MiR-125b-5p Is Involved in Sorafenib Resistance through Ataxin-1-Mediated Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Compounds
2.2. Cell Viability Analysis
2.3. MicroRNA Microarray Analysis
2.4. RT-PCR Analysis
2.5. Overexpression and Knockdown Experiments
2.6. In Silico Identification of miRNA Target Genes
2.7. miRNA Luciferase Reporter Assay
2.8. Promoter Assay
2.9. Western Blot Analysis
2.10. Wound Healing Assay
2.11. Cell Invasion Assay
2.12. Flow Cytometry Analysis
2.13. Tumor Xenograft Experiments
2.14. ATXN1 Expression from RNA-Chip Data
2.15. Statistical Analysis
3. Results
3.1. Differential miRNA Expression Profile between Sorafenib-Resistant PLC/PRF5-R1/R2 and PLC/PRF5 Cells
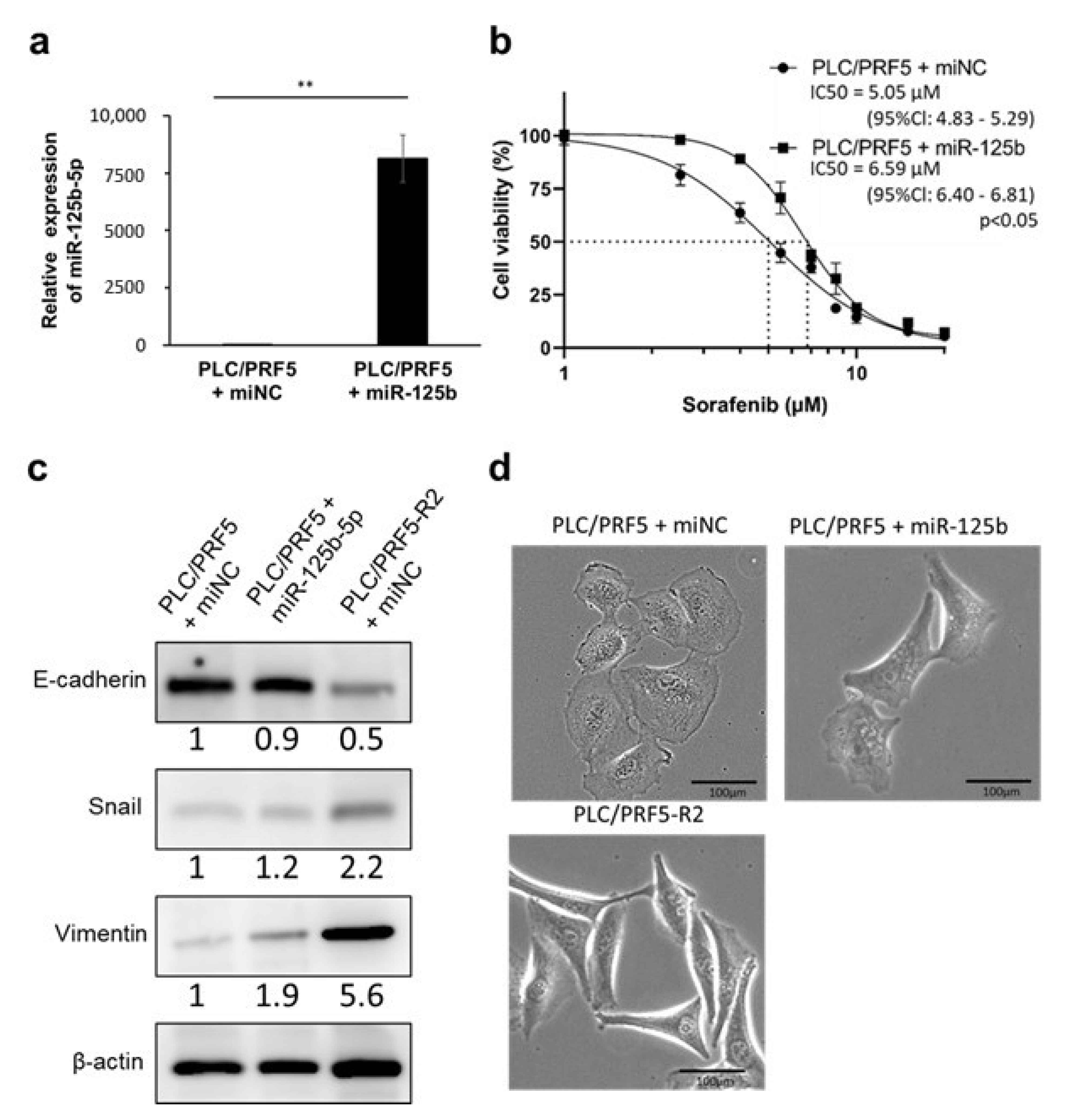
3.2. miR-125b-5p Confers Resistance to Sorafenib in PLC/PRF5 Cells
3.3. miR-125b-5p Induces EMT in PLC/PRF5 Cells
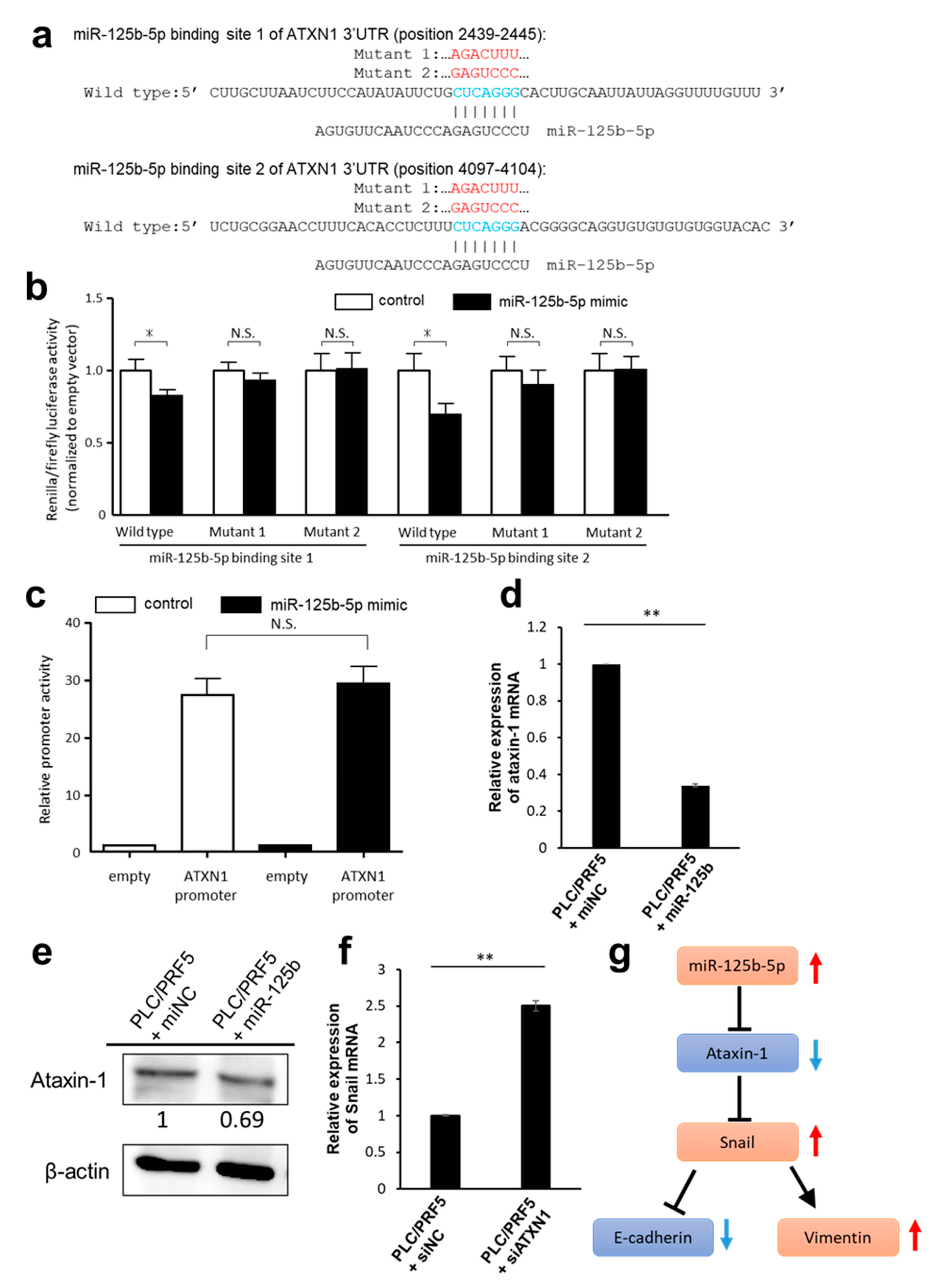
3.4. Predicted Target Genes of miR-125b-5p
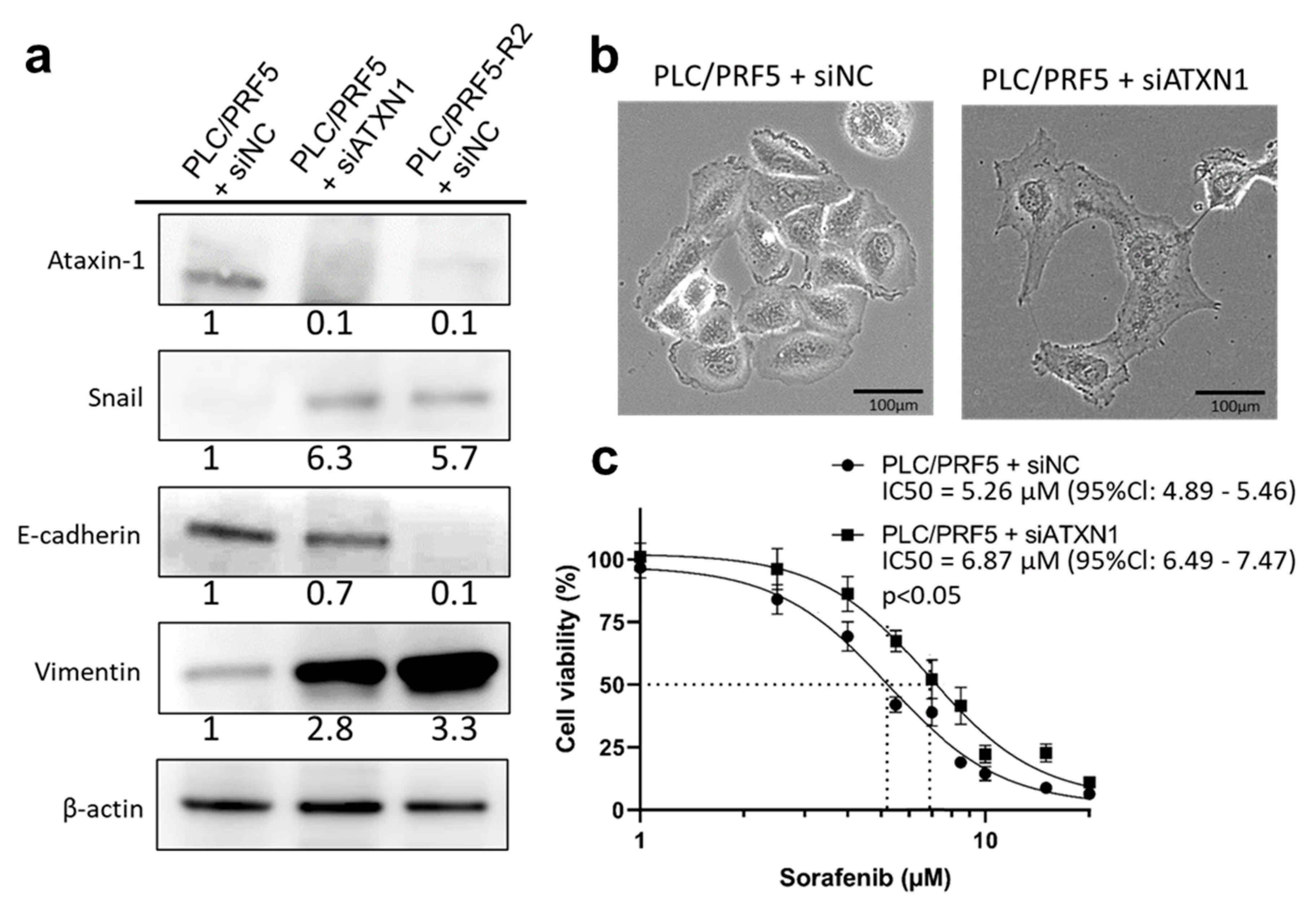
3.5. Downregulation of ATXN1 Induces EMT and Sorafenib Resistance
3.6. miR-125b-5p and ATXN1 Modulate Cell Migration and Invasion
3.7. miR-125b-5p and Downregulation of ATXN1 Confer Stemness Characteristic in HCC Cells
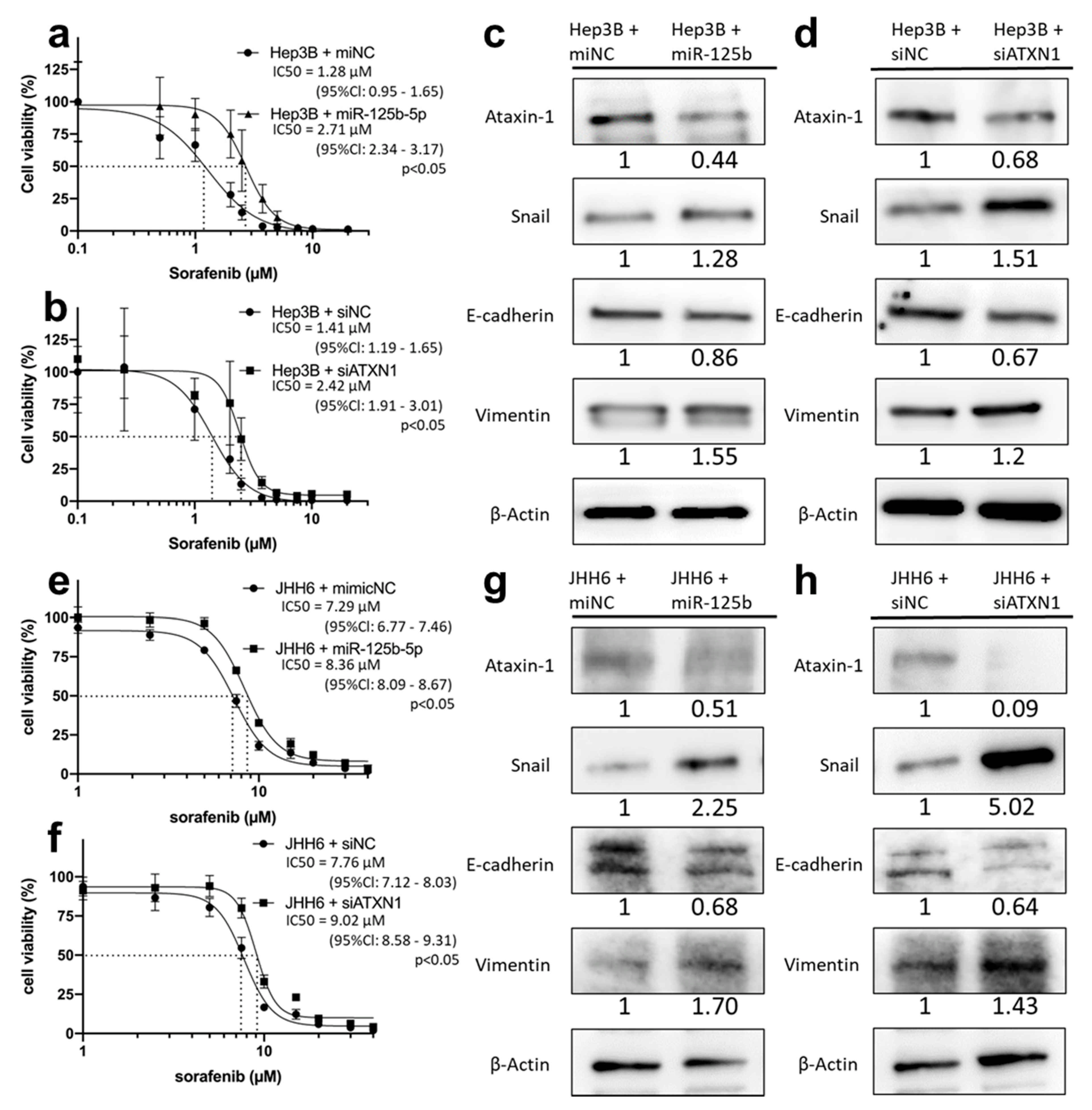
3.8. miR-125b-5p and siATXN1 Confer Drug Resistance by Inducing EMT in Various HCC Cell Lines
3.9. Overexpression of ATXN1 Confer Drug Resistance and EMT in HCC Cells
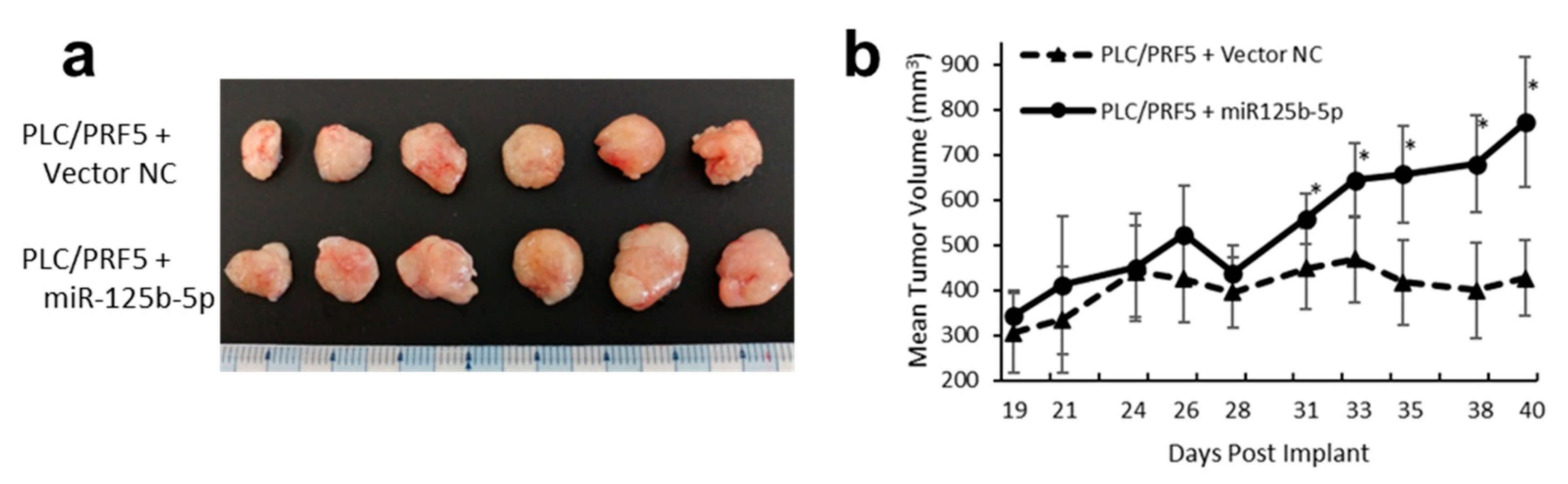
3.10. Overexpression of miR-125b-5p Confer Sorafenib Resistance In Vivo
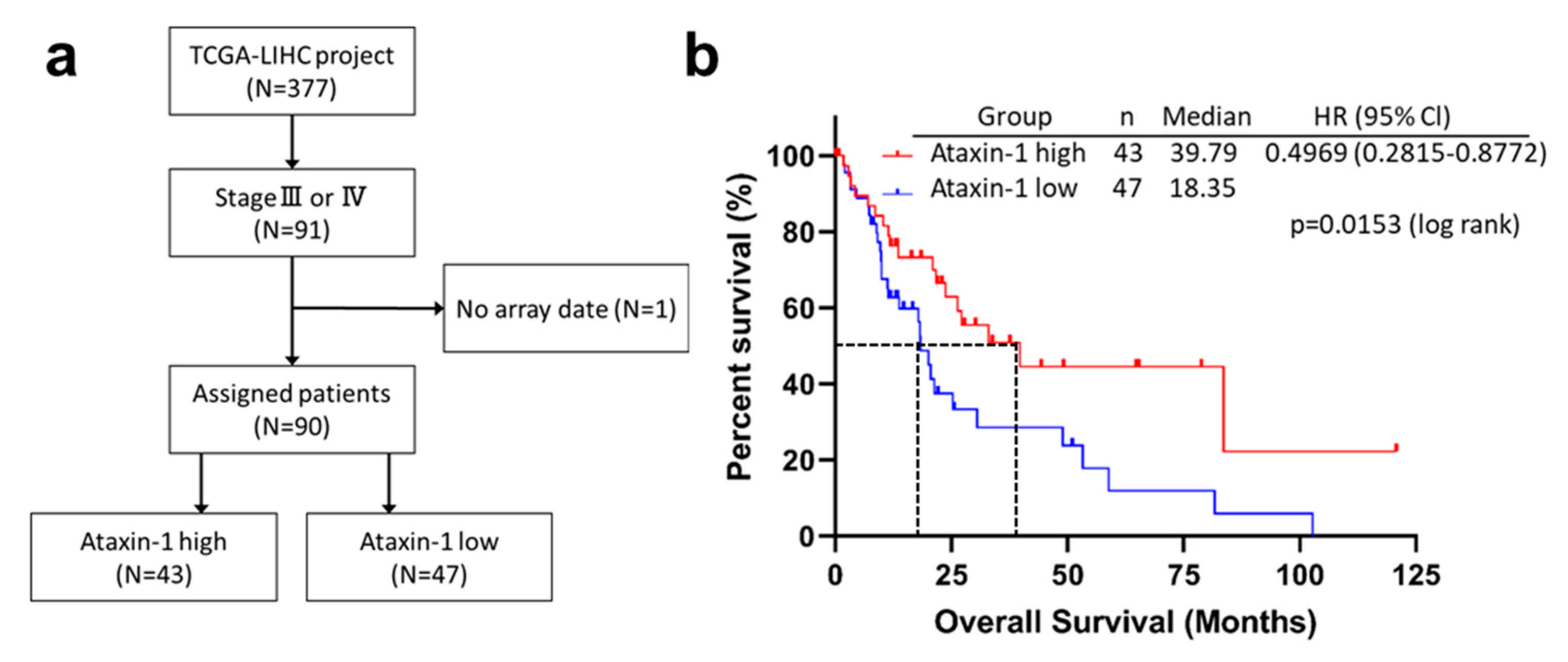
3.11. ATXN1 Expression was Significantly Associated with Survival in Patients with Advanced HCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Kang, Y.-K.; Chen, Z.; Tsao, C.-J.; Qin, S.; Kim, J.S.; Luo, R.; Feng, J.; Ye, S.; Yang, T.-S.; et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009, 10, 25–34. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocel-lular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- NCCN Guidelines Version 3. 2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/hepatobiliary.pdf (accessed on 23 August 2021).
- Tomonari, T.; Takeishi, S.; Taniguchi, T.; Tanaka, T.; Tanaka, H.; Fujimoto, S.; Kimura, T.; Okamoto, K.; Miyamoto, H.; Muguruma, N.; et al. MRP3 as a novel resistance factor for sorafenib in hepatocellular carcinoma. Oncotarget 2016, 7, 7207–7215. [Google Scholar] [CrossRef]
- Morgensztern, D.; McLeod, H.L. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anti-Cancer Drugs 2005, 16, 797–803. [Google Scholar] [CrossRef]
- Chen, K.-F.; Tai, W.-T.; Hsu, C.-Y.; Huang, J.-W.; Liu, C.-Y.; Chen, P.-J.; Kim, I.; Shiau, C.-W. Blockade of STAT3 activation by sorafenib derivatives through enhancing SHP-1 phosphatase activity. Eur. J. Med. Chem. 2012, 55, 220–227. [Google Scholar] [CrossRef]
- Méndez-Blanco, C.; Fondevila, F.; Palomo, A.G.; González-Gallego, J.; Mauriz, J.L. Sorafenib resistance in hepatocarcinoma: Role of hypoxia-inducible factors. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Zhu, Y.-J.; Zheng, B.; Wang, H.-Y.; Chen, L. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol. Sin. 2017, 38, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Lee, M.T.; Giraldez, A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in Zebrafish. Science 2012, 336, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Banzhaf-Strathmann, J.; Edbauer, D. Good guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell Commun. Signal. 2014, 12, 30. [Google Scholar] [CrossRef]
- Zhou, J.-N.; Zeng, Q.; Wang, H.-Y.; Zhang, B.; Li, S.-T.; Nan, X.; Cao, N.; Fu, C.-J.; Yan, X.-L.; Jia, Y.L.; et al. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology 2015, 62, 801–815. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in colorectal cancer: A culprit in metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef]
- Mestdagh, P.; Hartmann, N.; Baeriswyl, L.; Andreasen, D.; Bernard, N.; Chen, C.; Cheo, D.; D’Andrade, P.; DeMayo, M.; Dennis, L.; et al. Evaluation of quantitative mirnA expression platforms in the micrornA quality control (mirQC) study. Nat Methods 2014, 11, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Xia, S.; Liang, Y.; Ji, L.; Pan, Y.; Jiang, S.; Wan, Z.; Tao, L.; Chen, J.; Lin, C.; et al. LXR activation potentiates sorafenib sensitivity in HCC by activating microRNA-378a tran-scription. Theranostics 2020, 10, 8834–8850. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Van Malenstein, H.; Dekervel, J.; Verslype, C.; Van Cutsem, E.; Windmolders, P.; Nevens, F.; van Pelt, J. Long-term exposure to sorafenib of liver cancer cells induces resistance with epithelial-to-mesenchymal transition, increased invasion and risk of rebound growth. Cancer Lett. 2013, 329, 74–83. [Google Scholar] [CrossRef]
- Maheswaran, T.; Rushbrook, S.M. Epithelial-mesenchymal transition and the liver: Role in hepatocellular carcinoma and liver fibrosis. J. Gastroenterol. Hepatol. 2012, 27, 418–420. [Google Scholar] [CrossRef]
- Dazert, E.; Colombi, M.; Boldanova, T.; Moes, S.; Adametz, D.; Quagliata, L.; Roth, V.; Terracciano, L.; Heim, M.; Jenoe, P.; et al. Quantitative proteomics and phosphoproteomics on serial tumor biopsies from a sorafenib-treated HCC patient. Proc. Natl. Acad. Sci. USA 2016, 113, 1381–1386. [Google Scholar] [CrossRef]
- Lam, Y.C.; Bowman, A.; Jafar-Nejad, P.; Lim, J.; Richman, R.; Fryer, J.D.; Hyun, E.D.; Duvick, L.A.; Orr, H.; Botas, J.; et al. ATAXIN-1 interacts with the repressor capicua in its native complex to cause SCA1 neuropathology. Cell 2006, 127, 1335–1347. [Google Scholar] [CrossRef]
- McGranahan, N.; Favero, F.; De Bruin, E.C.; Birkbak, N.; Szallasi, Z.; Swanton, C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 2015, 7, 283ra54. [Google Scholar] [CrossRef] [PubMed]
- Asghari, M.; Abazari, M.F.; Bokharaei, H.; Aleagha, M.N.; Poortahmasebi, V.; Askari, H.; Torabinejad, S.; Ardalan, A.; Negaresh, N.; Ataei, A.; et al. Key genes and regulatory networks involved in the initiation, progression and invasion of colorectal cancer. Futur. Sci. OA 2018, 4, FSO278. [Google Scholar] [CrossRef] [PubMed]
- Takehara, M.; Sato, Y.; Kimura, T.; Noda, K.; Miyamoto, H.; Fujino, Y.; Miyoshi, J.; Nakamura, F.; Wada, H.; Bando, Y.; et al. Cancer-associated adipocytes promote pancreatic cancer progression through SAA1 expression. Cancer Sci. 2020, 111, 2883–2894. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, J.; Tanahashi, T.; Sato, Y.; Miyoshi, J.; Nakagawa, T.; Kimura, T.; Miyamoto, H.; Fujino, Y.; Nakamura, F.; Takehara, M.; et al. MicroRNA-296-5p Promotes Cell Invasion and Drug Resistance by Targeting Bcl2-Related Ovarian Killer, Leading to a Poor Prognosis in Pancreatic Cancer. Digestion. 2020, 101, 794–806. [Google Scholar] [CrossRef] [PubMed]
- miRDB. Available online: http://www.mirdb.org/ (accessed on 24 March 2020).
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef] [PubMed]
- TargetScanHuman Release 7.2. Available online: http://www.targetscan.orgvert_72/ (accessed on 24 March 2020).
- Tanaka, H.; Okamoto, K.; Sato, Y.; Tanaka, T.; Tomonari, T.; Nakamura, F.; Fujino, Y.; Mitsui, Y.; Miyamoto, H.; Muguruma, N.; et al. Synergistic anti-tumor activity of miriplatin and radiation through PUMA-mediated apoptosis in hepatocellular carcinoma. J. Gastroenterol. 2020, 55, 1072–1086. [Google Scholar] [CrossRef]
- Maehara, O.; Sato, F.; Natsuizaka, M.; Asano, A.; Kubota, Y.; Itoh, J.; Tsunematsu, S.; Terashita, K.; Tsukuda, Y.; Nakai, M.; et al. A pivotal role of Krüppel-like factor 5 in regulation of cancer stem-like cells in hepatocellular carcinoma. Cancer Biol. Ther. 2015, 16, 1453–1461. [Google Scholar] [CrossRef]
- Liu, L.; Cao, Y.; Chen, C.; Zhang, X.; McNabola, A.; Wilkie, D.; Wilhelm, S.; Lynch, M.; Carter, C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma Model PLC/PRF/5. Cancer Res. 2006, 66, 11851–11858. [Google Scholar] [CrossRef] [PubMed]
- GDC DatePortal. Available online: http://portal.gdc.cancer.gov/ (accessed on 24 March 2020).
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Grinchuk, O.V.; Yenamandra, S.P.; Iyer, R.; Singh, M.; Lee, H.K.; Lim, K.H.; Chow, P.K.; Kuznetsov, V.A. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol. Oncol. 2018, 12, 89–113. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Villanueva, A.; Kobayashi, M.; Peix, J.; Chiang, D.Y.; Camargo, A.; Gupta, S.; Moore, J.; Wrobel, M.J.; Lerner, J.; et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 1995–2004. [Google Scholar] [CrossRef]
- Kang, A.-R.; An, H.-T.; Ko, J.; Kang, S. Ataxin-1 regulates epithelial-mesenchymal transition of cervical cancer cells. Oncotarget 2017, 8, 18248–18259. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Kabakov, A.; Yakimova, A.; Matchuk, O. Molecular chaperones in cancer stem cells: Determinants of stemness and potential targets for antitumor therapy. Cells 2020, 9, 892. [Google Scholar] [CrossRef]
- Zhao, H.; Desai, V.; Wang, J.; Epstein, D.M.; Miglarese, M.; Buck, E. Epithelial–mesenchymal transition predicts sensitivity to the dual IGF-1R/IR inhibitor OSI-906 in hepatocellular carcinoma cell lines. Mol. Cancer Ther. 2012, 11, 503–513. [Google Scholar] [CrossRef][Green Version]
- Lu, Y.; Zhao, X.; Liu, Q.; Mingli, Y.; Graves-Deal, R.; Cao, Z.; Singh, B.; Franklin, J.L.; Wang, J.; Bhuminder, S.; et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat. Med. 2017, 23, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Shi, W.; Zhang, Y.; Wang, X.; Sun, S.; Song, Z.; Liu, M.; Zeng, Q.; Cui, S.; Qu, X. CXCL12/CXCR4 axis induced MIR-125b promotes invasion and confers 5-fluorouracil re-sistance through enhancing autophagy in colorectal cancer. Sci. Rep. 2017, 7, 42226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, S. Up-regulation of MIR-125b reverses epithelial-mesenchymal transition in paclitaxel-resistant lung cancer cells. Biol. Chem. 2015. [Google Scholar] [CrossRef]
- Zhao, D.; Zhai, B.; He, C.; Tan, G.; Jiang, X.; Pan, S.; Dong, X.; Wei, Z.; Ma, L.; Qiao, H.; et al. Upregulation of HIF-2α induced by sorafenib contributes to the resistance by activating the TGF-α/EGFR pathway in hepatocellular carcinoma cells. Cell. Signal. 2014, 26, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, M.; Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Shim, J.S. Targeting Epithelial–Mesenchymal Transition (EMT) to overcome drug resistance in cancer. Molecules 2016, 21, 965. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT CSCs drug resistance. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Biddle, A.; MacKenzie, I.C. Cancer stem cells and EMT in carcinoma. Cancer Metastasis Rev. 2012, 31, 285–293. [Google Scholar] [CrossRef]
- Lytle, N.K.; Barber, A.G.; Reya, T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 2018, 18, 669–680. [Google Scholar] [CrossRef]
- Kudo, M.; Ueshima, K.; Chan, S.; Minami, T.; Chishina, H.; Aoki, T.; Takita, M.; Hagiwara, S.; Minami, Y.; Ida, H.; et al. Lenvatinib as an initial treatment in patients with intermediate-stage hepatocellular carcinoma beyond up-to-seven criteria and child-pugh a liver function: A proof-of-concept study. Cancers 2019, 11, 1084. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirao, A.; Sato, Y.; Tanaka, H.; Nishida, K.; Tomonari, T.; Hirata, M.; Bando, M.; Kida, Y.; Tanaka, T.; Kawaguchi, T.; et al. MiR-125b-5p Is Involved in Sorafenib Resistance through Ataxin-1-Mediated Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Cancers 2021, 13, 4917. https://doi.org/10.3390/cancers13194917
Hirao A, Sato Y, Tanaka H, Nishida K, Tomonari T, Hirata M, Bando M, Kida Y, Tanaka T, Kawaguchi T, et al. MiR-125b-5p Is Involved in Sorafenib Resistance through Ataxin-1-Mediated Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Cancers. 2021; 13(19):4917. https://doi.org/10.3390/cancers13194917
Chicago/Turabian StyleHirao, Akihiro, Yasushi Sato, Hironori Tanaka, Kensei Nishida, Tetsu Tomonari, Misato Hirata, Masahiro Bando, Yoshifumi Kida, Takahiro Tanaka, Tomoyuki Kawaguchi, and et al. 2021. "MiR-125b-5p Is Involved in Sorafenib Resistance through Ataxin-1-Mediated Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma" Cancers 13, no. 19: 4917. https://doi.org/10.3390/cancers13194917
APA StyleHirao, A., Sato, Y., Tanaka, H., Nishida, K., Tomonari, T., Hirata, M., Bando, M., Kida, Y., Tanaka, T., Kawaguchi, T., Wada, H., Taniguchi, T., Okamoto, K., Miyamoto, H., Muguruma, N., Tanahashi, T., & Takayama, T. (2021). MiR-125b-5p Is Involved in Sorafenib Resistance through Ataxin-1-Mediated Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Cancers, 13(19), 4917. https://doi.org/10.3390/cancers13194917





