Optimization and Validation of a Novel Three-Dimensional Co-Culture System in Decellularized Human Liver Scaffold for the Study of Liver Fibrosis and Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Decellularization Protocols
2.2. Cell Lines, Culture Conditions and Cell Treatment
2.3. Repopulation and Culturing of Engineered Human Liver
2.4. Histology and Immunostaining Analysis
2.5. Viability Assessment
2.6. Gene Expression Analysis
2.7. Protein Extraction and Quantification
2.8. Western Blot
2.9. Protein Secretion Analysis
2.10. Flow Cytometry Analysis
2.11. Statistical Analysis
3. Results
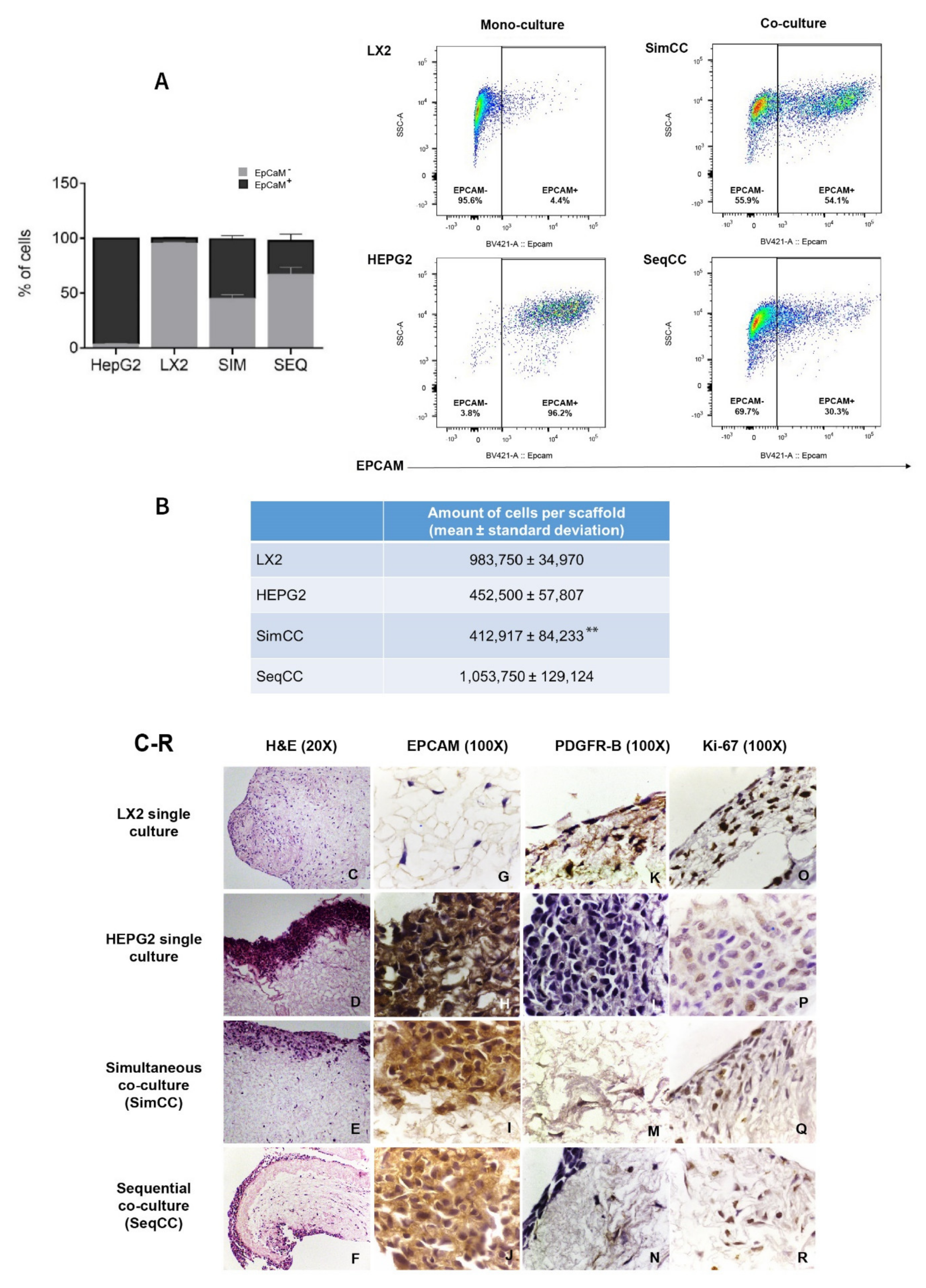
3.1. Optimization of LX2-HepG2 Cell Co-Culture Model in Human Liver 3D Scaffolds
3.2. Characterization of Cell Phenotype in Co-Cultures Versus Single Cultures in 3D ECM Scaffolds
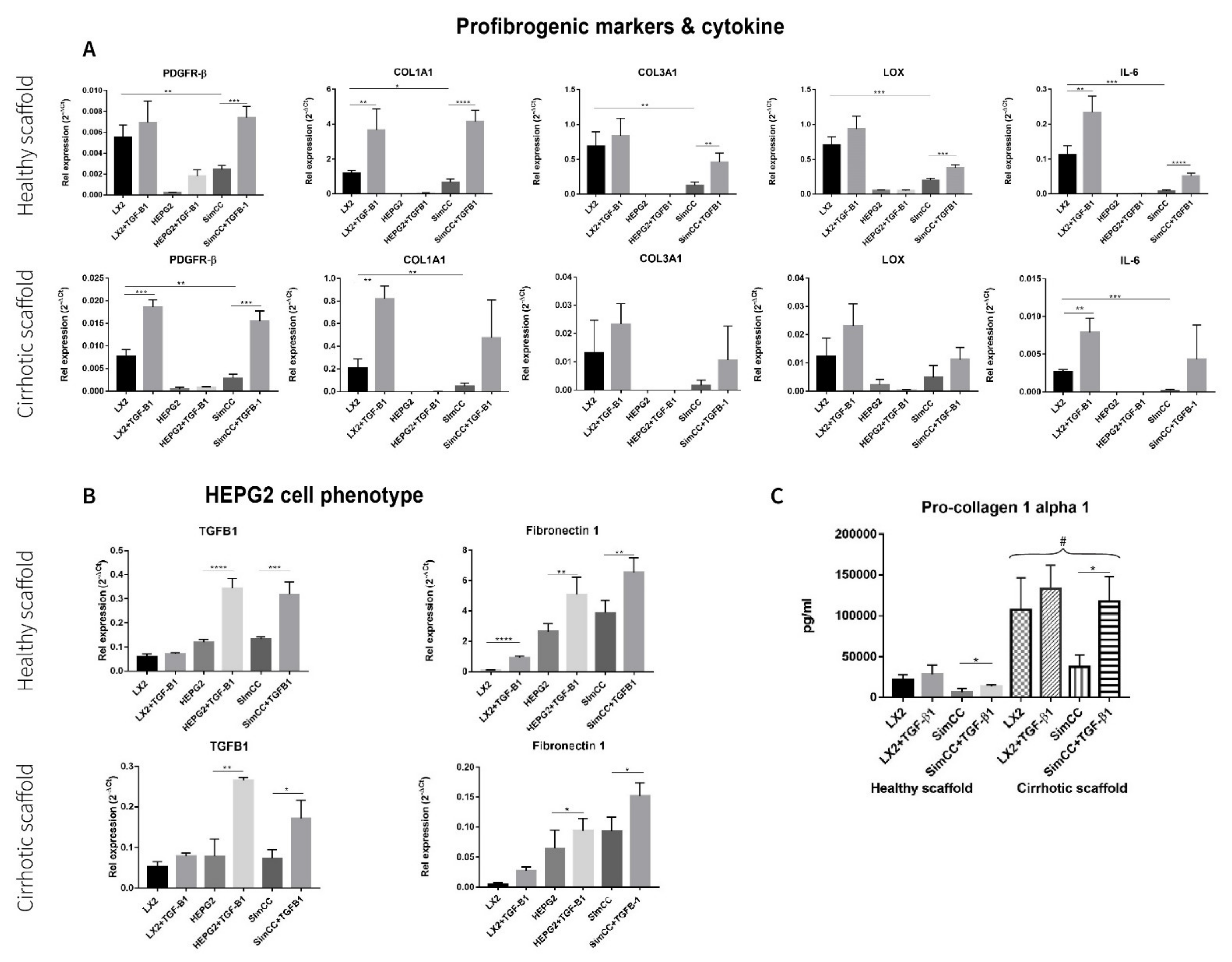
3.3. Response to TGF-β1 Treatment in Single and Simultaneous Co-Cultures in Healthy and Cirrhotic 3D Scaffolds
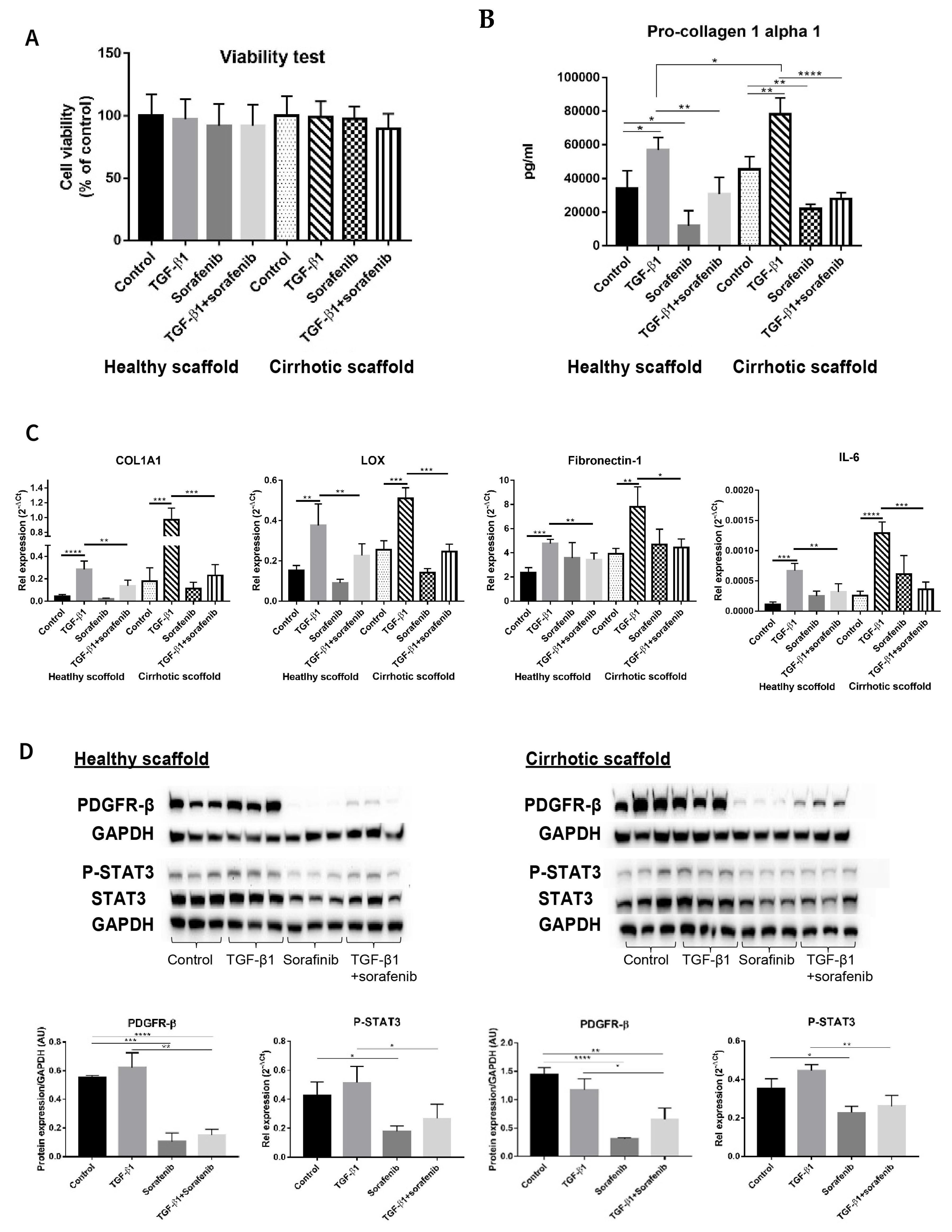
3.4. Evaluating Anti-Fibrotic Drug Response of Sorafenib in LX2-HepG2 Co-Culture Model in 3D Liver Scaffolds
3.5. Chemoresistance in LX2-HepG2 Co-Culture System in 3D Healthy and Cirrhotic Liver Scaffolds
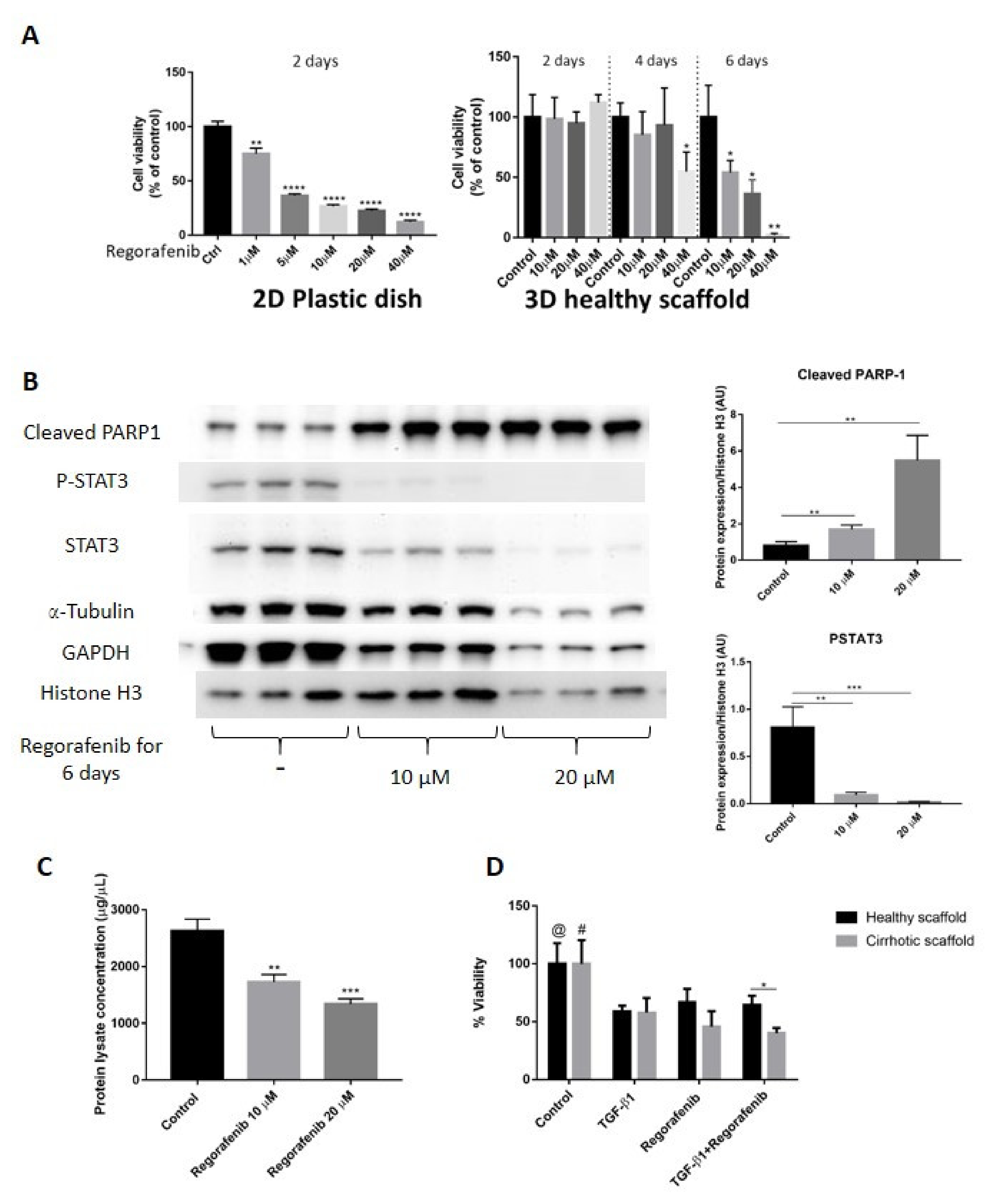
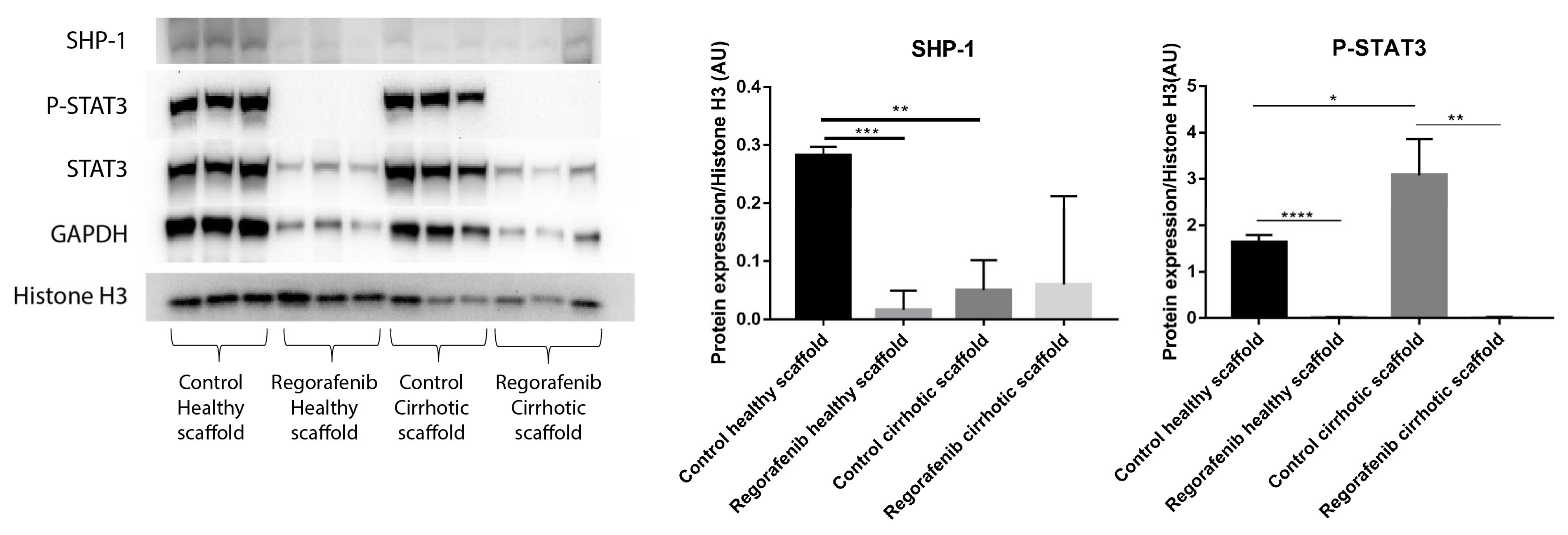
3.6. Regorafenib Inhibits TGF-β1-Induced EMT and STAT3 Phosphorylation in 3D Human Liver Scaffold Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amann, T.; Bataille, F.; Spruss, T.; Muhlbauer, M.; Gabele, E.; Scholmerich, J.; Kiefer, P.; Bosserhoff, A.K.; Hellerbrand, C. Activated hepatic stellate cells promote tumorigenicity of hepatocellular carcinoma. Cancer Sci. 2009, 100, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Khawar, I.A.; Park, J.K.; Jung, E.S.; Lee, M.A.; Chang, S.; Kuh, H.J. Three Dimensional Mixed-Cell Spheroids Mimic Stroma-Mediated Chemoresistance and Invasive Migration in hepatocellular carcinoma. Neoplasia 2018, 20, 800–812. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Gea, V.; Toffanin, S.; Friedman, S.L.; Llovet, J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology 2013, 144, 512–527. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, S.; Hamberger, F.; Ravichandra, A.; Miller, M.; Nair, A.; Affo, S.; Filliol, A.; Chin, L.; Savage, T.M.; Yin, D.; et al. Tumor restriction by type I collagen opposes tumor-promoting effects of cancer-associated fibroblasts. J. Clin. Investig. 2021, 131, e146987. [Google Scholar] [CrossRef]
- Bottcher, K.; Pinzani, M. Pathophysiology of liver fibrosis and the methodological barriers to the development of anti-fibrogenic agents. Adv. Drug Deliv. Rev. 2017, 121, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef] [Green Version]
- Nam, M.H.; Lee, H.S.; Seomun, Y.; Lee, Y.; Lee, K.W. Monocyte-endothelium-smooth muscle cell interaction in co-culture: Proliferation and cytokine productions in response to advanced glycation end products. Biochim. Biophys. Acta 2011, 1810, 907–912. [Google Scholar] [CrossRef]
- Mazza, G.; Al-Akkad, W.; Rombouts, K. Engineering in vitro models of hepatofibrogenesis. Adv. Drug Deliv. Rev. 2017, 121, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Leite, S.B.; Roosens, T.; El Taghdouini, A.; Mannaerts, I.; Smout, A.J.; Najimi, M.; Sokal, E.; Noor, F.; Chesne, C.; van Grunsven, L.A. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials 2016, 78, 1–10. [Google Scholar] [CrossRef]
- Prestigiacomo, V.; Weston, A.; Messner, S.; Lampart, F.; Suter-Dick, L. Pro-fibrotic compounds induce stellate cell activation, ECM-remodelling and Nrf2 activation in a human 3D-multicellular model of liver fibrosis. PLoS ONE 2017, 12, e0179995. [Google Scholar] [CrossRef]
- Mazza, G.; Telese, A.; Al-Akkad, W.; Frenguelli, L.; Levi, A.; Marrali, M.; Longato, L.; Thanapirom, K.; Vilia, M.G.; Lombardi, B.; et al. Cirrhotic Human Liver Extracellular Matrix 3D Scaffolds Promote Smad-Dependent TGF-beta1 Epithelial Mesenchymal Transition. Cells 2019, 9, 83. [Google Scholar] [CrossRef] [Green Version]
- Sawitza, I.; Kordes, C.; Reister, S.; Haussinger, D. The niche of stellate cells within rat liver. Hepatology 2009, 50, 1617–1624. [Google Scholar] [CrossRef]
- Yip, D.; Cho, C.H. A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing. Biochem. Biophys. Res. Commun. 2013, 433, 327–332. [Google Scholar] [CrossRef]
- Van Grunsven, L.A. 3D in vitro models of liver fibrosis. Adv. Drug Deliv. Rev. 2017, 121, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. Adv. Life Sci. R D 2017, 22, 456–472. [Google Scholar] [CrossRef] [Green Version]
- Mazza, G.; Al-Akkad, W.; Telese, A.; Longato, L.; Urbani, L.; Robinson, B.; Hall, A.; Kong, K.; Frenguelli, L.; Marrone, G.; et al. Rapid production of human liver scaffolds for functional tissue engineering by high shear stress oscillation-decellularization. Sci. Rep. 2017, 7, 5534. [Google Scholar] [CrossRef]
- Gissen, P.; Arias, I.M. Structural and functional hepatocyte polarity and liver disease. J. Hepatol. 2015, 63, 1023–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavri-Damelin, D.; Damelin, L.H.; Eaton, S.; Rees, M.; Selden, C.; Hodgson, H.J. Cells for bioartificial liver devices: The human hepatoma-derived cell line C3A produces urea but does not detoxify ammonia. Biotechnol. Bioeng. 2008, 99, 644–651. [Google Scholar] [CrossRef]
- Mazza, G.; Rombouts, K.; Rennie Hall, A.; Urbani, L.; Vinh Luong, T.; Al-Akkad, W.; Longato, L.; Brown, D.; Maghsoudlou, P.; Dhillon, A.P.; et al. Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci. Rep. 2015, 5, 13079. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Terrada, D.; Cheung, S.W.; Finegold, M.J.; Knowles, B.B. Hep G2 is a hepatoblastoma-derived cell line. Hum. Pathol. 2009, 40, 1512–1515. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hui, A.Y.; Albanis, E.; Arthur, M.J.; O’Byrne, S.M.; Blaner, W.S.; Mukherjee, P.; Friedman, S.L.; Eng, F.J. Human hepatic stellate cell lines, LX-1 and LX-2: New tools for analysis of hepatic fibrosis. Gut 2005, 54, 142–151. [Google Scholar] [CrossRef] [Green Version]
- Taimr, P.; Higuchi, H.; Kocova, E.; Rippe, R.A.; Friedman, S.; Gores, G.J. Activated stellate cells express the TRAIL receptor-2/death receptor-5 and undergo TRAIL-mediated apoptosis. Hepatology 2003, 37, 87–95. [Google Scholar] [CrossRef]
- Weiskirchen, R.; Weimer, J.; Meurer, S.K.; Kron, A.; Seipel, B.; Vater, I.; Arnold, N.; Siebert, R.; Xu, L.; Friedman, S.L.; et al. Genetic characteristics of the human hepatic stellate cell line LX-2. PLoS ONE 2013, 8, e75692. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Kim, S.H.; Kim, K.M.; Choi, E.K.; Kim, J.; Seo, H.R. Activated hepatic stellate cells play pivotal roles in hepatocellular carcinoma cell chemoresistance and migration in multicellular tumor spheroids. Sci. Rep. 2016, 6, 36750. [Google Scholar] [CrossRef] [Green Version]
- Bruix, J.; Qin, S.; Merle, P.; Granito, A.; Huang, Y.H.; Bodoky, G.; Pracht, M.; Yokosuka, O.; Rosmorduc, O.; Breder, V.; et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 389, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Marrone, G.; De Chiara, F.; Bottcher, K.; Levi, A.; Dhar, D.; Longato, L.; Mazza, G.; Zhang, Z.; Marrali, M.; Fernandez-Iglesias, A.; et al. The adenosine monophosphate-activated protein kinase-vacuolar adenosine triphosphatase-pH axis: A key regulator of the profibrogenic phenotype of human hepatic stellate cells. Hepatology 2018, 68, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, R.; Chen, J.; Liang, Y.; Lin, S.; Zhu, L.; Liang, X.; Cai, X. Sorafenib: A potential therapeutic drug for hepatic fibrosis and its outcomes. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 88, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.M.; Jha, R.K.; Li, B.; Chen, C.; Li, W.Z.; Zheng, J.B.; Wang, L.; Huanchen, S. Sorafenib inhibition of hepatic stellate cell proliferation in tumor microenvironment of hepatocellular carcinoma: A study of the sorafenib mechanisms. Cell Biochem. Biophys. 2014, 69, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, J.; Zhang, D.; Zhang, J.; Ma, J.; Jiang, H. New insights into the antifibrotic effects of sorafenib on hepatic stellate cells and liver fibrosis. J. Hepatol. 2010, 53, 132–144. [Google Scholar] [CrossRef]
- Thabut, D.; Routray, C.; Lomberk, G.; Shergill, U.; Glaser, K.; Huebert, R.; Patel, L.; Masyuk, T.; Blechacz, B.; Vercnocke, A.; et al. Complementary vascular and matrix regulatory pathways underlie the beneficial mechanism of action of sorafenib in liver fibrosis. Hepatology 2011, 54, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Su, T.H.; Shiau, C.W.; Jao, P.; Liu, C.H.; Liu, C.J.; Tai, W.T.; Jeng, Y.M.; Yang, H.C.; Tseng, T.C.; Huang, H.P.; et al. Sorafenib and its derivative SC-1 exhibit antifibrotic effects through signal transducer and activator of transcription 3 inhibition. Proc. Natl. Acad. Sci. USA 2015, 112, 7243–7248. [Google Scholar] [CrossRef] [Green Version]
- Mehmood, T.; Maryam, A.; Tian, X.; Khan, M.; Ma, T. Santamarine Inhibits NF-small ka, CyrillicB and STAT3 Activation and Induces Apoptosis in HepG2 Liver Cancer Cells via Oxidative Stress. J. Cancer 2017, 8, 3707–3717. [Google Scholar] [CrossRef]
- Weng, Z.; Luo, Y.; Yang, X.; Greenhaw, J.J.; Li, H.; Xie, L.; Mattes, W.B.; Shi, Q. Regorafenib impairs mitochondrial functions, activates AMP-activated protein kinase, induces autophagy, and causes rat hepatocyte necrosis. Toxicology 2015, 327, 10–21. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Liu, T.; Wu, J.C.; Luo, S.Z.; Chen, R.; Lu, L.G.; Xu, M.Y. STAT3 aggravates TGF-beta1-induced hepatic epithelial-to-mesenchymal transition and migration. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 98, 214–221. [Google Scholar] [CrossRef]
- Fan, L.C.; Shiau, C.W.; Tai, W.T.; Hung, M.H.; Chu, P.Y.; Hsieh, F.S.; Lin, H.; Yu, H.C.; Chen, K.F. SHP-1 is a negative regulator of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncogene 2017, 36, 5768–5769. [Google Scholar] [CrossRef] [Green Version]
- Caruso, S.; Calatayud, A.L.; Pilet, J.; La Bella, T.; Rekik, S.; Imbeaud, S.; Letouze, E.; Meunier, L.; Bayard, Q.; Rohr-Udilova, N.; et al. Analysis of Liver Cancer Cell Lines Identifies Agents With Likely Efficacy Against Hepatocellular Carcinoma and Markers of Response. Gastroenterology 2019, 157, 760–776. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, K.; Longato, L.; Marrone, G.; Mazza, G.; Ghemtio, L.; Hall, A.; Luong, T.V.; Caruso, S.; Viollet, B.; Zucman-Rossi, J.; et al. AICAR and compound C negatively modulate HCC-induced primary human hepatic stellate cell activation in vitro. American journal of physiology. Gastrointest. Liver Physiol. 2021, 320, G543–G556. [Google Scholar] [CrossRef]
- Barcena-Varela, M.; Caruso, S.; Llerena, S.; Alvarez-Sola, G.; Uriarte, I.; Latasa, M.U.; Urtasun, R.; Rebouissou, S.; Alvarez, L.; Jimenez, M.; et al. Dual Targeting of Histone Methyltransferase G9a and DNA-Methyltransferase 1 for the Treatment of Experimental Hepatocellular Carcinoma. Hepatology 2019, 69, 587–603. [Google Scholar] [CrossRef]
- Dou, C.; Liu, Z.; Tu, K.; Zhang, H.; Chen, C.; Yaqoob, U.; Wang, Y.; Wen, J.; van Deursen, J.; Sicard, D.; et al. P300 Acetyltransferase Mediates Stiffness-Induced Activation of Hepatic Stellate Cells into Tumor-Promoting Myofibroblasts. Gastroenterology 2018, 154, 2209–2221.e14. [Google Scholar] [CrossRef] [Green Version]
- Giannelli, G.; Villa, E.; Lahn, M. Transforming growth factor-beta as a therapeutic target in hepatocellular carcinoma. Cancer Res. 2014, 74, 1890–1894. [Google Scholar] [CrossRef] [Green Version]
- Reichl, P.; Haider, C.; Grubinger, M.; Mikulits, W. TGF-beta in epithelial to mesenchymal transition and metastasis of liver carcinoma. Curr. Pharm. Des. 2012, 18, 4135–4147. [Google Scholar] [CrossRef] [PubMed]
- Casini, A.; Pinzani, M.; Milani, S.; Grappone, C.; Galli, G.; Jezequel, A.M.; Schuppan, D.; Rotella, C.M.; Surrenti, C. Regulation of extracellular matrix synthesis by transforming growth factor beta 1 in human fat-storing cells. Gastroenterology 1993, 105, 245–253. [Google Scholar] [CrossRef]
- Bachem, M.G.; Meyer, D.; Melchior, R.; Sell, K.M.; Gressner, A.M. Activation of rat liver perisinusoidal lipocytes by transforming growth factors derived from myofibroblastlike cells. A potential mechanism of self perpetuation in liver fibrogenesis. J. Clin. Investig. 1992, 89, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Park, J.; Kim, Y.; Sohn, A.; Yeo, I.; Jong Yu, S.; Yoon, J.H.; Park, T.; Kim, Y. Serum fibronectin distinguishes the early stages of hepatocellular carcinoma. Sci. Rep. 2017, 7, 9449. [Google Scholar] [CrossRef] [Green Version]
- Hong, F.; Chou, H.; Fiel, M.I.; Friedman, S.L. Antifibrotic activity of sorafenib in experimental hepatic fibrosis: Refinement of inhibitory targets, dosing, and window of efficacy in vivo. Dig. Dis. Sci. 2013, 58, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Mejias, M.; Garcia-Pras, E.; Tiani, C.; Miquel, R.; Bosch, J.; Fernandez, M. Beneficial effects of sorafenib on splanchnic, intrahepatic, and portocollateral circulations in portal hypertensive and cirrhotic rats. Hepatology 2009, 49, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Reiberger, T.; Angermayr, B.; Schwabl, P.; Rohr-Udilova, N.; Mitterhauser, M.; Gangl, A.; Peck-Radosavljevic, M. Sorafenib attenuates the portal hypertensive syndrome in partial portal vein ligated rats. J. Hepatol. 2009, 51, 865–873. [Google Scholar] [CrossRef]
- Svinka, J.; Mikulits, W.; Eferl, R. STAT3 in hepatocellular carcinoma: New perspectives. Hepatic Oncol. 2014, 1, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Nunez Lopez, O.; Bohanon, F.J.; Wang, X.; Ye, N.; Corsello, T.; Rojas-Khalil, Y.; Chen, H.; Chen, H.; Zhou, J.; Radhakrishnan, R.S. STAT3 Inhibition Suppresses Hepatic Stellate Cell Fibrogenesis: HJC0123, a Potential Therapeutic Agent for Liver Fibrosis. RSC Adv. 2016, 6, 100652–100663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Hori, Y.; Yamamoto, T.; Urashima, T.; Ohara, Y.; Tanaka, H. 3D spheroid cultures improve the metabolic gene expression profiles of HepaRG cells. Biosci. Rep. 2015, 35, e00208. [Google Scholar] [CrossRef]
- Takai, A.; Fako, V.; Dang, H.; Forgues, M.; Yu, Z.; Budhu, A.; Wang, X.W. Three-dimensional Organotypic Culture Models of Human Hepatocellular Carcinoma. Sci. Rep. 2016, 6, 21174. [Google Scholar] [CrossRef] [Green Version]
- Miyauchi, Y.; Yasuchika, K.; Fukumitsu, K.; Ishii, T.; Ogiso, S.; Minami, T.; Kojima, H.; Yamaoka, R.; Katayama, H.; Kawai, T.; et al. A novel three-dimensional culture system maintaining the physiological extracellular matrix of fibrotic model livers accelerates progression of hepatocellular carcinoma cells. Sci. Rep. 2017, 7, 9827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Liu, Y.; Xie, H.G.; Zhao, S.; Xu, X.X.; Fan, L.X.; Guo, X.; Lu, T.; Sun, G.W.; Ma, X.J. Role of three-dimensional matrix stiffness in regulating the chemoresistance of hepatocellular carcinoma cells. Biotechnol. Appl. Biochem. 2015, 62, 556–562. [Google Scholar] [CrossRef]
- Wilhelm, S.M.; Dumas, J.; Adnane, L.; Lynch, M.; Carter, C.A.; Schutz, G.; Thierauch, K.H.; Zopf, D. Regorafenib (BAY 73-4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer 2011, 129, 245–255. [Google Scholar] [CrossRef]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef] [Green Version]
- Wakefield, L.M.; Hill, C.S. Beyond TGFbeta: Roles of other TGFbeta superfamily members in cancer. Nature reviews. Cancer 2013, 13, 328–341. [Google Scholar] [CrossRef]
- Fan, L.C.; Teng, H.W.; Shiau, C.W.; Tai, W.T.; Hung, M.H.; Yang, S.H.; Jiang, J.K.; Chen, K.F. Regorafenib (Stivarga) pharmacologically targets epithelial-mesenchymal transition in colorectal cancer. Oncotarget 2016, 7, 64136–64147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tai, W.T.; Chu, P.Y.; Shiau, C.W.; Chen, Y.L.; Li, Y.S.; Hung, M.H.; Chen, L.J.; Chen, P.L.; Su, J.C.; Lin, P.Y.; et al. STAT3 mediates regorafenib-induced apoptosis in hepatocellular carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 5768–5776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, L.Z.; Ding, K.; Wang, Z.R.; Ding, C.H.; Lei, S.J.; Liu, J.P.; Yin, C.; Hu, P.F.; Ding, J.; Chen, W.S.; et al. SHP-1 Acts as a Tumor Suppressor in Hepatocarcinogenesis and HCC Progression. Cancer Res. 2018, 78, 4680–4691. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thanapirom, K.; Caon, E.; Papatheodoridi, M.; Frenguelli, L.; Al-Akkad, W.; Zhenzhen, Z.; Vilia, M.G.; Pinzani, M.; Mazza, G.; Rombouts, K. Optimization and Validation of a Novel Three-Dimensional Co-Culture System in Decellularized Human Liver Scaffold for the Study of Liver Fibrosis and Cancer. Cancers 2021, 13, 4936. https://doi.org/10.3390/cancers13194936
Thanapirom K, Caon E, Papatheodoridi M, Frenguelli L, Al-Akkad W, Zhenzhen Z, Vilia MG, Pinzani M, Mazza G, Rombouts K. Optimization and Validation of a Novel Three-Dimensional Co-Culture System in Decellularized Human Liver Scaffold for the Study of Liver Fibrosis and Cancer. Cancers. 2021; 13(19):4936. https://doi.org/10.3390/cancers13194936
Chicago/Turabian StyleThanapirom, Kessarin, Elisabetta Caon, Margarita Papatheodoridi, Luca Frenguelli, Walid Al-Akkad, Zhang Zhenzhen, Maria Giovanna Vilia, Massimo Pinzani, Giuseppe Mazza, and Krista Rombouts. 2021. "Optimization and Validation of a Novel Three-Dimensional Co-Culture System in Decellularized Human Liver Scaffold for the Study of Liver Fibrosis and Cancer" Cancers 13, no. 19: 4936. https://doi.org/10.3390/cancers13194936
APA StyleThanapirom, K., Caon, E., Papatheodoridi, M., Frenguelli, L., Al-Akkad, W., Zhenzhen, Z., Vilia, M. G., Pinzani, M., Mazza, G., & Rombouts, K. (2021). Optimization and Validation of a Novel Three-Dimensional Co-Culture System in Decellularized Human Liver Scaffold for the Study of Liver Fibrosis and Cancer. Cancers, 13(19), 4936. https://doi.org/10.3390/cancers13194936






