Therapeutic Effects of Inhibition of Sphingosine-1-Phosphate Signaling in HIF-2α Inhibitor-Resistant Clear Cell Renal Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Therapy Studies and Tumor Monitoring
2.3. Cell Culture, HIF-2α Mutagenesis and Retroviral Infections
2.4. Soft Agar Colony Forming Assay
2.5. Immunohistochemistry
2.6. Western Blotting
2.7. RNA-Sequencing, Gene-Set Enrichment Analysis and sc-RNA-Sequencing
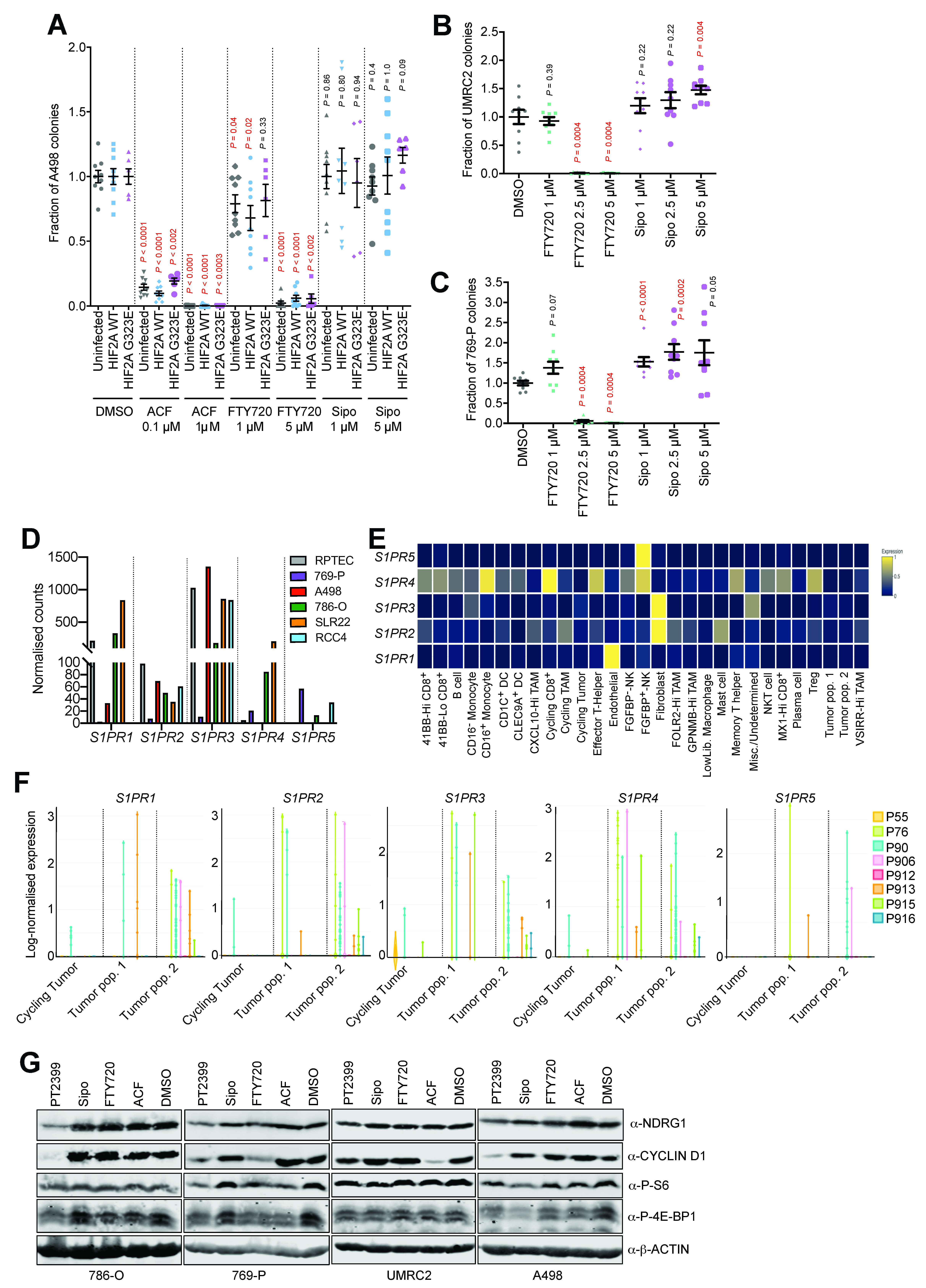
3. Results and Discussion
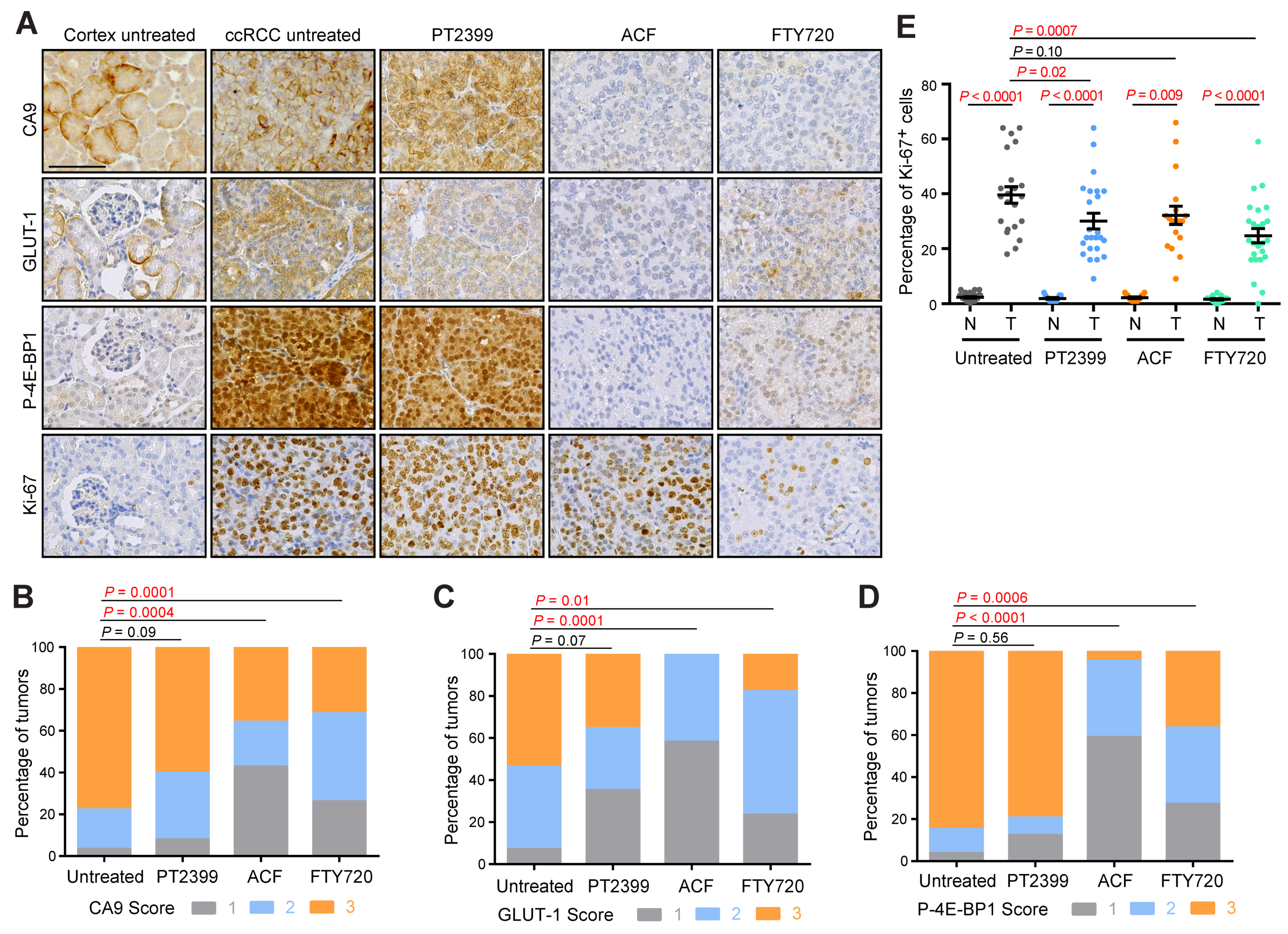
3.1. Evidence of Sphingosine Pathway Activation in Mouse and Human ccRCC
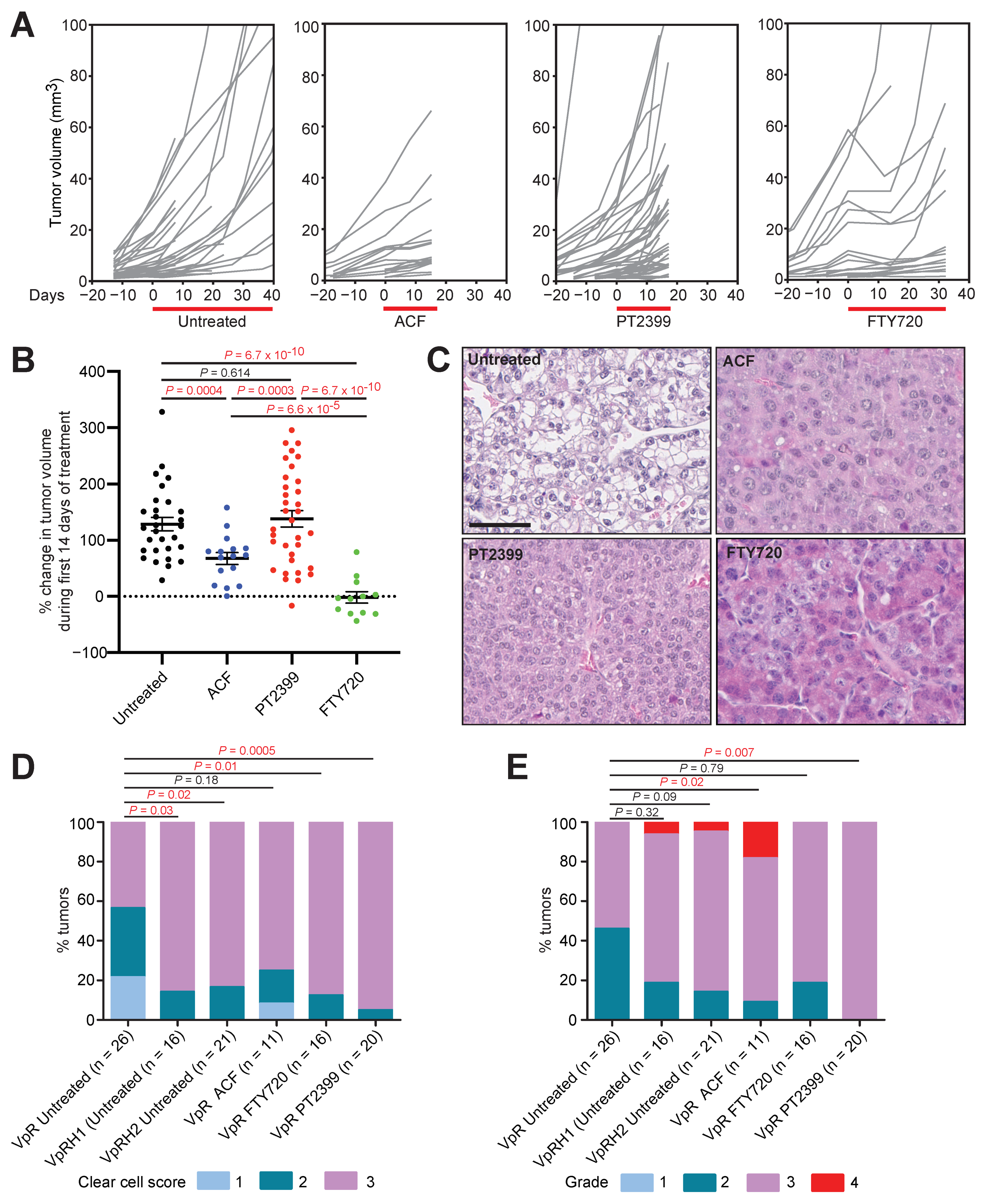
3.2. Acriflavine and FTY720 Show Antitumor Effects in HIF-2α-Resistant Mouse ccRCC
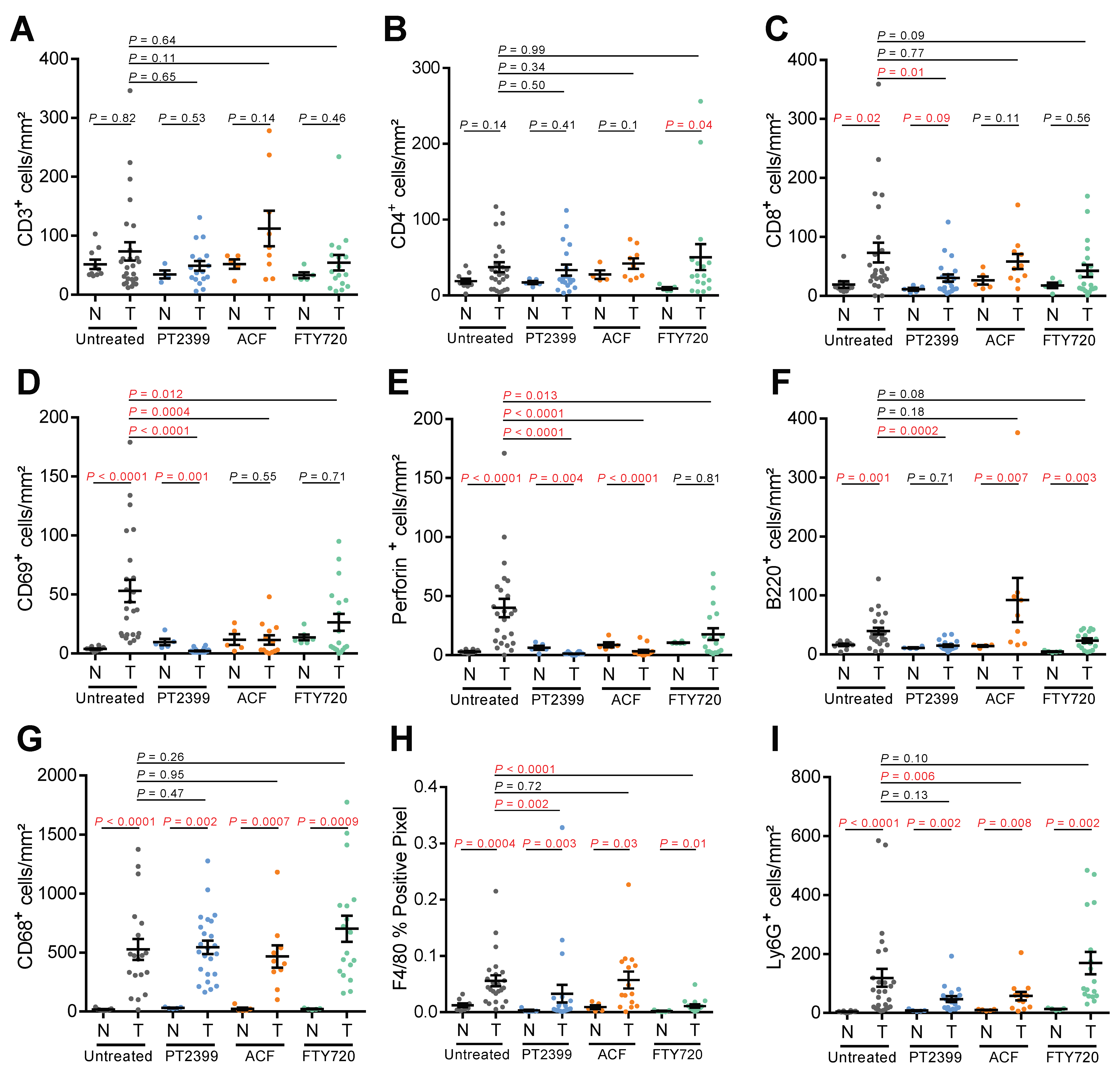
3.3. Impact of Pharmacologic HIF-Inhibition on the Tumor Immune Microenvironment
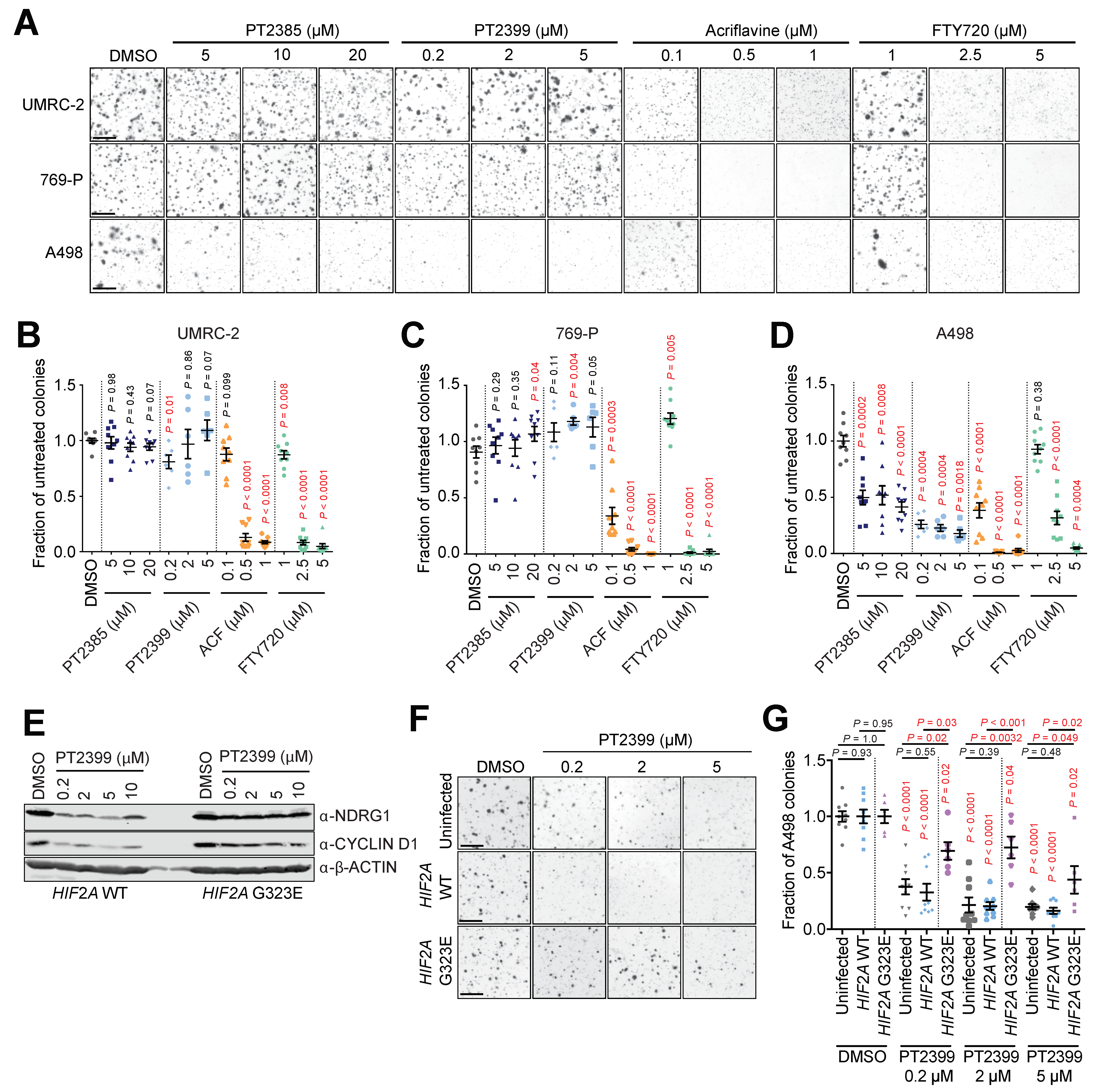
3.4. Acriflavine and FTY720 Inhibit Growth of HIF-2α-Resistant Human ccRCC Cell Lines
3.5. Multiple S1P Receptors Contribute to ccRCC Tumor Cell Proliferation
3.6. HIF-α-Dependent and -Independent Effects of ACF and FTY720
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frew, I.J.; Moch, H. A Clearer View of the Molecular Complexity of Clear Cell Renal Cell Carcinoma. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Hoefflin, R.; Harlander, S.; Schäfer, S.; Metzger, P.; Kuo, F.; Schönenberger, D.; Adlesic, M.; Peighambari, A.; Seidel, P.; Chen, C.; et al. HIF-1α and HIF-2α Differently Regulate Tumour Development and Inflammation of Clear Cell Renal Cell Carcinoma in Mice. Nat. Commun. 2020, 11, 4111. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Beroukhim, R.; Schumacher, S.E.; Zhou, J.; Chang, M.; Signoretti, S.; Kaelin, W.G. Genetic and Functional Studies Implicate HIF1 as a 14q Kidney Cancer Suppressor Gene. Cancer Discov. 2011, 1, 222–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raval, R.R.; Lau, K.W.; Tran, M.G.B.; Sowter, H.M.; Mandriota, S.J.; Li, J.-L.; Pugh, C.W.; Maxwell, P.H.; Harris, A.L.; Ratcliffe, P.J. Contrasting Properties of Hypoxia-Inducible Factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-Associated Renal Cell Carcinoma. Mol. Cell. Biol. 2005, 25, 5675–5686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, K.; Klco, J.; Nakamura, E.; Lechpammer, M.; Kaelin, W.G. Inhibition of HIF Is Necessary for Tumor Suppression by the von Hippel-Lindau Protein. Cancer Cell 2002, 1, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Du, X.; Rizzi, J.P.; Liberzon, E.; Chakraborty, A.A.; Gao, W.; Carvo, I.; Signoretti, S.; Bruick, R.K.; Josey, J.A.; et al. On-Target Efficacy of a HIF-2α Antagonist in Preclinical Kidney Cancer Models. Nature 2016, 539, 107–111. [Google Scholar] [CrossRef] [Green Version]
- Wallace, E.M.; Rizzi, J.P.; Han, G.; Wehn, P.M.; Cao, Z.; Du, X.; Cheng, T.; Czerwinski, R.M.; Dixon, D.D.; Goggin, B.S.; et al. A Small-Molecule Antagonist of HIF2α Is Efficacious in Preclinical Models of Renal Cell Carcinoma. Cancer Res. 2016, 76, 5491–5500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Wang, K.; Rizzi, J.P.; Huang, H.; Grina, J.A.; Schlachter, S.T.; Wang, B.; Wehn, P.M.; Yang, H.; Dixon, D.D.; et al. 3-[(1S,2S,3R)-2,3-Difluoro-1-Hydroxy-7-Methylsulfonylindan-4-Yl]Oxy-5-Fluorobenzonitrile (PT2977), a Hypoxia-Inducible Factor 2α (HIF-2α) Inhibitor for the Treatment of Clear Cell Renal Cell Carcinoma. J. Med. Chem. 2019, 62, 6876–6893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choueiri, T.K.; Kaelin, W.G. Targeting the HIF2–VEGF Axis in Renal Cell Carcinoma. Nat. Med. 2020, 26, 1519–1530. [Google Scholar] [CrossRef]
- Chen, W.; Hill, H.; Christie, A.; Kim, M.S.; Holloman, E.; Pavia-Jimenez, A.; Homayoun, F.; Ma, Y.; Patel, N.; Yell, P.; et al. Targeting Renal Cell Carcinoma with a HIF-2 Antagonist. Nature 2016, 539, 112–117. [Google Scholar] [CrossRef] [Green Version]
- Courtney, K.D.; Infante, J.R.; Lam, E.T.; Figlin, R.A.; Rini, B.I.; Brugarolas, J.; Zojwalla, N.J.; Lowe, A.M.; Wang, K.; Wallace, E.M.; et al. Phase I Dose-Escalation Trial of PT2385, a First-in-Class Hypoxia-Inducible Factor-2α Antagonist in Patients with Previously Treated Advanced Clear Cell Renal Cell Carcinoma. J. Clin. Oncol. 2018, 36, 867–874. [Google Scholar] [CrossRef]
- Choueiri, T.K. Phase I/II Study of the Oral HIF-2 α Inhibitor MK-6482 in Patients with Advanced Clear Cell Renal Cell Carcinoma (RCC). J. Clin. Oncol. 2020, 38, 611. [Google Scholar] [CrossRef]
- Courtney, K.D.; Ma, Y.; Diaz de Leon, A.; Christie, A.; Xie, Z.; Woolford, L.; Singla, N.; Joyce, A.; Hill, H.; Madhuranthakam, A.J.; et al. HIF-2 Complex Dissociation, Target Inhibition, and Acquired Resistance with PT2385, a First-in-Class HIF-2 Inhibitor, in Patients with Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2020, 26, 793–803. [Google Scholar] [CrossRef]
- Harlander, S.; Schönenberger, D.; Toussaint, N.C.; Prummer, M.; Catalano, A.; Brandt, L.; Moch, H.; Wild, P.J.; Frew, I.J. Combined Mutation in Vhl, Trp53 and Rb1 Causes Clear Cell Renal Cell Carcinoma in Mice. Nat. Med. 2017, 23, 869–877. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Zhang, H.; Qian, D.Z.; Rey, S.; Liu, J.O.; Semenza, G.L. Acriflavine Inhibits HIF-1 Dimerization, Tumor Growth, and Vascularization. Proc. Natl. Acad. Sci. USA 2009, 106, 17910–17915. [Google Scholar] [CrossRef] [Green Version]
- Shay, J.E.S.; Imtiyaz, H.Z.; Sivanand, S.; Durham, A.C.; Skuli, N.; Hsu, S.; Mucaj, V.; Eisinger-Mathason, T.S.K.; Krock, B.L.; Giannoukos, D.N.; et al. Inhibition of Hypoxia-Inducible Factors Limits Tumor Progression in a Mouse Model of Colorectal Cancer. Carcinogenesis 2014, 35, 1067–1077. [Google Scholar] [CrossRef]
- Yin, T.; He, S.; Shen, G.; Wang, Y. HIF-1 Dimerization Inhibitor Acriflavine Enhances Antitumor Activity of Sunitinib in Breast Cancer Model. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2015, 22, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Mangraviti, A.; Raghavan, T.; Volpin, F.; Skuli, N.; Gullotti, D.; Zhou, J.; Asnaghi, L.; Sankey, E.; Liu, A.; Wang, Y.; et al. HIF-1α- Targeting Acriflavine Provides Long Term Survival and Radiological Tumor Response in Brain Cancer Therapy. Sci. Rep. 2017, 7, 14978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gstalder, C.; Ader, I.; Cuvillier, O. FTY720 (Fingolimod) Inhibits HIF1 and HIF2 Signaling, Promotes Vascular Remodeling, and Chemosensitizes in Renal Cell Carcinoma Animal Model. Mol. Cancer Ther. 2016, 15, 2465–2474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouquerel, P.; Gstalder, C.; Müller, D.; Laurent, J.; Brizuela, L.; Sabbadini, R.A.; Malavaud, B.; Pyronnet, S.; Martineau, Y.; Ader, I.; et al. Essential Role for SphK1/S1P Signaling to Regulate Hypoxia-Inducible Factor 2α Expression and Activity in Cancer. Oncogenesis 2016, 5, e209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyne, N.J.; Pyne, S. Sphingosine 1-Phosphate and Cancer. Nat. Rev. Cancer 2010, 10, 489–503. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, X.; Bullock, A.J.; Callea, M.; Shah, H.; Song, J.; Moreno, K.; Visentin, B.; Deutschman, D.; Alsop, D.C.; et al. Anti-S1P Antibody as a Novel Therapeutic Strategy for VEGFR TKI-Resistant Renal Cancer. Clin. Cancer Res. 2015, 21, 1925–1934. [Google Scholar] [CrossRef] [Green Version]
- Salama, M.F.; Carroll, B.; Adada, M.; Pulkoski-Gross, M.; Hannun, Y.A.; Obeid, L.M. A Novel Role of Sphingosine Kinase-1 in the Invasion and Angiogenesis of VHL Mutant Clear Cell Renal Cell Carcinoma. FASEB J. 2015, 29, 2803–2813. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Simon, J.M.; Xie, H.; Hu, L.; Wang, J.; Zurlo, G.; Fan, C.; Ptacek, T.S.; Herring, L.; Tan, X.; et al. Genome-Wide Screening Identifies SFMBT1 as an Oncogenic Driver in Cancer with VHL Loss. Mol. Cell 2020, 77, 1294–1306.e5. [Google Scholar] [CrossRef]
- Pchejetski, D.; Bohler, T.; Brizuela, L.; Sauer, L.; Doumerc, N.; Golzio, M.; Salunkhe, V.; Teissié, J.; Malavaud, B.; Waxman, J.; et al. FTY720 (Fingolimod) Sensitizes Prostate Cancer Cells to Radiotherapy by Inhibition of Sphingosine Kinase-1. Cancer Res. 2010, 70, 8651–8661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, K.G.; Tonelli, F.; Li, Z.; Lu, X.; Bittman, R.; Pyne, S.; Pyne, N.J. FTY720 Analogues as Sphingosine Kinase 1 Inhibitors: Enzyme Inhibition Kinetics, Allosterism, Proteasomal Degradation, and Actin Rearrangement in MCF-7 Breast Cancer Cells. J. Biol. Chem. 2011, 286, 18633–18640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Bartz, S.; Mao, M.; Li, L.; Kaelin, W.G. The Hypoxia-Inducible Factor 2alpha N-Terminal and C-Terminal Transactivation Domains Cooperate to Promote Renal Tumorigenesis in Vivo. Mol. Cell. Biol. 2007, 27, 2092–2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoma, C.R.; Frew, I.J.; Hoerner, C.R.; Montani, M.; Moch, H.; Krek, W. PVHL and GSK3β Are Components of a Primary Cilium-Maintenance Signalling Network. Nat. Cell Biol. 2007, 9, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open Source Software for Digital Pathology Image Analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoefflin, R.; Lahrmann, B.; Warsow, G.; Hübschmann, D.; Spath, C.; Walter, B.; Chen, X.; Hofer, L.; Macher-Goeppinger, S.; Tolstov, Y.; et al. Spatial Niche Formation but Not Malignant Progression Is a Driving Force for Intratumoural Heterogeneity. Nat. Commun. 2016, 7, ncomms11845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating High-Throughput Genomic Analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally Applicable Gene Set Enrichment for Pathway Analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.-G.; Yan, G.-R.; He, Q.-Y. DOSE: An R/Bioconductor Package for Disease Ontology Semantic and Enrichment Analysis. Bioinforma. Oxf. Engl. 2015, 31, 608–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, K.; He, M.X.; Bakouny, Z.; Kanodia, A.; Napolitano, S.; Wu, J.; Grimaldi, G.; Braun, D.A.; Cuoco, M.S.; Mayorga, A.; et al. Tumor and Immune Reprogramming during Immunotherapy in Advanced Renal Cell Carcinoma. Cancer Cell 2021, 39, 649–661.e5. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Doedens, A.L.; Phan, A.T.; Stradner, M.H.; Fujimoto, J.K.; Nguyen, J.V.; Yang, E.; Johnson, R.S.; Goldrath, A.W. Hypoxia-Inducible Factors Enhance the Effector Responses of CD8+ T Cells to Persistent Antigen. Nat. Immunol. 2013, 14, 1173–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clever, D.; Roychoudhuri, R.; Constantinides, M.G.; Askenase, M.H.; Sukumar, M.; Klebanoff, C.A.; Eil, R.L.; Hickman, H.D.; Yu, Z.; Pan, J.H.; et al. Oxygen Sensing by T Cells Establishes an Immunologically Tolerant Metastatic Niche. Cell 2016, 166, 1117–1131.e14. [Google Scholar] [CrossRef] [Green Version]
- Finlay, D.K.; Rosenzweig, E.; Sinclair, L.V.; Feijoo-Carnero, C.; Hukelmann, J.L.; Rolf, J.; Panteleyev, A.A.; Okkenhaug, K.; Cantrell, D.A. PDK1 Regulation of MTOR and Hypoxia-Inducible Factor 1 Integrate Metabolism and Migration of CD8+ T Cells. J. Exp. Med. 2012, 209, 2441–2453. [Google Scholar] [CrossRef] [Green Version]
- Palazon, A.; Tyrakis, P.A.; Macias, D.; Veliça, P.; Rundqvist, H.; Fitzpatrick, S.; Vojnovic, N.; Phan, A.T.; Loman, N.; Hedenfalk, I.; et al. An HIF-1α/VEGF-A Axis in Cytotoxic T Cells Regulates Tumor Progression. Cancer Cell 2017, 32, 669–683.e5. [Google Scholar] [CrossRef] [Green Version]
- Veliça, P.; Cunha, P.P.; Vojnovic, N.; Foskolou, I.P.; Bargiela, D.; Gojkovic, M.; Rundqvist, H.; Johnson, R.S. Modified Hypoxia-Inducible Factor Expression in CD8+ T Cells Increases Antitumor Efficacy. Cancer Immunol. Res. 2021, 9, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Han, J.W.; Choi, Y.J.; Rha, M.-S.; Koh, J.Y.; Kim, K.H.; Kim, C.G.; Lee, Y.J.; Kim, A.R.; Park, J.; et al. Functions of Human Liver CD69+CD103-CD8+ T Cells Depend on HIF-2α Activity in Healthy and Pathologic Livers. J. Hepatol. 2020, 72, 1170–1181. [Google Scholar] [CrossRef]
- Rini, B.I. Results from a Phase I Expansion Cohort of the First-in-Class Oral HIF-2α Inhibitor PT2385 in Combination with Nivolumab in Patients with Previously Treated Advanced RCC. J. Clin. Oncol 2019, 37, 558. [Google Scholar] [CrossRef]
- Wu, D.; Su, X.; Lu, J.; Li, S.; Hood, B.L.; Vasile, S.; Potluri, N.; Diao, X.; Kim, Y.; Khorasanizadeh, S.; et al. Bidirectional Modulation of HIF-2 Activity through Chemical Ligands. Nat. Chem. Biol. 2019, 15, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Kappos, L.; Bar-Or, A.; Cree, B.A.C.; Fox, R.J.; Giovannoni, G.; Gold, R.; Vermersch, P.; Arnold, D.L.; Arnould, S.; Scherz, T.; et al. Siponimod versus Placebo in Secondary Progressive Multiple Sclerosis (EXPAND): A Double-Blind, Randomised, Phase 3 Study. Lancet Lond. Engl. 2018, 391, 1263–1273. [Google Scholar] [CrossRef]
- Nagahashi, M.; Takabe, K.; Terracina, K.P.; Soma, D.; Hirose, Y.; Kobayashi, T.; Matsuda, Y.; Wakai, T. Sphingosine-1-Phosphate Transporters as Targets for Cancer Therapy. BioMed Res. Int. 2014, 2014, 651727. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoefflin, R.; Harlander, S.; Abhari, B.A.; Peighambari, A.; Adlesic, M.; Seidel, P.; Zodel, K.; Haug, S.; Göcmen, B.; Li, Y.; et al. Therapeutic Effects of Inhibition of Sphingosine-1-Phosphate Signaling in HIF-2α Inhibitor-Resistant Clear Cell Renal Cell Carcinoma. Cancers 2021, 13, 4801. https://doi.org/10.3390/cancers13194801
Hoefflin R, Harlander S, Abhari BA, Peighambari A, Adlesic M, Seidel P, Zodel K, Haug S, Göcmen B, Li Y, et al. Therapeutic Effects of Inhibition of Sphingosine-1-Phosphate Signaling in HIF-2α Inhibitor-Resistant Clear Cell Renal Cell Carcinoma. Cancers. 2021; 13(19):4801. https://doi.org/10.3390/cancers13194801
Chicago/Turabian StyleHoefflin, Rouven, Sabine Harlander, Behnaz A. Abhari, Asin Peighambari, Mojca Adlesic, Philipp Seidel, Kyra Zodel, Stefan Haug, Burulca Göcmen, Yong Li, and et al. 2021. "Therapeutic Effects of Inhibition of Sphingosine-1-Phosphate Signaling in HIF-2α Inhibitor-Resistant Clear Cell Renal Cell Carcinoma" Cancers 13, no. 19: 4801. https://doi.org/10.3390/cancers13194801
APA StyleHoefflin, R., Harlander, S., Abhari, B. A., Peighambari, A., Adlesic, M., Seidel, P., Zodel, K., Haug, S., Göcmen, B., Li, Y., Lahrmann, B., Grabe, N., Heide, D., Boerries, M., Köttgen, A., Heikenwalder, M., & Frew, I. J. (2021). Therapeutic Effects of Inhibition of Sphingosine-1-Phosphate Signaling in HIF-2α Inhibitor-Resistant Clear Cell Renal Cell Carcinoma. Cancers, 13(19), 4801. https://doi.org/10.3390/cancers13194801






