HPV16 Load Is a Potential Biomarker to Predict Risk of High-Grade Cervical Lesions in High-Risk HPV-Infected Women: A Large Longitudinal French Hospital-Based Cohort Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Cytological and Histological Data
2.3. High-Risk HPV DNA Testing
2.4. Data Analyses
3. Results
3.1. Population
3.2. Pathological and Virological Characteristics at First, Visit
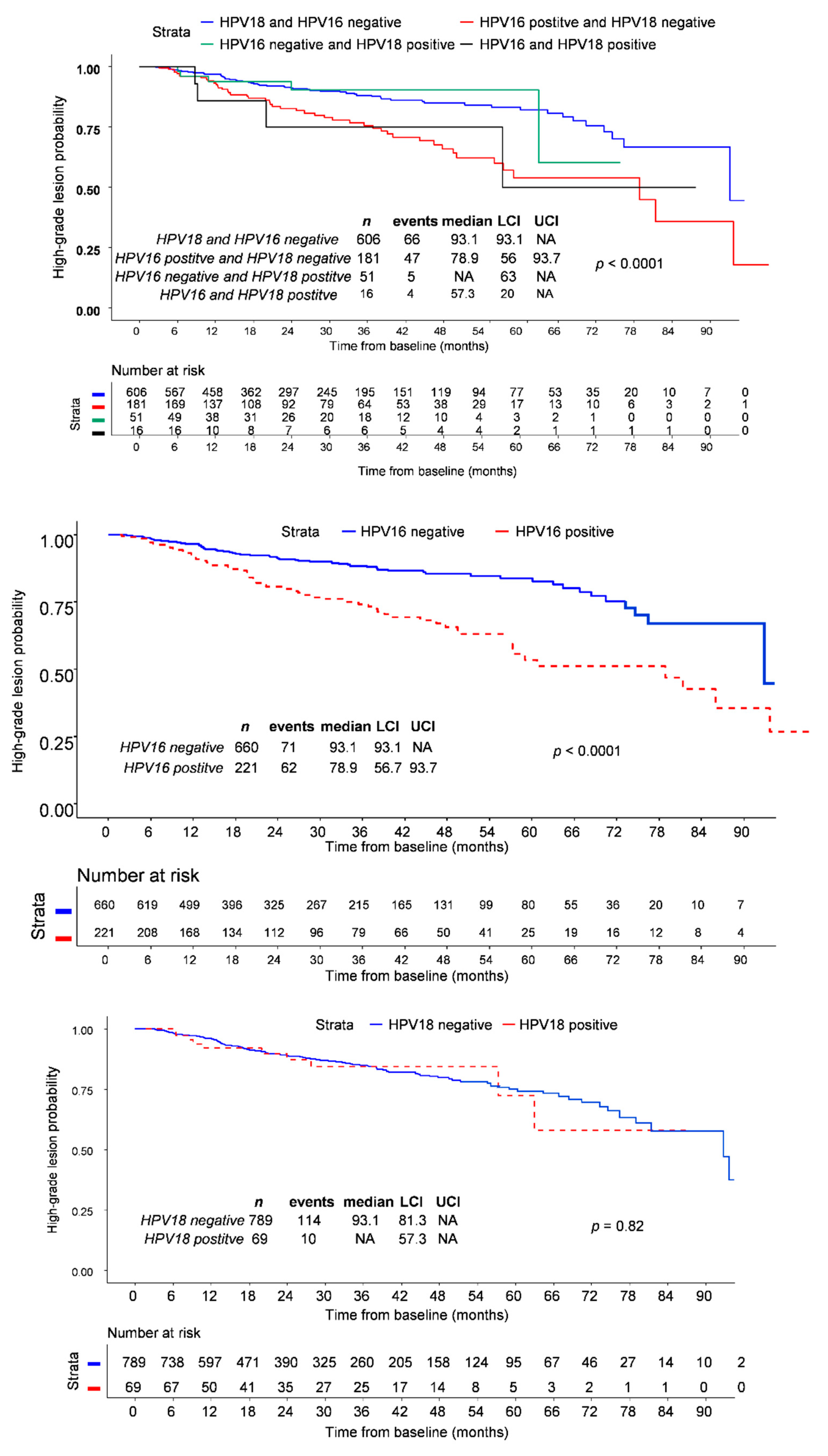
3.3. Follow-Up and Time to Lesion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bosch, F.X.; de Sanjosé, S. Chapter 1: Human papillomavirus and cervical cancer—Burden and assessment of causality. J. Natl. Cancer Inst. Monogr. 2003, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Wallin, K.L.; Wiklund, F.; Angström, T.; Bergman, F.; Stendahl, U.; Wadell, G.; Hallmans, G.; Dillner, J. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N. Engl. J. Med. 1999, 341, 1633–1638. [Google Scholar] [CrossRef]
- Dalstein, V.; Riethmuller, D.; Prétet, J.-L.; Le Bail Carval, K.; Sautière, J.-L.; Carbillet, J.-P.; Kantelip, B.; Schaal, J.-P.; Mougin, C. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: A longitudinal French cohort study. Int. J. Cancer 2003, 106, 396–403. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part B: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- Muñoz, N.; Bosch, F.X.; de Sanjosé, S.; Herrero, R.; Castellsagué, X.; Shah, K.V.; Snijders, P.J.F.; Meijer, C.J.L.M. International agency for research on cancer multicenter cervical cancer study group epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, P.; Howell-Jones, R.; Li, N.; Bruni, L.; de Sanjosé, S.; Franceschi, S.; Clifford, G.M. Human papillomavirus types in 115,789 HPV-positive women: A meta-analysis from cervical infection to cancer. Int. J. Cancer 2012, 131, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Prétet, J.-L.; Jacquard, A.-C.; Carcopino, X.; Charlot, J.-F.; Bouhour, D.; Kantelip, B.; Soubeyrand, B.; Leocmach, Y.; Mougin, C.; Riethmuller, D.; et al. Human papillomavirus (HPV) genotype distribution in invasive cervical cancers in France: EDITH study. Int. J. Cancer 2008, 122, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.J.; Castle, P.E.; Lorincz, A.T.; Wacholder, S.; Sherman, M.; Scott, D.R.; Rush, B.B.; Glass, A.G.; Schiffman, M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. Natl. Cancer Inst. 2005, 97, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- ASCUS-LSIL Triage Study (ALTS) Group. Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am. J. Obstet. Gynecol. 2003, 188, 1383–1392. [Google Scholar] [CrossRef]
- Ronco, G.; Giorgi Rossi, P. Role of HPV DNA testing in modern gynaecological practice. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 47, 107–118. [Google Scholar] [CrossRef]
- Koliopoulos, G.; Nyaga, V.N.; Santesso, N.; Bryant, A.; Martin-Hirsch, P.P.; Mustafa, R.A.; Schünemann, H.; Paraskevaidis, E.; Arbyn, M. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 2017, 8, CD008587. [Google Scholar] [CrossRef] [PubMed]
- Chrysostomou, A.C.; Stylianou, D.C.; Constantinidou, A.; Kostrikis, L.G. Cervical cancer screening programs in Europe: The transition towards HPV vaccination and population-based HPV testing. Viruses 2018, 10, 729. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.T.; Castle, P.E.; Behrens, C.M.; Sharma, A.; Wright, T.C.; Cuzick, J. Athena HPV study group comparison of cervical cancer screening strategies incorporating different combinations of cytology, HPV testing, and genotyping for HPV 16/18: Results from the ATHENA HPV study. Am. J. Obstet. Gynecol. 2013, 208, 184.e1–184.e11. [Google Scholar] [CrossRef]
- Wright, T.C.; Stoler, M.H.; Sharma, A.; Zhang, G.; Behrens, C.; Wright, T.L. ATHENA (Addressing THE Need for Advanced HPV Diagnostics) study group evaluation of HPV-16 and HPV-18 genotyping for the triage of women with high-risk HPV+ cytology-negative results. Am. J. Clin. Pathol. 2011, 136, 578–586. [Google Scholar] [CrossRef]
- Wentzensen, N.; Arbyn, M.; Berkhof, J.; Bower, M.; Canfell, K.; Einstein, M.; Farley, C.; Monsonego, J.; Franceschi, S. Eurogin 2016 Roadmap: How HPV knowledge is changing screening practice. Int. J. Cancer 2017, 140, 2192–2200. [Google Scholar] [CrossRef]
- Schiffman, M.; Burk, R.D.; Boyle, S.; Raine-Bennett, T.; Katki, H.A.; Gage, J.C.; Wentzensen, N.; Kornegay, J.R.; Aldrich, C.; Tam, T.; et al. A study of genotyping for management of human papillomavirus-positive, cytology-negative cervical screening results. J. Clin. Microbiol. 2015, 53, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Stoler, M.H.; Baker, E.; Boyle, S.; Aslam, S.; Ridder, R.; Huh, W.K.; Wright, T.C. Approaches to triage optimization in HPV primary screening: Extended genotyping and p16/Ki-67 dual-stained cytology-retrospective insights from ATHENA. Int. J. Cancer 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhoef, V.M.J.; Bosgraaf, R.P.; van Kemenade, F.J.; Rozendaal, L.; Heideman, D.A.M.; Hesselink, A.T.; Bekkers, R.L.M.; Steenbergen, R.D.M.; Massuger, L.F.A.G.; Melchers, W.J.G.; et al. Triage by methylation-marker testing versus cytology in women who test HPV-positive on self-collected cervicovaginal specimens (PROHTECT-3): A randomised controlled non-inferiority trial. Lancet Oncol. 2014, 15, 315–322. [Google Scholar] [CrossRef]
- Kelly, H.; Benavente, Y.; Pavon, M.A.; De Sanjose, S.; Mayaud, P.; Lorincz, A.T. Performance of DNA methylation assays for detection of high-grade cervical intraepithelial neoplasia (CIN2+): A systematic review and meta-analysis. Br. J. Cancer 2019, 121, 954–965. [Google Scholar] [CrossRef]
- Monnier-Benoit, S.; Dalstein, V.; Riethmuller, D.; Lalaoui, N.; Mougin, C.; Prétet, J.L. Dynamics of HPV16 DNA load reflect the natural history of cervical HPV-associated lesions. J. Clin. Virol. 2006, 35, 270–277. [Google Scholar] [CrossRef]
- van Duin, M.; Snijders, P.J.F.; Schrijnemakers, H.F.J.; Voorhorst, F.J.; Rozendaal, L.; Nobbenhuis, M.A.E.; van den Brule, A.J.C.; Verheijen, R.H.M.; Helmerhorst, T.J.; Meijer, C.J.L.M. Human papillomavirus 16 load in normal and abnormal cervical scrapes: An indicator of CIN II/III and viral clearance. Int. J. Cancer 2002, 98, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Moberg, M.; Gustavsson, I.; Wilander, E.; Gyllensten, U. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br. J. Cancer 2005, 92, 891–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalstein, V.; Riethmuller, D.; Sautière, J.L.; Prétet, J.L.; Kantelip, B.; Schaal, J.P.; Mougin, C. Detection of cervical precancer and cancer in a hospital population; Benefits of testing for human papillomavirus. Eur. J. Cancer Oxf. Engl. 1990 2004, 40, 1225–1232. [Google Scholar] [CrossRef]
- Jacquin, E.; Saunier, M.; Mauny, F.; Schwarz, E.; Mougin, C.; Prétet, J.-L. Real-time duplex PCR for simultaneous HPV 16 and HPV 18 DNA quantitation. J. Virol. Methods 2013, 193, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Hughes, J.P.; Castle, P.E.; Edelstein, Z.R.; Wang, C.; Galloway, D.A.; Koutsky, L.A.; Kiviat, N.B.; Schiffman, M. Viral load in the natural history of human papillomavirus type 16 infection: A nested case-control study. J. Infect. Dis. 2011, 203, 1425–1433. [Google Scholar] [CrossRef] [Green Version]
- Carcopino, X.; Henry, M.; Mancini, J.; Giusiano, S.; Boubli, L.; Olive, D.; Tamalet, C. Significance of HPV 16 and 18 viral load quantitation in women referred for colposcopy. J. Med. Virol. 2012, 84, 306–313. [Google Scholar] [CrossRef]
- Carcopino, X.; Henry, M.; Mancini, J.; Giusiano, S.; Boubli, L.; Olive, D.; Tamalet, C. Two years outcome of women infected with high risk HPV having normal colposcopy following low-grade or equivocal cytological abnormalities: Are HPV16 and 18 viral load clinically useful predictive markers? J. Med. Virol. 2012, 84, 964–972. [Google Scholar] [CrossRef]
- Josefsson, A.M.; Magnusson, P.K.; Ylitalo, N.; Sørensen, P.; Qwarforth-Tubbin, P.; Andersen, P.K.; Melbye, M.; Adami, H.O.; Gyllensten, U.B. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: A nested case-control study. Lancet Lond. Engl. 2000, 355, 2189–2193. [Google Scholar] [CrossRef]
- Depuydt, C.E.; Thys, S.; Beert, J.; Jonckheere, J.; Salembier, G.; Bogers, J.J. Linear viral load increase of a single HPV-type in women with multiple HPV infections predicts progression to cervical cancer. Int. J. Cancer 2016, 139, 2021–2032. [Google Scholar] [CrossRef]
- Depuydt, C.E.; Criel, A.M.; Benoy, I.H.; Arbyn, M.; Vereecken, A.J.; Bogers, J.J. Changes in type-specific human papillomavirus load predict progression to cervical cancer. J. Cell. Mol. Med. 2012, 16, 3096–3104. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, N.F.; Platt, R.W.; Duarte-Franco, E.; Costa, M.C.; Sobrinho, J.P.; Prado, J.C.M.; Ferenczy, A.; Rohan, T.E.; Villa, L.L.; Franco, E.L. Human papillomavirus infection and time to progression and regression of cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 2003, 95, 1336–1343. [Google Scholar] [CrossRef] [Green Version]
- Moscicki, A.B.; Hills, N.; Shiboski, S.; Powell, K.; Jay, N.; Hanson, E.; Miller, S.; Clayton, L.; Farhat, S.; Broering, J.; et al. Risks for incident human papillomavirus infection and low-grade squamous intraepithelial lesion development in young females. JAMA 2001, 285, 2995–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gravitt, P.E.; Burk, R.D.; Lorincz, A.; Herrero, R.; Hildesheim, A.; Sherman, M.E.; Bratti, M.C.; Rodriguez, A.C.; Helzlsouer, K.J.; Schiffman, M. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol. Biomark. Prev. Publ. 2003, 12, 477–484. [Google Scholar]
- Origoni, M.; Carminati, G.; Rolla, S.; Clementi, M.; Sideri, M.; Sandri, M.T.; Candiani, M. Human papillomavirus viral load expressed as relative light units (RLU) correlates with the presence and grade of preneoplastic lesions of the uterine cervix in atypical squamous cells of undetermined significance (ASCUS) cytology. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2401–2406. [Google Scholar] [CrossRef]
- Luo, H.; Du, H.; Belinson, J.L.; Wu, R. Evaluation of alternately combining HPV viral load and 16/18 genotyping in secondary screening algorithms. PLoS ONE 2019, 14, e0220200. [Google Scholar] [CrossRef]
- Heard, I.; Tondeur, L.; Arowas, L.; Falguières, M.; Demazoin, M.-C.; Favre, M. Human papillomavirus types distribution in organised cervical cancer screening in France. PLoS ONE 2013, 8, e79372. [Google Scholar] [CrossRef]
- Lagos, M.; Van De Wyngard, V.; Poggi, H.; Cook, P.; Viviani, P.; Barriga, M.I.; Pruyas, M.; Ferreccio, C. HPV16/18 genotyping for the triage of HPV positive women in primary cervical cancer screening in Chile. Infect. Agent. Cancer 2015, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- Cuzick, J.; Wheeler, C. Need for expanded HPV genotyping for cervical screening. Papillomavirus Res. Amst. Neth. 2016, 2, 112–115. [Google Scholar] [CrossRef] [Green Version]
- Poljak, M.; Oštrbenk Valenčak, A.; Gimpelj Domjanič, G.; Xu, L.; Arbyn, M. Commercially available molecular tests for human papillomaviruses: A global overview. Clin. Microbiol. Infect. Off. 2020. [Google Scholar] [CrossRef]

| Characteristics | Study Population (n = 885) n (%) |
|---|---|
| Age (year) | |
| Mean ± SD | 37.4 ± 11.7 |
| Median (q1–q3) | 34.5 (28.5–44.5) |
| Parity | |
| 0 | 260 (35.0) |
| 1 | 191 (25.7) |
| 2–3 | 260 (35.0) |
| >3 | 33 (4.4) |
| NA | 141 |
| Contraception | |
| Oral contraception | 381 (47.7) |
| Local contraception | 138 (17.3) |
| Intra-uterine device | 155 (19.4) |
| Menopause | 124 (15.5) |
| NA | 87 |
| Immunocompromised | |
| Transplantation | 21 (15.8) |
| HIV infection | 34 (25.6) |
| Haemopathy and cancer | 33 (24.8) |
| Auto-immune disease | 44 (33.1) |
| Chronic renal failure | 1 (0.7) |
| NA | 752 |
| Tobacco smoking history | |
| Current smoker | 284 (32.1) |
| Never / Ex-smoker | 601 (67.9) |
| Sexually Transmitted Infection | |
| HIV * | 34 (47.2) |
| Chlamydia | 24 (33.33) |
| HSV2 | 16 (22.2) |
| Syphilis | 2 (2.8) |
| HBV | 2 (2.8) |
| NA | 813 |
| Variables | Study Population (n = 885) n (%) |
|---|---|
| Cytology | |
| Unsatisfactory | 66 (7.5) |
| NILM | 621 (70.3) |
| ASC-US | 51 (5.7) |
| LSIL | 146 (16.5) |
| NA | 1 |
| Histology | |
| Normal | 45 (5.9) |
| CIN1 | 50 (5.7) |
| NA | 790 (89.3) |
| hc2 | |
| Mean RLU/PC (pg/mL) ± SD | 326 ± 597 |
| Median RLU/PC (pg/mL) (q1–q3) | 53 (8–319) |
| hc2 RLU/PC distribution | |
| 1–10 pg/mL | 262 (29.6) |
| 11–100 pg/mL | 267 (30.2) |
| >100 pg/mL | 356 (40.2) |
| HPV16 | |
| Negative | 660 (74.9) |
| Positive* | 221 (25.9) |
| Mean (Log10GE/103 cells) ± SD | 3.1 ± 1.3 |
| Median (q1–q3) (Log10GE/103 cells) | 3.2 (2.1–4) |
| Mean (Log10GE/mL) ± SD | 7.1 ± 7.9 |
| Median (q1–q3) (Log10GE/mL) | 5.8 (5–6.6) |
| NA | |
| HPV18 | |
| Negative | 789 (92) |
| Positive * | 69 (8.4) |
| Mean (Log10 GE/103 cells) ± SD | 3.5 ± 1.5 |
| Median (q1–q3) (Log10GE/ 103 cells) | 3.6 (2.7–4.6) |
| Mean (Log10GE/ mL) ± SD | 6.2 ± 1.4 |
| Median (q1–q3) (Log10GE/mL) | 6.2 (4.3–7.1) |
| NA | 27 |
| Variables | n (Events) a | HR | 95% CI | p-Value |
|---|---|---|---|---|
| Parity | ||||
| 0 | 260 (44) | 1 | - | 0.44 |
| 1 | 191 (32) | 0.87 | 0.55–1.37 | - |
| 2–3 | 258 (45) | 0.86 | 0.63–1.45 | - |
| >3 | 33 (2) | 0.32 | 0.08–1.33 | - |
| Contraception | ||||
| Yes b | 536 (95) | 1 | - | 0.0223 |
| No c | 262 (30) | 0.62 | 0.41–0.93 | - |
| Immunodepression | ||||
| No | 737 (112) | 1 | - | 0.75 |
| Yes | 145 (23) | 0.93 | 0.59–1.46 | - |
| Tobacco smoking | ||||
| No | 598 (85) | 1 | - | 0.0582 |
| Yes | 284 (50) | 1.4 | 0.99–1.99 | - |
| Cytology | ||||
| Normal | 621 (76) | 1 | - | <0.0001 |
| Abnormal | 263 (58) | 2.16 | 1.54–3.05 | - |
| Hc2 | ||||
| 1–100 pg/mL | 529 (61) | 1 | <0.0001 | |
| >100 pg/mL | 356 (74) | 1.98 | 1.41–2.78 | - |
| HPV16 | ||||
| Absence | 660 (71) | 1 | - | <0.0001 |
| Presence | 221 (62) | 2.39 | 1.70–3.37 | - |
| HPV16 load | ||||
| Absence and Log10 HPV16 GE/103 cells < 3.2 | 780 (91) | 1 | <0.0001 | |
| Log10 HPV16 GE/103 cells ≥ 3.2 | 101 (42) | 3.09 | 2.14–4.48 | - |
| HPV18 | ||||
| Absence | 789 (114) | 1 | - | 0.83 |
| Presence | 69 (10) | 1.7 | 0.56–2.05 | - |
| Log10 HPV18 GE/103 cells | ||||
| <5.2 | 59 (7) | 1 | - | 0.0968 |
| ≥5.2 | 10 (3) | 3.19 | 0.81–12.51 | - |
| Variables | n (Events) | HR | 95% CI | p-Value | Weight |
|---|---|---|---|---|---|
| - | 880 (132) | - | - | - | - |
| Hc2 | |||||
| 1–100 pg/mL | 527 (60) | 1 | - | 0.05 | 0 |
| >100 pg/mL | 353 (72) | 1.44 | 1.00–2.08 | - | 1 |
| Cytology | |||||
| Normal | 618 (75) | 1 | - | <0.0001 | 0 |
| Abnormal | 262 (57) | 2.02 | 1.43–2.87 | - | 2 |
| HPV16 | |||||
| Absence and Log10 HPV16 GE/103 cells < 3.2 | 780 (91) | 1 | - | <0.0001 | 0 |
| Log10 HPV16 GE/103 cells ≥ 3.2 | 100 (41) | 2.67 | 1.80–3.97 | - | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baumann, A.; Henriques, J.; Selmani, Z.; Meurisse, A.; Lepiller, Q.; Vernerey, D.; Valmary-Degano, S.; Paget-Bailly, S.; Riethmuller, D.; Ramanah, R.; et al. HPV16 Load Is a Potential Biomarker to Predict Risk of High-Grade Cervical Lesions in High-Risk HPV-Infected Women: A Large Longitudinal French Hospital-Based Cohort Study. Cancers 2021, 13, 4149. https://doi.org/10.3390/cancers13164149
Baumann A, Henriques J, Selmani Z, Meurisse A, Lepiller Q, Vernerey D, Valmary-Degano S, Paget-Bailly S, Riethmuller D, Ramanah R, et al. HPV16 Load Is a Potential Biomarker to Predict Risk of High-Grade Cervical Lesions in High-Risk HPV-Infected Women: A Large Longitudinal French Hospital-Based Cohort Study. Cancers. 2021; 13(16):4149. https://doi.org/10.3390/cancers13164149
Chicago/Turabian StyleBaumann, Antoine, Julie Henriques, Zohair Selmani, Aurélia Meurisse, Quentin Lepiller, Dewi Vernerey, Séverine Valmary-Degano, Sophie Paget-Bailly, Didier Riethmuller, Rajeev Ramanah, and et al. 2021. "HPV16 Load Is a Potential Biomarker to Predict Risk of High-Grade Cervical Lesions in High-Risk HPV-Infected Women: A Large Longitudinal French Hospital-Based Cohort Study" Cancers 13, no. 16: 4149. https://doi.org/10.3390/cancers13164149
APA StyleBaumann, A., Henriques, J., Selmani, Z., Meurisse, A., Lepiller, Q., Vernerey, D., Valmary-Degano, S., Paget-Bailly, S., Riethmuller, D., Ramanah, R., Mougin, C., & Prétet, J.-L. (2021). HPV16 Load Is a Potential Biomarker to Predict Risk of High-Grade Cervical Lesions in High-Risk HPV-Infected Women: A Large Longitudinal French Hospital-Based Cohort Study. Cancers, 13(16), 4149. https://doi.org/10.3390/cancers13164149






