MEM: An Algorithm for the Reliable Detection of Microsatellite Instability (MSI) on a Small NGS Panel in Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
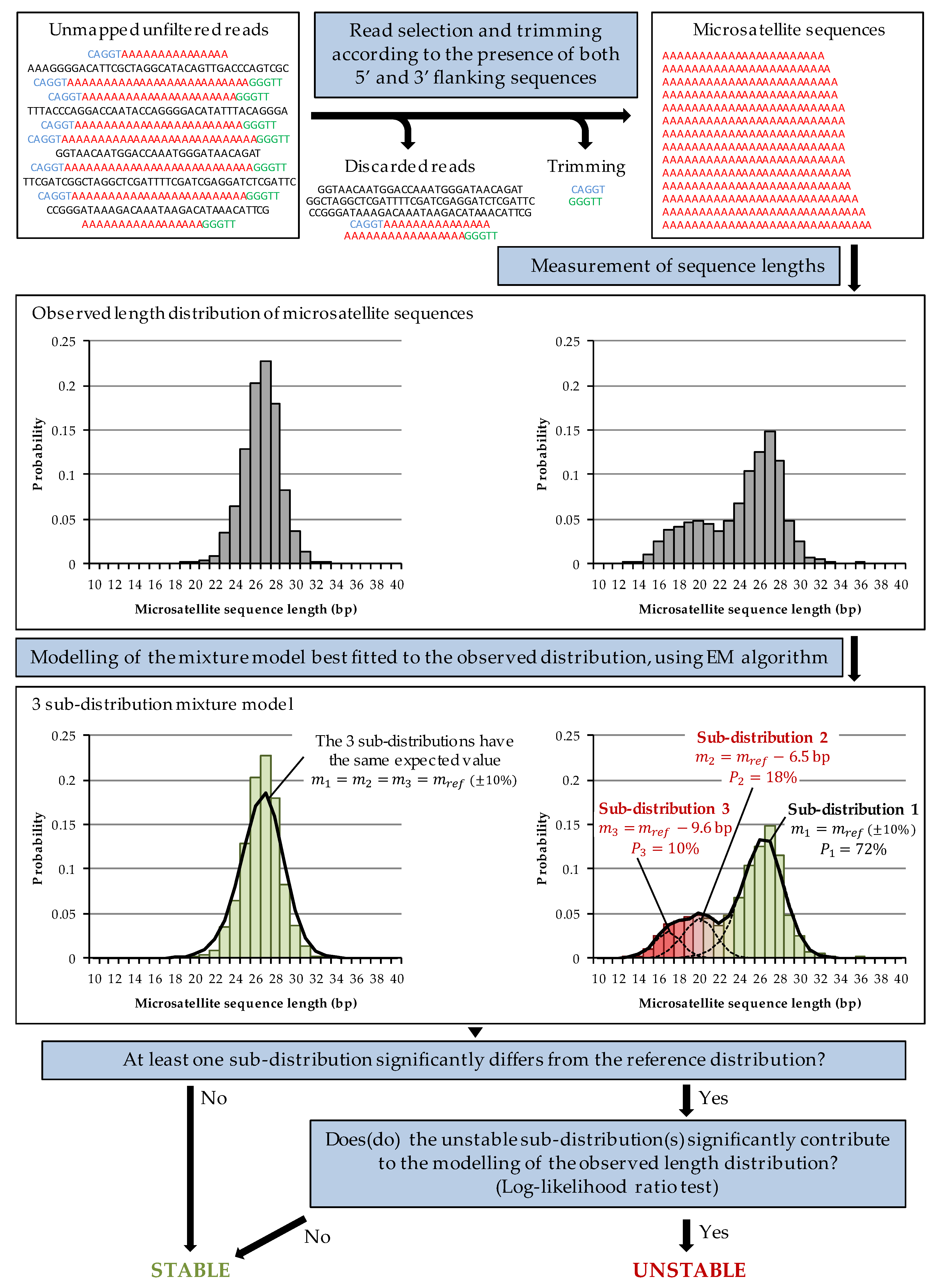
2.1. MEM Algorithm
2.2. Evaluation of the Limit of Detection
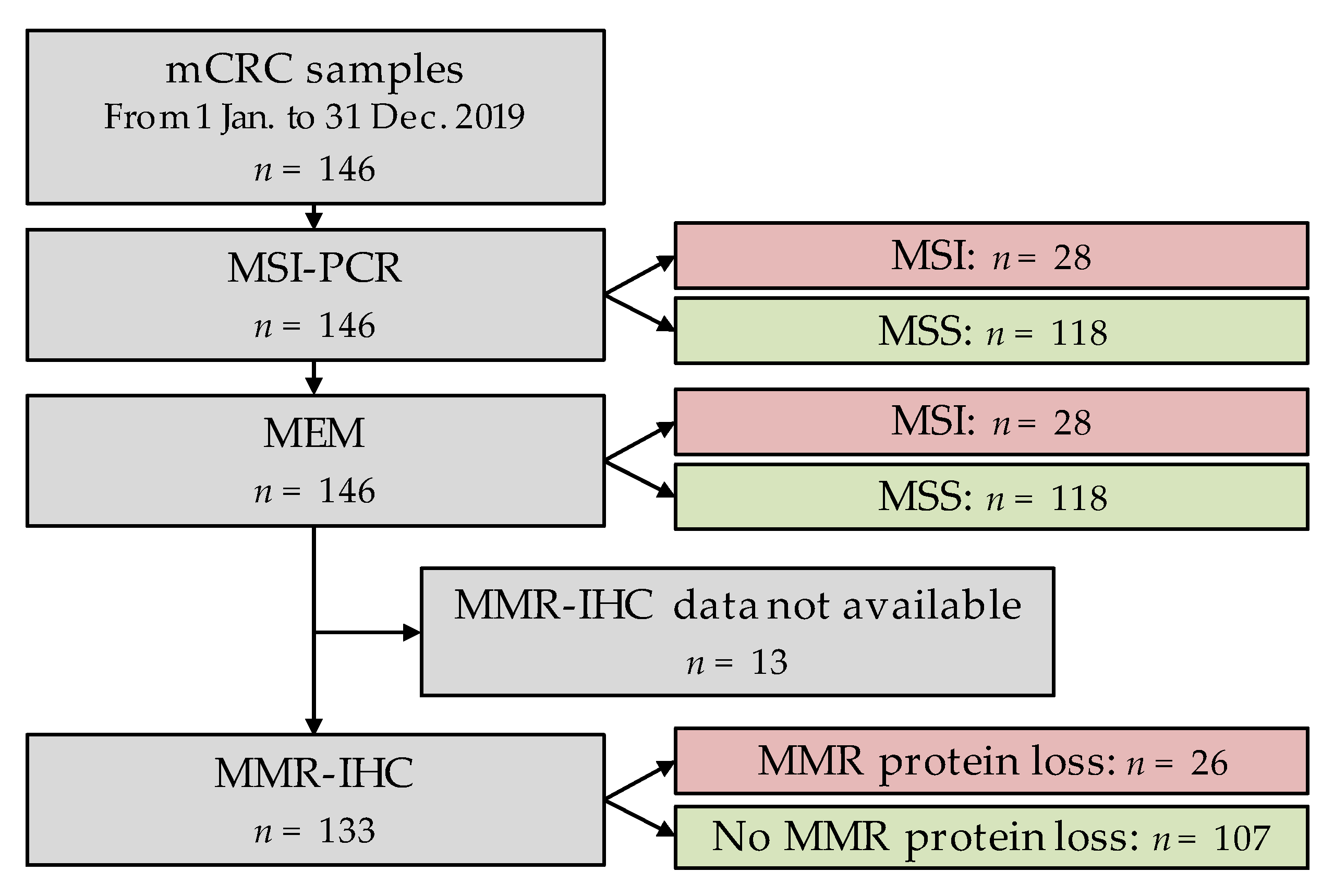
2.3. Validation on Tumour Samples
2.4. Ethical Aspects
3. Results
3.1. Limit of Detection
3.2. MEM Algorithm vs. MSI-PCR Comparison
3.3. MEM vs. MMR-IHC Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. MEM Algorithm
Appendix A.1. Determination of the Length Distribution of Microsatellite Sequences
- : probability of occurrence of a microsatellite sequence of length
- : number of trimmed sequences of length
- : maximum length
Appendix A.2. Modelling of Length Distribution Using a Mixture Model
Appendix A.3. Expectation-Maximisation Algorithm
Appendix A.4. Interpretation
References
- Strand, M.; Prolla, T.A.; Liskay, R.M.; Petes, T.D. Destabilization of Tracts of Simple Repetitive DNA in Yeast by Mutations Affecting DNA Mismatch Repair. Nature 1993, 365, 274–276. [Google Scholar] [CrossRef]
- Schlötterer, C. Genome Evolution: Are Microsatellites Really Simple Sequences? Curr. Biol. 1998, 8, R132–R134. [Google Scholar] [CrossRef] [Green Version]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.-Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hause, R.J.; Pritchard, C.C.; Shendure, J.; Salipante, S.J. Classification and Characterization of Microsatellite Instability across 18 Cancer Types. Nat. Med. 2016, 22, 1342–1350. [Google Scholar] [CrossRef]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; de la Chapelle, A.; Rüschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.-Y.; Andre, F.; et al. ESMO Recommendations on Microsatellite Instability Testing for Immunotherapy in Cancer, and Its Relationship with PD-1/PD-L1 Expression and Tumour Mutational Burden: A Systematic Review-Based Approach. Ann. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef] [Green Version]
- Goel, A.; Nagasaka, T.; Hamelin, R.; Boland, C.R. An Optimized Pentaplex PCR for Detecting DNA Mismatch Repair-Deficient Colorectal Cancers. PLoS ONE 2010, 5, e9393. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [Green Version]
- André, T.; Shiu, K.-K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.-P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef]
- Marcus, L.; Lemery, S.J.; Keegan, P.; Pazdur, R. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin. Cancer Res. 2019, 25, 3753–3758. [Google Scholar] [CrossRef] [Green Version]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in Patients with Metastatic DNA Mismatch Repair-Deficient or Microsatellite Instability-High Colorectal Cancer (CheckMate 142): An Open-Label, Multicentre, Phase 2 Study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.-J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Abdullaev, S.; André, T.; Lei, M.; Lenz, H.-J.; Novotny, J.; Paulson, A.S.; Tejpar, S.; Yamazaki, K.; Ledeine, J.-M. A Phase III Study of Nivolumab (NIVO), NIVO + Ipilimumab (IPI), or Chemotherapy (CT) for Microsatellite Instability-High (MSI-H)/Mismatch Repair-Deficient (DMMR) Metastatic Colorectal Cancer (MCRC): Checkmate 8HW. JCO 2020, 38, TPS266. [Google Scholar] [CrossRef]
- Hampel, H.; Pearlman, R.; Beightol, M.; Zhao, W.; Jones, D.; Frankel, W.L.; Goodfellow, P.J.; Yilmaz, A.; Miller, K.; Bacher, J.; et al. Assessment of Tumor Sequencing as a Replacement for Lynch Syndrome Screening and Current Molecular Tests for Patients With Colorectal Cancer. JAMA Oncol. 2018, 4, 1–8. [Google Scholar] [CrossRef]
- Salipante, S.J.; Scroggins, S.M.; Hampel, H.L.; Turner, E.H.; Pritchard, C.C. Microsatellite Instability Detection by next Generation Sequencing. Clin. Chem. 2014, 60, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Hempelmann, J.A.; Scroggins, S.M.; Pritchard, C.C.; Salipante, S.J. MSIplus for Integrated Colorectal Cancer Molecular Testing by Next-Generation Sequencing. J. Mol. Diagn. 2015, 17, 705–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kautto, E.A.; Bonneville, R.; Miya, J.; Yu, L.; Krook, M.A.; Reeser, J.W.; Roychowdhury, S. Performance Evaluation for Rapid Detection of Pan-Cancer Microsatellite Instability with MANTIS. Oncotarget 2017, 8, 7452–7463. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.N.; McPherson, J.R.; Cutcutache, I.; Teh, B.T.; Tan, P.; Rozen, S.G. MSIseq: Software for Assessing Microsatellite Instability from Catalogs of Somatic Mutations. Sci. Rep. 2015, 5, 13321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, B.; Ye, K.; Zhang, Q.; Lu, C.; Xie, M.; McLellan, M.D.; Wendl, M.C.; Ding, L. MSIsensor: Microsatellite Instability Detection Using Paired Tumor-Normal Sequence Data. Bioinformatics 2014, 30, 1015–1016. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Huang, Y.; Fang, X.; Liu, C.; Deng, W.; Zhong, C.; Xu, J.; Xu, D.; Yuan, Y. A Novel and Reliable Method to Detect Microsatellite Instability in Colorectal Cancer by Next-Generation Sequencing. J. Mol. Diagn. 2018, 20, 225–231. [Google Scholar] [CrossRef] [Green Version]
- Cortes-Ciriano, I.; Lee, S.; Park, W.-Y.; Kim, T.-M.; Park, P.J. A Molecular Portrait of Microsatellite Instability across Multiple Cancers. Nat. Commun. 2017, 8, 15180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Soong, T.D.; Elemento, O. A Novel Approach for Characterizing Microsatellite Instability in Cancer Cells. PLoS ONE 2013, 8, e63056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, X.; Xiao, X.; Zhang, F.-R.; Yan, X.; Feng, X.; Zhao, Z.; Guan, Y.; Wang, J. Accurately Estimating the Length Distributions of Genomic Micro-Satellites by Tumor Purity Deconvolution. BMC Bioinform. 2020, 21, 82. [Google Scholar] [CrossRef] [Green Version]
- Ratovomanana, T.; Cohen, R.; Svrcek, M.; Renaud, F.; Cervera, P.; Siret, A.; Letourneur, Q.; Buhard, O.; Bourgoin, P.; Guillerm, E.; et al. Performance of Next Generation Sequencing for the Detection of Microsatellite Instability in Colorectal Cancer with Deficient DNA Mismatch Repair. Gastroenterology 2021. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, J.A.; Park, H.E.; Han, H.; Kim, Y.; Bae, J.M.; Kim, J.H.; Cho, N.-Y.; Kim, H.-P.; Kim, T.-Y.; et al. Targeted Next-Generation Sequencing-Based Detection of Microsatellite Instability in Colorectal Carcinomas. PLoS ONE 2021, 16, e0246356. [Google Scholar] [CrossRef]
- Zhao, L.; Shan, G.; Li, L.; Yu, Y.; Cheng, G.; Zheng, X. A Robust Method for the Rapid Detection of Microsatellite Instability in Colorectal Cancer. Oncol. Lett. 2020, 20, 1982–1988. [Google Scholar] [CrossRef]
- Zheng, K.; Wan, H.; Zhang, J.; Shan, G.; Chai, N.; Li, D.; Fang, N.; Liu, L.; Zhang, J.; Du, R.; et al. A Novel NGS-Based Microsatellite Instability (MSI) Status Classifier with 9 Loci for Colorectal Cancer Patients. J. Transl. Med. 2020, 18, 215. [Google Scholar] [CrossRef]
- Suraweera, N.; Duval, A.; Reperant, M.; Vaury, C.; Furlan, D.; Leroy, K.; Seruca, R.; Iacopetta, B.; Hamelin, R. Evaluation of Tumor Microsatellite Instability Using Five Quasimonomorphic Mononucleotide Repeats and Pentaplex PCR. Gastroenterology 2002, 123, 1804–1811. [Google Scholar] [CrossRef]
- Dedeurwaerdere, F.; Claes, K.B.; Van Dorpe, J.; Rottiers, I.; Van der Meulen, J.; Breyne, J.; Swaerts, K.; Martens, G. Comparison of Microsatellite Instability Detection by Immunohistochemistry and Molecular Techniques in Colorectal and Endometrial Cancer. Sci. Rep. 2021, 11, 12880. [Google Scholar] [CrossRef]
- Saul, M.; Poorman, K.; Tae, H.; Vanderwalde, A.; Stafford, P.; Spetzler, D.; Korn, W.M.; Gatalica, Z.; Swensen, J. Population Bias in Somatic Measurement of Microsatellite Instability Status. Cancer Med. 2020, 9, 6452–6460. [Google Scholar] [CrossRef] [PubMed]
| MSI-PCR | Cohen’s Kappa | |||
|---|---|---|---|---|
| MSI | MSS | |||
| MEM | MSI | 28 | 0 | 1 |
| MSS | 0 | 118 | ||
| MSI-PCR | Cohen’s Kappa | ||||
|---|---|---|---|---|---|
| Unstable | Stable | ||||
| BAT-25 | MEM | Unstable | 28 | 0 | 1 |
| Stable | 0 | 118 | |||
| BAT-26 | MEM | Unstable | 27 | 0 | 1 |
| Stable | 0 | 118 | |||
| NR-21 | MEM | Unstable | 28 | 0 | 1 |
| Stable | 0 | 118 | |||
| NR-24 | MEM | Unstable | 26 | 0 | 1 |
| Stable | 0 | 120 | |||
| MSI-PCR | ||||
|---|---|---|---|---|
| MSI | MSS | |||
| NR-27 | MEM | Unstable | 26 | 2 |
| Stable | 1 | 116 | ||
| MMR-IHC | Cohen’s Kappa | |||
|---|---|---|---|---|
| Loss of Expression of ≥1 MMR Protein | No Loss of Expression | |||
| MEM | MSI | 26 | 0 | 0.931 |
| MSS | 3 | 104 | ||
| MSI-NGS Algorithm | Normal Sample Required | Microsatellite Count | Validation Cohort | Sensitivity | Specificity |
|---|---|---|---|---|---|
| mSINGS/ MSIplus | No | N/A (TGS) | Zhao [27]: n = 113 | 88.9% | 99.0% |
| 15 to 2957 | Salipante [16]: n = 108 | 96.4–100% | 97.2–100% | ||
| 2539 | Kautto [18]: n = 275 | 76.1% | 99.7% | ||
| 3154 | Lee [26]: n = 117 | 92.5% | 100% | ||
| 230 | Lee [26]: n = 117 | 95.0% | 100% | ||
| 23 | Lee [26]: n = 117 | 95.0% | 93.7% | ||
| 11 | Hempelmann [17]: n = 81 | 97.1% | 100% | ||
| MSIsensor | Yes | N/A (WES) | Niu [20]: n = 242 | 98.6% | 98.2% |
| 2539 | Kautto [18]: n = 275 | 96.5% | 98.7% | ||
| N/A (WES) | Ratovmanana [25]: n = 333 | 85.5–90.2% | 95.5–100% | ||
| 441 | Ratovmanana [25]: n = 152 | 97.1% | 73.3% | ||
| MANTIS | Yes | 2539 | Kautto [18]: n = 275 | 97.2% | 99.7% |
| N/A (TGS) | Zhao [27]: n = 113 | 88.9% | 86.5% | ||
| MSI-ColonCore | Yes | 90 | Zhu [21]: n = 91 | 97.9% | 100% |
| ELMSI | No | 20 to 100 | Wang [24]: simulated data | 70.0–82.1% | N/A |
| MSICare | Yes | N/A (WES) | Ratovmanana [25]: n = 333 | 96.1–100% | 97.0–100% |
| 441 | Ratovmanana [25]: n = 152 | 99.3% | 100% | ||
| mSILICO | No | 3154 | Lee [26]: n = 117 | 100% | 100% |
| 230 | Lee [26]: n = 117 | 100% | 77.2% | ||
| 23 | Lee [26]: n = 117 | 95.0% | 100% | ||
| NovoPM-MSI | Yes | 19 | Zhao [27]: n = 113 | 88.9% | 97.1% |
| USCI-MSI | Yes | 9 | Zheng [28]: n = 64 | 100% | 100% |
| MEM | No | 5 | (present study): n = 146 | 100% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herbreteau, G.; Airaud, F.; Pierre-Noël, E.; Vallée, A.; Bézieau, S.; Théoleyre, S.; Blons, H.; Garinet, S.; Denis, M.G. MEM: An Algorithm for the Reliable Detection of Microsatellite Instability (MSI) on a Small NGS Panel in Colorectal Cancer. Cancers 2021, 13, 4203. https://doi.org/10.3390/cancers13164203
Herbreteau G, Airaud F, Pierre-Noël E, Vallée A, Bézieau S, Théoleyre S, Blons H, Garinet S, Denis MG. MEM: An Algorithm for the Reliable Detection of Microsatellite Instability (MSI) on a Small NGS Panel in Colorectal Cancer. Cancers. 2021; 13(16):4203. https://doi.org/10.3390/cancers13164203
Chicago/Turabian StyleHerbreteau, Guillaume, Fabrice Airaud, Elise Pierre-Noël, Audrey Vallée, Stéphane Bézieau, Sandrine Théoleyre, Hélène Blons, Simon Garinet, and Marc Guillaume Denis. 2021. "MEM: An Algorithm for the Reliable Detection of Microsatellite Instability (MSI) on a Small NGS Panel in Colorectal Cancer" Cancers 13, no. 16: 4203. https://doi.org/10.3390/cancers13164203
APA StyleHerbreteau, G., Airaud, F., Pierre-Noël, E., Vallée, A., Bézieau, S., Théoleyre, S., Blons, H., Garinet, S., & Denis, M. G. (2021). MEM: An Algorithm for the Reliable Detection of Microsatellite Instability (MSI) on a Small NGS Panel in Colorectal Cancer. Cancers, 13(16), 4203. https://doi.org/10.3390/cancers13164203







