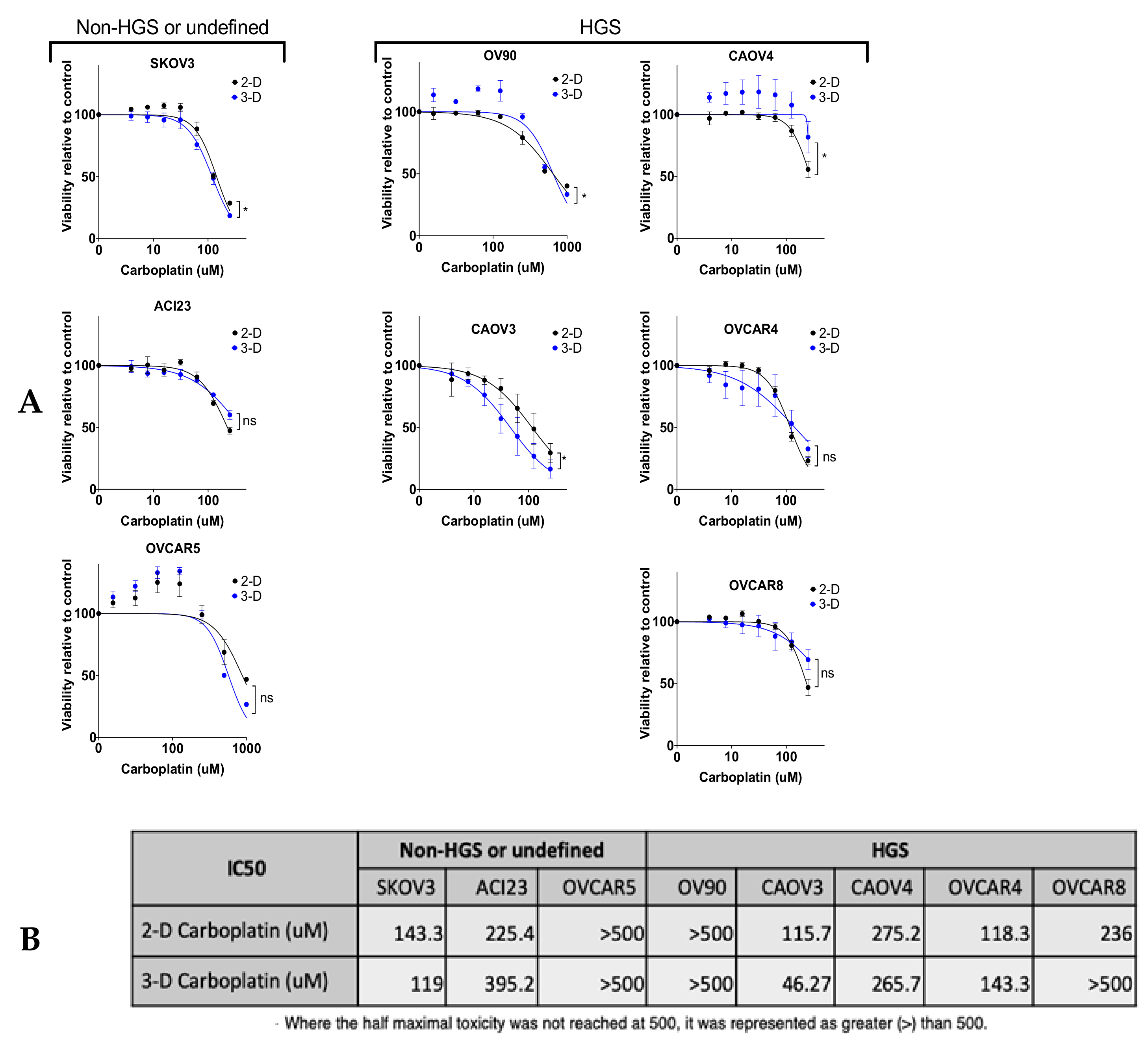
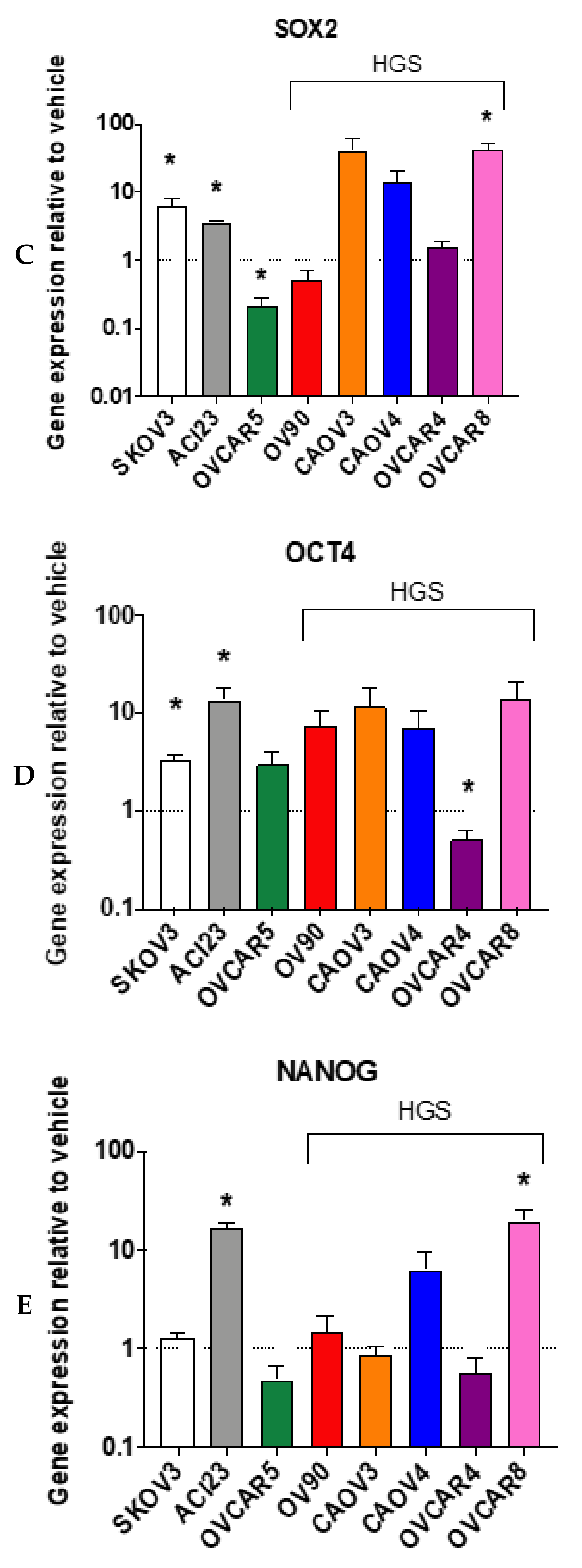
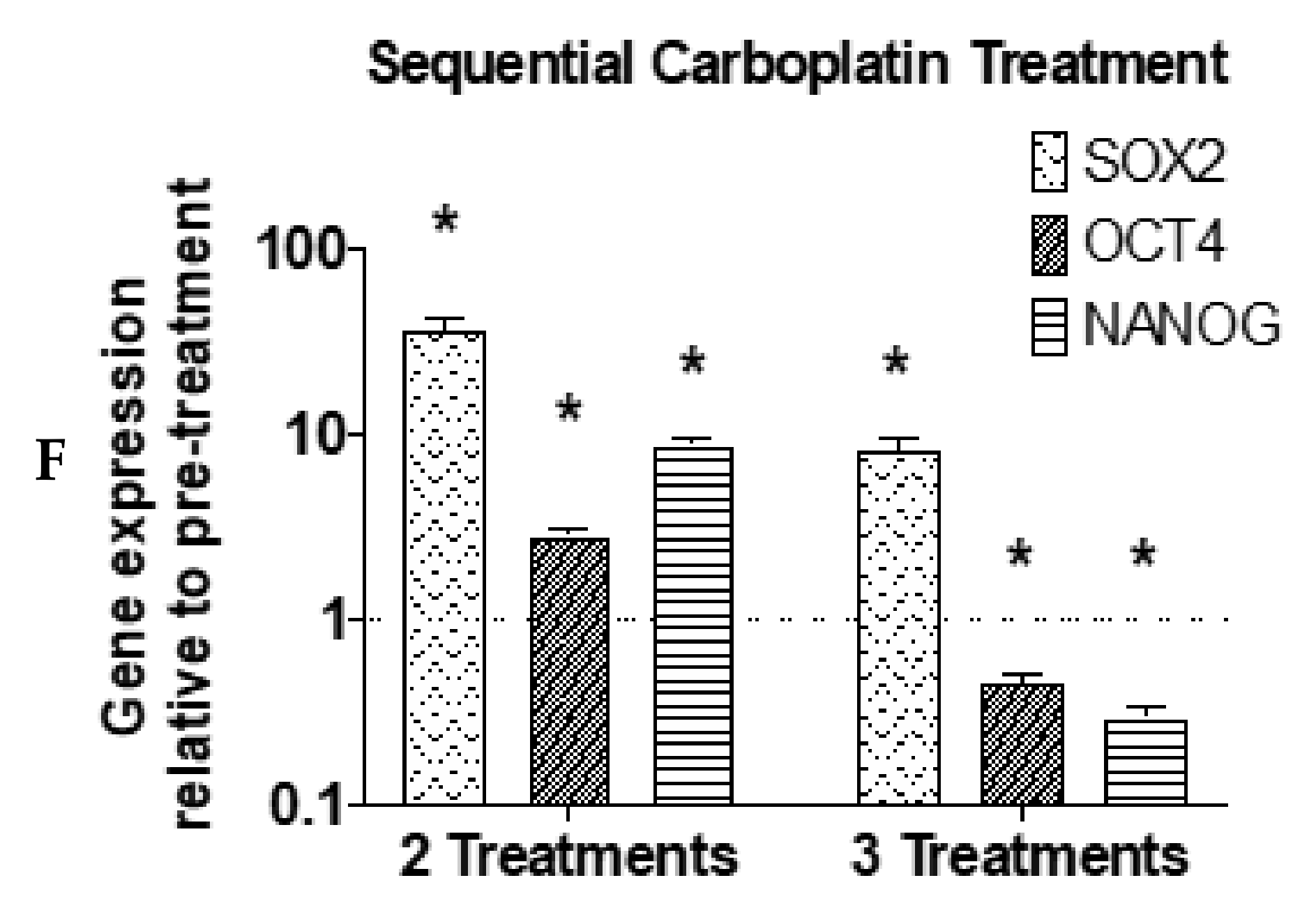
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, I.J.; Lui, P.P.; Obajdin, J.; Riccio, F.; Stroukov, W.; Willis, T.L.; Spagnoli, F.; Watt, F.M. Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence. Stem Cell Rep. 2019, 12, 1190–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Kong, J.; Brat, D.J. Cancer stem cell division: When the rules of asymmetry are broken. Stem Cells Dev. 2015, 24, 405–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.K.; Seo, E.J.; Choi, E.J.; Lee, S.I.; Kwon, Y.W.; Jang, I.H.; Kim, S.C.; Kim, K.H.; Suh, D.S.; Seong-Jang, K.; et al. Crucial role of HMGA1 in the self-renewal and drug resistance of ovarian cancer stem cells. Exp. Mol. Med. 2016, 48, e255. [Google Scholar] [CrossRef] [Green Version]
- Lathia, J.; Liu, H.; Matei, D. The Clinical Impact of Cancer Stem Cells. Oncologist 2019. [Google Scholar] [CrossRef] [Green Version]
- Terraneo, N.; Jacob, F.; Dubrovska, A.; Grünberg, J. Novel Therapeutic Strategies for Ovarian Cancer Stem Cells. Front. Oncol. 2020, 10, 319. [Google Scholar] [CrossRef] [Green Version]
- House, C.D.; Hernandez, L.; Annunziata, C.M. In vitro enrichment of ovarian cancer tumor-initiating cells. J. Vis. Exp. 2015. [Google Scholar] [CrossRef] [Green Version]
- Silva, I.A.; Bai, S.; McLean, K.; Yang, K.; Griffith, K.; Thomas, D.; Ginestier, C.; Johnston, C.; Kueck, A.; Reynolds, R.K.; et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011, 71, 3991–4001. [Google Scholar] [CrossRef] [Green Version]
- Hatina, J.; Boesch, M.; Sopper, S.; Kripnerova, M.; Wolf, D.; Reimer, D.; Marth, C.; Zeimet, A.G. Ovarian Cancer Stem Cell Heterogeneity. Adv. Exp. Med. Biol. 2019, 1139, 201–221. [Google Scholar] [CrossRef] [PubMed]
- Kryczek, I.; Liu, S.; Roh, M.; Vatan, L.; Szeliga, W.; Wei, S.; Banerjee, M.; Mao, Y.; Kotarski, J.; Wicha, M.S.; et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int. J. Cancer 2012, 130, 29–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bareiss, P.M.; Paczulla, A.; Wang, H.; Schairer, R.; Wiehr, S.; Kohlhofer, U.; Rothfuss, O.C.; Fischer, A.; Perner, S.; Staebler, A.; et al. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013, 73, 5544–5555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chefetz, I.; Grimley, E.; Yang, K.; Hong, L.; Vinogradova, E.V.; Suciu, R.; Kovalenko, I.; Karnak, D.; Morgan, C.A.; Chtcherbinine, M.; et al. A Pan-ALDH1A Inhibitor Induces Necroptosis in Ovarian Cancer Stem-like Cells. Cell Rep. 2019, 26, 3061–3075. [Google Scholar] [CrossRef] [Green Version]
- Lupia, M.; Cavallaro, U. Ovarian cancer stem cells: Still an elusive entity? Mol. Cancer 2017, 16, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 16061. [Google Scholar] [CrossRef]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354. [Google Scholar] [CrossRef] [Green Version]
- Samardzija, C.; Quinn, M.; Findlay, J.K.; Ahmed, N. Attributes of Oct4 in stem cell biology: Perspectives on cancer stem cells of the ovary. J. Ovarian Res. 2012, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.; Hou, Y.; Huang, Z.; Cai, J.; Wang, Z. SOX2 is required to maintain cancer stem cells in ovarian cancer. Cancer Sci. 2017, 108, 719–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parte, S.C.; Batra, S.K.; Kakar, S.S. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J. Ovarian Res. 2018, 11, 69. [Google Scholar] [CrossRef]
- House, C.D.; Jordan, E.; Hernandez, L.; Ozaki, M.; James, J.M.; Kim, M.; Kruhlak, M.J.; Batchelor, E.; Elloumi, F.; Cam, M.C.; et al. NFκB Promotes Ovarian Tumorigenesis via Classical Pathways That Support Proliferative Cancer Cells and Alternative Pathways That Support ALDH. Cancer Res. 2017, 77, 6927–6940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrich, D.G.; Lessey, B.A.; Savaris, R.F. Comparison of HSCORE assessment of endometrial beta3 integrin subunit expression with digital HSCORE using computerized image analysis (ImageJ). Anal. Quant. Cytopathol. Histpathol. 2013, 35, 210–216. [Google Scholar] [PubMed]
- Gao, Q.; Yang, Z.; Xu, S.; Li, X.; Yang, X.; Jin, P.; Liu, Y.; Zhou, X.; Zhang, T.; Gong, C.; et al. Heterotypic CAF-tumor spheroids promote early peritoneal metastatis of ovarian cancer. J. Exp. Med. 2019, 216, 688–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- cBioPortal for Cancer Genomics. Available online: www.cbioportal.org (accessed on 6 August 2020).
- Harrington, B.S.; Ozaki, M.K.; Caminear, M.W.; Hernandez, L.F.; Jordan, E.; Kalinowski, N.J.; Goldlust, I.S.; Guha, R.; Ferrer, M.; Thomas, C.; et al. Drugs Targeting Tumor-Initiating Cells Prolong Survival in a Post-Surgery, Post-Chemotherapy Ovarian Cancer Relapse Model. Cancers (Basel) 2020, 12, 1645. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef]
- Chen, M.W.; Yang, S.T.; Chien, M.H.; Hua, K.T.; Wu, C.J.; Hsiao, S.M.; Lin, H.; Hsiao, M.; Su, J.L.; Wei, L.H. The STAT3-miRNA-92-Wnt Signaling Pathway Regulates Spheroid Formation and Malignant Progression in Ovarian Cancer. Cancer Res. 2017, 77, 1955–1967. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.R.; Hoover, S.B.; Ross, M.M.; Zhou, W.; Meani, F.; Edwards, J.B.; Spehalski, E.I.; Risinger, J.I.; Alvord, W.G.; Quiñones, O.A.; et al. Serum S100A6 concentration predicts peritoneal tumor burden in mice with epithelial ovarian cancer and is associated with advanced stage in patients. PLoS ONE 2009, 4, e7670. [Google Scholar] [CrossRef]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef] [Green Version]
- Raghavan, S.; Mehta, P.; Ward, M.R.; Bregenzer, M.E.; Fleck, E.M.A.; Tan, L.; McLean, K.; Buckanovich, R.J.; Mehta, G. Personalized Medicine-Based Approach to Model Patterns of Chemoresistance and Tumor Recurrence Using Ovarian Cancer Stem Cell Spheroids. Clin. Cancer Res. 2017, 23, 6934–6945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgeois, D.L.; Kabarowski, K.A.; Porubsky, V.L.; Kreeger, P.K. High-grade serous ovarian cancer cell lines exhibit heterogeneous responses to growth factor stimulation. Cancer Cell. Int. 2015, 15, 112. [Google Scholar] [CrossRef] [PubMed]
- Beaufort, C.M.; Helmijr, J.C.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van IJcken, W.F.; Heine, A.A.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.; Brocker, C.; Thompson, D.C.; Black, W.; Vasiliou, K.; Nebert, D.W.; Vasiliou, V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genom. 2011, 5, 283–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Rizzino, A.; Wuebben, E.L. Sox2/Oct4: A delicately balanced partnership in pluripotent stem cells and embryogenesis. Biochim. Biophys. Acta 2016, 1859, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Raviv, A.; Esposito, D.; Flanders, K.C.; Daniel, C.; Nghiem, B.T.; Garfield, S.; Lim, L.; Mannan, P.; Robles, A.I.; et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Rep. 2015, 4, 155–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias, K.M.; Emori, M.M.; Papp, E.; MacDuffie, E.; Konecny, G.E.; Velculescu, V.E.; Drapkin, R. Beyond genomics: Critical evaluation of cell line utility for ovarian cancer research. Gynecol. Oncol. 2015, 139, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, L.; Kim, M.K.; Lyle, L.T.; Bunch, K.P.; House, C.D.; Ning, F.; Noonan, A.M.; Annunziata, C.M. Characterization of ovarian cancer cell lines as in vivo models for preclinical studies. Gynecol. Oncol. 2016, 142, 332–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.A.; Kwon, H.; Cho, H.; Chung, J.Y.; Hewitt, S.M.; Kim, J.H. ALDH1A2 Is a Candidate Tumor Suppressor Gene in Ovarian Cancer. Cancers (Basel) 2019, 11, 1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirst, J.; Pathak, H.B.; Hyter, S.; Pessetto, Z.Y.; Ly, T.; Graw, S.; Koestler, D.C.; Krieg, A.J.; Roby, K.F.; Godwin, A.K. Licofelone Enhances the Efficacy of Paclitaxel in Ovarian Cancer by Reversing Drug Resistance and Tumor Stem-like Properties. Cancer Res. 2018, 78, 4370–4385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgos-Ojeda, D.; Rueda, B.R.; Buckanovich, R.J. Ovarian cancer stem cell markers: Prognostic and therapeutic implications. Cancer Lett. 2012, 322, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, E.; Long, B.; Sullivan, P.; McClellan, S.; Finan, M.A.; Reed, E.; Shevde, L.; Rocconi, R.P. CD44+/CD24- ovarian cancer cells demonstrate cancer stem cell properties and correlate to survival. Clin. Exp. Metastasis 2012, 29, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zeng, J.; Liang, B.; Zhao, Z.; Sun, L.; Cao, D.; Yang, J.; Shen, K. Ovarian cancer cells with the CD117 phenotype are highly tumorigenic and are related to chemotherapy outcome. Exp. Mol. Pathol. 2011, 91, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, J.; Chen, D.; Yang, J.; Yang, C.; Zhang, Y.; Zhang, H.; Dou, J. Evaluation of characteristics of CD44+CD117+ ovarian cancer stem cells in three dimensional basement membrane extract scaffold versus two dimensional monocultures. BMC Cell Biol. 2013, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Zhang, P.; Wang, H.; Hou, D.; Li, W.; Xiao, G.; Li, C. Inhibitory effects of metformin at low concentration on epithelial-mesenchymal transition of CD44(+)CD117(+) ovarian cancer stem cells. Stem Cell Res. Ther. 2015, 6, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cioffi, M.; D’Alterio, C.; Camerlingo, R.; Tirino, V.; Consales, C.; Riccio, A.; Ieranò, C.; Cecere, S.C.; Losito, N.S.; Greggi, S.; et al. Identification of a distinct population of CD133(+)CXCR4(+) cancer stem cells in ovarian cancer. Sci. Rep. 2015, 5, 10357. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Chang, D.Y.; Mercado-Uribe, I.; Liu, J. Sex-determining region Y-box 2 expression predicts poor prognosis in human ovarian carcinoma. Hum. Pathol. 2012, 43, 1405–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bååth, M.; Westbom-Fremer, S.; Martin de la Fuente, L.; Ebbesson, A.; Davis, J.; Malander, S.; Måsbäck, A.; Kannisto, P.; Hedenfalk, I. SOX2 is a promising predictor of relapse and death in advanced stage high-grade serous ovarian cancer patients with residual disease after debulking surgery. Mol. Cell. Oncol. 2020, 7, 1805094. [Google Scholar] [CrossRef]
- Card, D.A.; Hebbar, P.B.; Li, L.; Trotter, K.W.; Komatsu, Y.; Mishina, Y.; Archer, T.K. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008, 28, 6426–6438. [Google Scholar] [CrossRef] [Green Version]
- Friman, E.T.; Deluz, C.; Meireles-Filho, A.C.; Govindan, S.; Gardeux, V.; Deplancke, B.; Suter, D.M. Dynamic regulation of chromatin accessibility by pluripotency transcription factors across the cell cycle. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Strebinger, D.; Deluz, C.; Friman, E.T.; Govindan, S.; Alber, A.B.; Suter, D.M. Endogenous fluctuations of OCT4 and SOX2 bias pluripotent cell fate decisions. Mol. Syst. Biol. 2019, 15, e9002. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, M.; Gilbert, S.F.; Waters, J.A.; Lujano-Olazaba, O.; Lara, J.; Alexander, L.J.; Green, S.E.; Burkeen, G.A.; Patrus, O.; Sarwar, Z.; et al. Characterization of SOX2, OCT4 and NANOG in Ovarian Cancer Tumor-Initiating Cells. Cancers 2021, 13, 262. https://doi.org/10.3390/cancers13020262
Robinson M, Gilbert SF, Waters JA, Lujano-Olazaba O, Lara J, Alexander LJ, Green SE, Burkeen GA, Patrus O, Sarwar Z, et al. Characterization of SOX2, OCT4 and NANOG in Ovarian Cancer Tumor-Initiating Cells. Cancers. 2021; 13(2):262. https://doi.org/10.3390/cancers13020262
Chicago/Turabian StyleRobinson, Mikella, Samuel F. Gilbert, Jennifer A. Waters, Omar Lujano-Olazaba, Jacqueline Lara, Logan J. Alexander, Samuel E. Green, Gregory A. Burkeen, Omid Patrus, Zinia Sarwar, and et al. 2021. "Characterization of SOX2, OCT4 and NANOG in Ovarian Cancer Tumor-Initiating Cells" Cancers 13, no. 2: 262. https://doi.org/10.3390/cancers13020262
APA StyleRobinson, M., Gilbert, S. F., Waters, J. A., Lujano-Olazaba, O., Lara, J., Alexander, L. J., Green, S. E., Burkeen, G. A., Patrus, O., Sarwar, Z., Holmberg, R., Wang, C., & House, C. D. (2021). Characterization of SOX2, OCT4 and NANOG in Ovarian Cancer Tumor-Initiating Cells. Cancers, 13(2), 262. https://doi.org/10.3390/cancers13020262






