RAD51D Aberrant Splicing in Breast Cancer: Identification of Splicing Regulatory Elements and Minigene-Based Evaluation of 53 DNA Variants
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Variant and Transcript Annotations
2.2. Bioinformatics
2.3. Minigene Construction and Site-Directed Mutagenesis
2.4. Functional Assays
2.5. ACMG/AMP-Like Classification of 37 RAD51D Variants Detected in BRIDGES Samples
3. Results
3.1. Bioinformatics Analysis and Functional Assays
3.2. Transcript Analysis
3.3. ESE Mapping
3.4. ACMG/AMP-Like Classification of 37 RAD51D Variants Identified in the BRIDGES Cohort
4. Discussion
4.1. Splice-Site Variants
4.2. SRE-Spliceogenic Variants
4.3. ACMG/AMP-Based Classification of Spliceogenic Variants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sullivan, M.R.; Bernstein, K.A. RAD-ical New Insights into RAD51 Regulation. Genes 2018, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Yang, X.; Song, H.; Leslie, G.; Engel, C.; Hahnen, E.; Auber, B.; Horváth, J.; Kast, K.; Niederacher, D.; Turnbull, C.; et al. Ovarian and breast cancer risks associated with pathogenic variants in RAD51C and RAD51D. J. Natl. Cancer Inst. 2020, 37, 1–9. [Google Scholar] [CrossRef]
- Loveday, C.; Turnbull, C.; Ramsay, E.; Hughes, D.; Ruark, E.; Frankum, J.R.; Bowden, G.; Kalmyrzaev, B.; Warren-Perry, M.; Snape, K.; et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat. Genet. 2011, 43, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Dorling, L.; Carvalho, S.; Allen, J.; González-Neira, A.; Luccarini, C.; Wahlström, C.; Pooley, K.A.; Parsons, M.T.; Fortuno, C.; Wang, Q.; et al. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar]
- Hu, C.; Hart, S.N.; Gnanaolivu, R.; Huang, H.; Lee, K.Y.; Na, J.; Gao, C.; Lilyquist, J.; Yadav, S.; Boddicker, N.J.; et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021, 384, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Manning, K.S.; Cooper, T.A. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017, 18, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef]
- Lopez-Bigas, N.; Audit, B.; Ouzounis, C.; Parra, G.; Guigo, R.; López-Bigas, N.; Audit, B.; Ouzounis, C.; Parra, G.; Guigó, R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005, 579, 1900–1903. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, C.; De Leeneer, K.; Esteban-Cardeñosa, E.M.; Avila Cobos, F.; Lastra, E.; Abella, L.E.; de la Cruz, V.; Lobatón, C.D.; Claes, K.B.; Durán, M.; et al. Germline Genetic Findings Which May Impact Therapeutic Decisions in Families with a Presumed Predisposition for Hereditary Breast and Ovarian Cancer. Cancers 2020, 12, 2151. [Google Scholar] [CrossRef]
- Gelli, E.; Colombo, M.; Pinto, A.; De Vecchi, G.; Foglia, C.; Amitrano, S.; Morbidoni, V.; Imperatore, V.; Manoukian, S.; Baldassarri, M.; et al. Usefulness and Limitations of Comprehensive Characterization of mRNA Splicing Profiles in the Definition of the Clinical Relevance of BRCA1/2 Variants of Uncertain Significance. Cancers 2019, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Morbidoni, V.; Baschiera, E.; Forzan, M.; Fumini, V.; Ali, D.S.; Giorgi, G.; Buson, L.; Desbats, M.A.; Cassina, M.; Clementi, M.; et al. Hybrid Minigene Assay: An Efficient Tool to Characterize mRNA Splicing Profiles of NF1 Variants. Cancers 2021, 13, 999. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, C.; Krieger, S.; Vezain, M.; Rousselin, A.; Tournier, I.; Martins, A.; Berthet, P.; Chevrier, A.; Dugast, C.; Layet, V.; et al. Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J. Med. Genet. 2008, 45, 438–446. [Google Scholar] [CrossRef]
- Whiley, P.J.; de la Hoya, M.; Thomassen, M.; Becker, A.; Brandão, R.; Pedersen, I.S.; Montagna, M.; Menéndez, M.; Quiles, F.; Gutiérrez-Enríquez, S.; et al. Comparison of mRNA splicing assay protocols across multiple laboratories: Recommendations for best practice in standardized clinical testing. Clin. Chem. 2014, 60, 341–352. [Google Scholar] [CrossRef]
- Caputo, S.M.; Léone, M.; Damiola, F.; Ehlen, A.; Carreira, A.; Gaidrat, P.; Martins, A.; Brandão, R.D.; Peixoto, A.; Vega, A.; et al. Full in-frame exon 3 skipping of BRCA2 confers high risk of breast and/or ovarian cancer. Oncotarget 2018, 9, 17334–17348. [Google Scholar] [CrossRef]
- Sanoguera-Miralles, L.; Valenzuela-Palomo, A.; Bueno-Martínez, E.; Llovet, P.; Díez-Gómez, B.; Caloca, M.J.; Pérez-Segura, P.; Fraile-Bethencourt, E.; Colmena, M.; Carvalho, S.; et al. Comprehensive Functional Characterization and Clinical Interpretation of 20 Splice-Site Variants of the RAD51C Gene. Cancers 2020, 12, 3771. [Google Scholar] [CrossRef]
- Lopez-Perolio, I.; Leman, R.; Behar, R.; Lattimore, V.; Pearson, J.F.; Castéra, L.; Martins, A.; Vaur, D.; Goardon, N.; Davy, G.; et al. Alternative splicing and ACMG-AMP-2015-based classification of PALB2 genetic variants: An ENIGMA report. J. Med. Genet. 2019, 56, 453–460. [Google Scholar] [CrossRef]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Houdayer, C.; Caux-Moncoutier, V.; Krieger, S.; Barrois, M.; Bonnet, F.; Bourdon, V.; Bronner, M.; Buisson, M.; Coulet, F.; Gaildrat, P.; et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum. Mutat. 2012, 33, 1228–1238. [Google Scholar] [CrossRef]
- Erkelenz, S.; Theiss, S.; Otte, M.; Widera, M.; Peter, J.O.; Schaal, H. Genomic HEXploring allows landscaping of novel potential splicing regulatory elements. Nucleic Acids Res. 2014, 42, 10681–10697. [Google Scholar] [CrossRef] [PubMed]
- Raponi, M.; Kralovicova, J.; Copson, E.; Divina, P.; Eccles, D.; Johnson, P.; Baralle, D.; Vorechovsky, I. Prediction of single-nucleotide substitutions that result in exon skipping: Identification of a splicing silencer in BRCA1 exon 6. Hum. Mutat. 2011, 32, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Shang, S.; Kalachikov, S.M.; Morozova, I.; Yu, L.; Russo, J.J.; Ju, J.; Chasin, L.A. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res. 2011, 21, 1360–1374. [Google Scholar] [CrossRef] [PubMed]
- Fraile-Bethencourt, E.; Díez-Gómez, B.; Velásquez-Zapata, V.; Acedo, A.; Sanz, D.J.; Velasco, E.A. Functional classification of DNA variants by hybrid minigenes: Identification of 30 spliceogenic variants of BRCA2 exons 17 and 18. PLoS Genet. 2017, 13, e1006691. [Google Scholar] [CrossRef] [PubMed]
- Acedo, A.; Hernández-Moro, C.; Curiel-García, Á.; Díez-Gómez, B.; Velasco, E.A. Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons. Hum. Mutat. 2015, 36, 210–221. [Google Scholar] [CrossRef]
- De Garibay, G.R.; Acedo, A.; García-Casado, Z.; Gutiérrez-Enríquez, S.; Tosar, A.; Romero, A.; Garre, P.; Llort, G.; Thomassen, M.; Díez, O.; et al. Capillary electrophoresis analysis of conventional splicing assays: IARC analytical and clinical classification of 31 BRCA2 genetic variants. Hum. Mutat. 2014, 35, 53–57. [Google Scholar] [CrossRef]
- Acedo, A.; Sanz, D.J.; Durán, M.; Infante, M.; Pérez-Cabornero, L.; Miner, C.; Velasco, E.A. Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes. Breast Cancer Res. 2012, 14, R87. [Google Scholar] [CrossRef]
- Montalban, G.; Bonache, S.; Moles-Fernández, A.; Gadea, N.; Tenés, A.; Torres-Esquius, S.; Carrasco, E.; Balmaña, J.; Diez, O.; Gutiérrez-Enríquez, S. Incorporation of semi-quantitative analysis of splicing alterations for the clinical interpretation of variants in BRCA1 and BRCA2 genes. Hum. Mutat. 2019, 40, 2296–2317. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Abou Tayoun, A.N.; Pesaran, T.; DiStefano, M.T.; Oza, A.; Rehm, H.L.; Biesecker, L.G.; Harrison, S.M.; ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI). Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018, 39, 1517–1524. [Google Scholar] [CrossRef]
- Brnich, S.E.; Abou Tayoun, A.N.; Couch, F.J.; Cutting, G.R.; Greenblatt, M.S.; Heinen, C.D.; Kanavy, D.M.; Luo, X.; McNulty, S.M.; Starita, L.M.; et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2020, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Krempely, K.; Roberts, M.E.; Anderson, M.J.; Carneiro, F.; Chao, E.; Dixon, K.; Figueiredo, J.; Ghosh, R.; Huntsman, D.; et al. Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline CDH1 sequence variants. Hum. Mutat. 2018, 39, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Brandão, R.D.; Mensaert, K.; López-Perolio, I.; Tserpelis, D.; Xenakis, M.; Lattimore, V.; Walker, L.C.; Kvist, A.; Vega, A.; Gutiérrez-Enríquez, S.; et al. Targeted RNA-seq successfully identifies normal and pathogenic splicing events in breast/ovarian cancer susceptibility and Lynch syndrome genes. Int. J. Cancer 2019, 145, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Fairbrother, W.G.; Holste, D.; Burge, C.B.; Sharp, P.A. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004, 2, E268. [Google Scholar] [CrossRef] [PubMed]
- Keren, H.; Lev-Maor, G.; Ast, G. Alternative splicing and evolution: Diversification, exon definition and function. Nat. Rev. Genet. 2010, 11, 345–355. [Google Scholar] [CrossRef]
- Canson, D.; Glubb, D.; Spurdle, A.B. Variant effect on splicing regulatory elements, branchpoint usage, and pseudoexonization: Strategies to enhance bioinformatic prediction using hereditary cancer genes as exemplars. Hum. Mutat. 2020, 41, 1705–1721. [Google Scholar] [CrossRef]
- Soukarieh, O.; Gaildrat, P.; Hamieh, M.; Drouet, A.; Baert-Desurmont, S.; Frébourg, T.; Tosi, M.; Martins, A. Exonic Splicing Mutations Are More Prevalent than Currently Estimated and Can Be Predicted by Using In Silico Tools. PLoS Genet. 2016, 12, e1005756. [Google Scholar]
- Konstanta, I.; Fostira, F.; Apostolou, P.; Stratikos, E.; Kalfakakou, D.; Pampanos, A.; Kollia, P.; Papadimitriou, C.; Konstantopoulou, I.; Yannoukakos, D. Contribution of RAD51D germline mutations in breast and ovarian cancer in Greece. J. Hum. Genet. 2018, 63, 1149–1158. [Google Scholar] [CrossRef] [PubMed]
- Van Marcke, C.; Collard, A.; Vikkula, M.; Duhoux, F.P. Prevalence of pathogenic variants and variants of unknown significance in patients at high risk of breast cancer: A systematic review and meta-analysis of gene-panel data. Crit. Rev. Oncol. Hematol. 2018, 132, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Rhine, C.L.; Neil, C.; Glidden, D.T.; Cygan, K.J.; Fredericks, A.M.; Wang, J.; Walton, N.A.; Fairbrother, W.G. Future directions for high-throughput splicing assays in precision medicine. Hum. Mutat. 2019, 40, 1225–1234. [Google Scholar] [CrossRef]
- Alenezi, W.M.; Fierheller, C.T.; Recio, N.; Tonin, P.N. Literature review of BARD1 as a cancer predisposing gene with a focus on breast and ovarian cancers. Genes 2020, 11, 856. [Google Scholar] [CrossRef]
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016, 2, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Pavanello, M.; Chan, I.H.Y.; Ariff, A.; Pharoah, P.D.P.; Gayther, S.A.; Ramus, S.J. Rare germline genetic variants and the risks of epithelial ovarian cancer. Cancers 2020, 12, 3046. [Google Scholar] [CrossRef] [PubMed]
- Kyo, S.; Ishikawa, N.; Nakamura, K.; Nakayama, K. The fallopian tube as origin of ovarian cancer: Change of diagnostic and preventive strategies. Cancer Med. 2020, 9, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Goren, A.; Ast, G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008, 24, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Sanz, D.J.; Acedo, A.; Infante, M.; Durán, M.; Pérez-Cabornero, L.; Esteban-Cardeñosa, E.; Lastra, E.; Pagani, F.; Miner, C.; Velasco, E.A. A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin. Cancer Res. 2010, 16, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Fraile-Bethencourt, E.; Valenzuela-Palomo, A.; Díez-Gómez, B.; Acedo, A.; Velasco, E.A. Identification of Eight Spliceogenic Variants in BRCA2 Exon 16 by Minigene Assays. Front. Genet. 2018, 9, 188. [Google Scholar] [CrossRef]
- Fraile-Bethencourt, E.; Valenzuela-Palomo, A.; Díez-Gómez, B.; Goina, E.; Acedo, A.; Buratti, E.; Velasco, E.A. Mis-splicing in breast cancer: Identification of pathogenic BRCA2 variants by systematic minigene assays. J. Pathol. 2019, 248, 409–420. [Google Scholar] [CrossRef]
- Fraile-Bethencourt, E.; Valenzuela-Palomo, A.; Díez-Gómez, B.; Caloca, M.J.; Gómez-Barrero, S.; Velasco, E.A. Minigene Splicing Assays Identify 12 Spliceogenic Variants of BRCA2 Exons 14 and 15. Front. Genet. 2019, 10, 503. [Google Scholar] [CrossRef]
- Reid, S.; Schindler, D.; Hanenberg, H.; Barker, K.; Hanks, S.; Kalb, R.; Neveling, K.; Kelly, P.; Seal, S.; Freund, M.; et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 2007, 39, 162–164. [Google Scholar] [CrossRef]
- Montalban, G.; Fraile-Bethencourt, E.; López-Perolio, I.; Pérez-Segura, P.; Infante, M.; Durán, M.; Alonso-Cerezo, M.C.; López-Fernández, A.; Diez, O.; de la Hoya, M.; et al. Characterization of spliceogenic variants located in regions linked to high levels of alternative splicing: BRCA2 c.7976+5G>T as a case study. Hum. Mutat. 2018, 39, 1155–1160. [Google Scholar] [CrossRef]
- Landrith, T.; Li, B.; Cass, A.A.; Conner, B.R.; LaDuca, H.; McKenna, D.B.; Maxwell, K.N.; Domchek, S.; Morman, N.A.; Heinlen, C.; et al. Splicing profile by capture RNA-seq identifies pathogenic germline variants in tumor suppressor genes. NPJ Precis. Oncol. 2020, 4, 4. [Google Scholar] [CrossRef]
- Cáceres, J.F.; Kornblihtt, A.R. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002, 18, 186–193. [Google Scholar] [CrossRef]
- Goina, E.; Skoko, N.; Pagani, F. Binding of DAZAP1 and hnRNPA1/A2 to an Exonic Splicing Silencer in a Natural BRCA1 Exon 18 Mutant. Mol. Cell. Biol. 2008, 28, 3850–3860. [Google Scholar] [CrossRef] [PubMed]
- Kralovicova, J.; Hwang, G.; Asplund, A.C.; Churbanov, A.; Smith, C.I.E.; Vorechovsky, I. Compensatory signals associated with the activation of human GC 5’ splice sites. Nucleic Acids Res. 2011, 39, 7077–7091. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Buratti, E.; Baralle, F.E. Functional properties and evolutionary splicing constraints on a composite exonic regulatory element of splicing in CFTR exon 12. Nucleic Acids Res. 2010, 38, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Pagani, F.; Stuani, C.; Tzetis, M.; Kanavakis, E.; Efthymiadou, A.; Doudounakis, S.; Casals, T.; Baralle, F.E. New type of disease causing mutations: The example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003, 12, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Dal Mas, A.; Fortugno, P.; Donadon, I.; Levati, L.; Castiglia, D.; Pagani, F. Exon-Specific U1s Correct SPINK 5 Exon 11 Skipping Caused by a Synonymous Substitution that Affects a Bifunctional Splicing Regulatory Element. Hum. Mutat. 2015, 36, 504–512. [Google Scholar] [CrossRef]
- Tubeuf, H.; Charbonnier, C.; Soukarieh, O.; Blavier, A.; Lefebvre, A.; Dauchel, H.; Frebourg, T.; Gaildrat, P.; Martins, A. Large-scale comparative evaluation of user-friendly tools for predicting variant-induced alterations of splicing regulatory elements. Hum. Mutat. 2020, 41, 1811–1829. [Google Scholar] [CrossRef]
- Dosil, V.; Tosar, A.; Cañadas, C.; Pérez-Segura, P.; Díaz-Rubio, E.; Caldés, T.; de la Hoya, M. Alternative splicing and molecular characterization of splice site variants: BRCA1 c.591C>T as a case study. Clin. Chem. 2010, 56, 53–61. [Google Scholar] [CrossRef]
- Kim, Y.M.; Choi, B.-S. Structural and functional characterization of the N-terminal domain of human Rad51D. Int. J. Biochem. Cell Biol. 2011, 43, 416–422. [Google Scholar] [CrossRef]
- Miller, K.A. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 2004, 32, 169–178. [Google Scholar] [CrossRef]
- Shin, D.S.; Pellegrini, L.; Daniels, D.; Yelent, B.; Craig, L.; Bates, D.; Yu, D.S.; Shivji, M.K.; Hitomi, C.; Arvai, A.S.; et al. Full-length archaeal Rad51 structure and mutants: Mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003, 22, 4566–4576. [Google Scholar] [CrossRef]
- Chang, Y.-W.; Ko, T.-P.; Lee, C.-D.; Chang, Y.-C.; Lin, K.-A.; Chang, C.-S.; Wang, A.H.-J.; Wang, T.-F. Three New Structures of Left-Handed RadA Helical Filaments: Structural Flexibility of N-Terminal Domain Is Critical for Recombinase Activity. PLoS ONE 2009, 4, e4890. [Google Scholar] [CrossRef]
- Tavtigian, S.V.; on behalf of the ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI); Greenblatt, M.S.; Harrison, S.M.; Nussbaum, R.L.; Prabhu, S.A.; Boucher, K.M.; Biesecker, L.G. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018, 20, 1054–1060. [Google Scholar] [CrossRef]
- Baldock, R.A.; Pressimone, C.A.; Baird, J.M.; Khodakov, A.; Luong, T.; Grundy, M.K.; Smith, C.M.; Karpenshif, Y.; Bratton-Palmer, D.S.; Prakash, R.; et al. RAD51D splice variants and cancer-associated mutations reveal XRCC2 interaction to be critical for homologous recombination. DNA Repair 2019, 76, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Gruver, A.M.; Miller, K.A.; Rajesh, C.; Smiraldo, P.G.; Kaliyaperumal, S.; Balder, R.; Stiles, K.M.; Albala, J.S.; Pittman, D.L. The ATPase motif in RAD51D is required for resistance to DNA interstrand crosslinking agents and interaction with RAD51C. Mutagenesis 2005, 20, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Wiese, C.; Hinz, J.M.; Tebbs, R.S.; Nham, P.B.; Urbin, S.S.; Collins, D.W.; Thompson, L.H.; Schild, D. Disparate requirements for the Walker A and B ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res. 2006, 34, 2833–2843. [Google Scholar] [CrossRef] [PubMed]
- Whiffin, N.; Minikel, E.; Walsh, R.; O’Donnell-Luria, A.H.; Karczewski, K.; Ing, A.Y.; Barton, P.J.R.; Funke, B.; A Cook, S.; MacArthur, D.; et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet. Med. 2017, 19, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Polley, E.C.; Yadav, S.; Lilyquist, J.; Shimelis, H.; Na, J.; Hart, S.N.; E Goldgar, D.; Shah, S.; Pesaran, T.; et al. The Contribution of Germline Predisposition Gene Mutations to Clinical Subtypes of Invasive Breast Cancer From a Clinical Genetic Testing Cohort. J. Natl. Cancer Inst. 2020, 112, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Lilyquist, J.; LaDuca, H.; Polley, E.; Davis, B.T.; Shimelis, H.; Hu, C.; Hart, S.N.; Dolinsky, J.S.; Couch, F.J.; Goldgar, D.E. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested on multi-gene panels compared to reference controls. Gynecol. Oncol. 2017, 147, 375–380. [Google Scholar] [CrossRef] [PubMed]
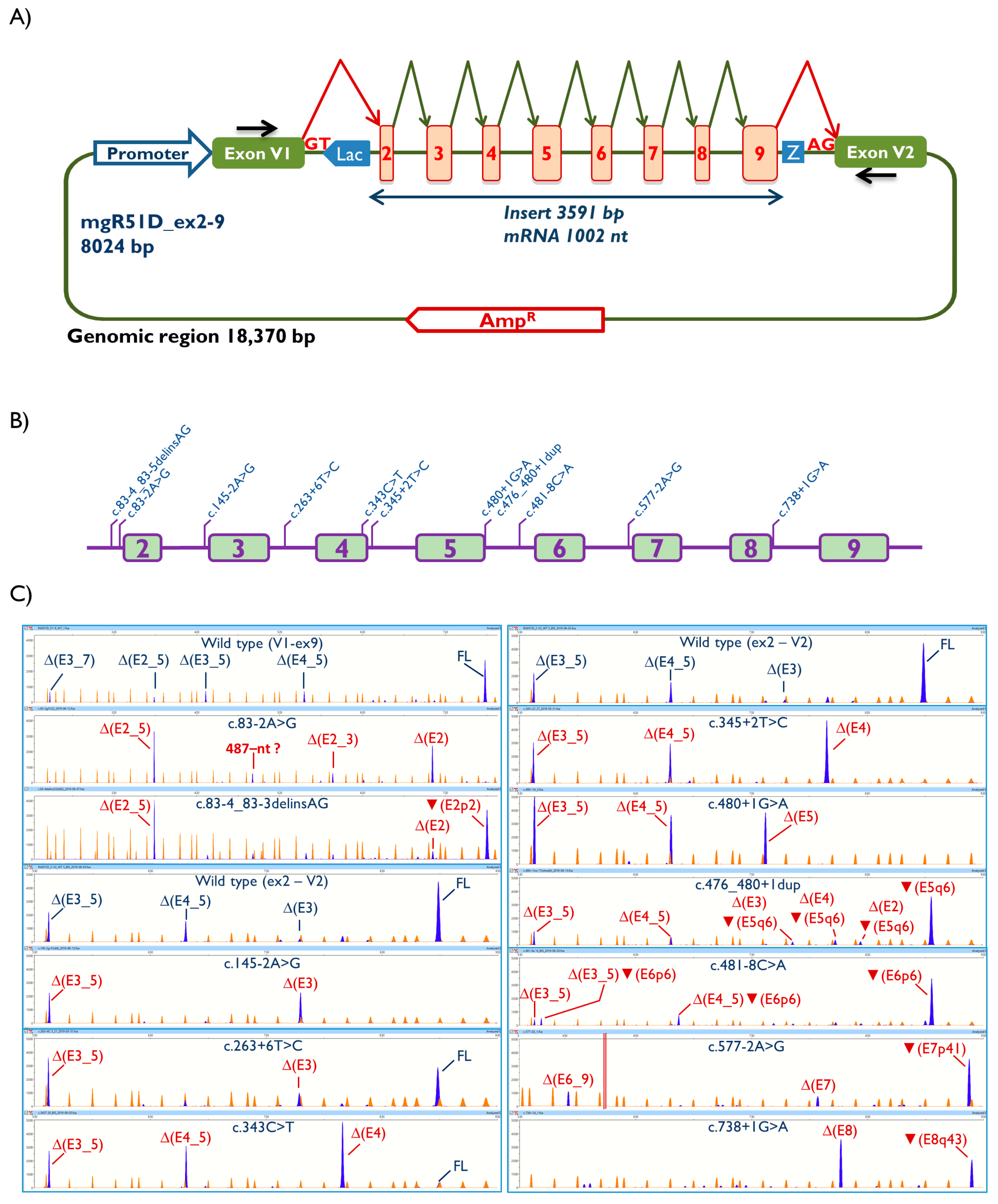
| Variant (HGVS) 1 | Bioinformatics (MaxEnt Scan) 2 | Transcripts 3 | |||
|---|---|---|---|---|---|
| Canonical | PTC | In-Frame | Uncharacterized | ||
| mgR51D_ex2-9 | Primers V1-ex9 | ||||
| Wild type | - | 57.8% ± 5.6 | ∆(E4_5) (13.8% ± 1.3); ∆(E3_7) (9.9% ± 2.6); ∆(E2_5) (5.2% ± 0.8) | ∆(E3_5) (13.3% ± 2.4) | - |
| c.83-2A>G | [−] 3′SS (8.52→0.56) | - | ∆(E2_5) (41.9% ± 1.1); ∆(E2) (39.6% ± 0.9); ∆(E2_3) (9.6% ± 0.1) | - | 487-nt (9.0% ± 0.2) |
| c.83-4_83-3delinsAG | [−] 3′SS [+] 3′SS (5.42) 2-nt upstream | - | ▼(E2p2) (54.2% ± 2.0); ∆(E2_5) (38.6% ± 2.1); ∆(E2) (7.2% ± 0.4) | - | - |
| mgR51D_ex2-9 | Primers ex2-V2 | - | - | ||
| Wild type | 73.1% ± 5.6 | ∆(E4_5) (9.4% ± 3) | ∆(E3_5) (17.5% ± 5.2) | - | |
| c.145-2A>G | [−] 3′SS (2.43) | - | ∆(E3) (55.5% ± 0.6) | ∆(E3_5) (44.5% ± 0.6) | - |
| c.263+6T>C | 5′SS: 7.44→4.86 | 49.0% ± 1.2 | ∆(E3) (15.6% ± 0.3) | ∆(E3_5) (35.4% ± 1.2) | - |
| c.343C>T | 5′SS: 7.79→4.36 | 3.4% ± 0.7 | ∆(E4) (45.7% ± 1.0); ∆(E4_5) (28.1% ± 0.6) | ∆(E3_5) (22.7% ± 1.2) | - |
| c.345+2T>C | [−] 5′SS (7.79→0.04) | - | ∆(E4) (49.6% ± 1.4); ∆(E4_5) (26.4% ± 1.0) | ∆(E3_5) (24.0% ± 0.7) | - |
| c.480+1G>A | [−] 5′SS (11.08→2.9) | - | ∆(E4_5) (29.2% ± 0.4) | ∆(E3_5) (41.2% ± 0.7); ∆(E5) (29.6% ± 0.4) | - |
| c.476_480+1dup | [−] 5′SS (11.08→0.5) | - | ∆(E4_5) (8.6% ± 0.4); (∆(E4) ▼(E5q6)) (6.3% ± 0.1); (∆(E2) ▼(E5q6)) (4.3% ± 0.1); (∆(E3) ▼(E5q6)) (3.6% ± 0.4) | ▼(E5q6) (60.4% ± 4.3); ∆(E3_5) (16.7% ± 5.0) | - |
| c.481-8C>A | 3′SS: 8.21→1.75 [+] 3′SS (11.06) 6-nt upstream | 0.4% ± 0.1 | (∆(E4_5) ▼(E6p6)) (13.4% ± 0.2) | ▼(E6p6) (70.0% ± 0.8); (∆(E3_5) ▼(E6p6)) (8.8% ± 0.3); ∆(E3_5) (7.5% ± 0.3) | - |
| c.577-2A>G | [−] 3′SS (10.36→2.41) | - | ▼(E7p41) (65.5% ± 0.5); ∆(E7) (14.3% ± 0.2) | ∆(E6_9) (20.2% ± 0.8) | - |
| c.738+1G>A 4 | [−] 5′SS (6.13→−2.05) | - | ▼(E8q43) (27.9% ± 0.1); ∆(E8) (51.1% ± 0.2); ∆(E4_7) (10.9% ± 0.1) | - | 1363-nt (10.1% ± 0.0) |
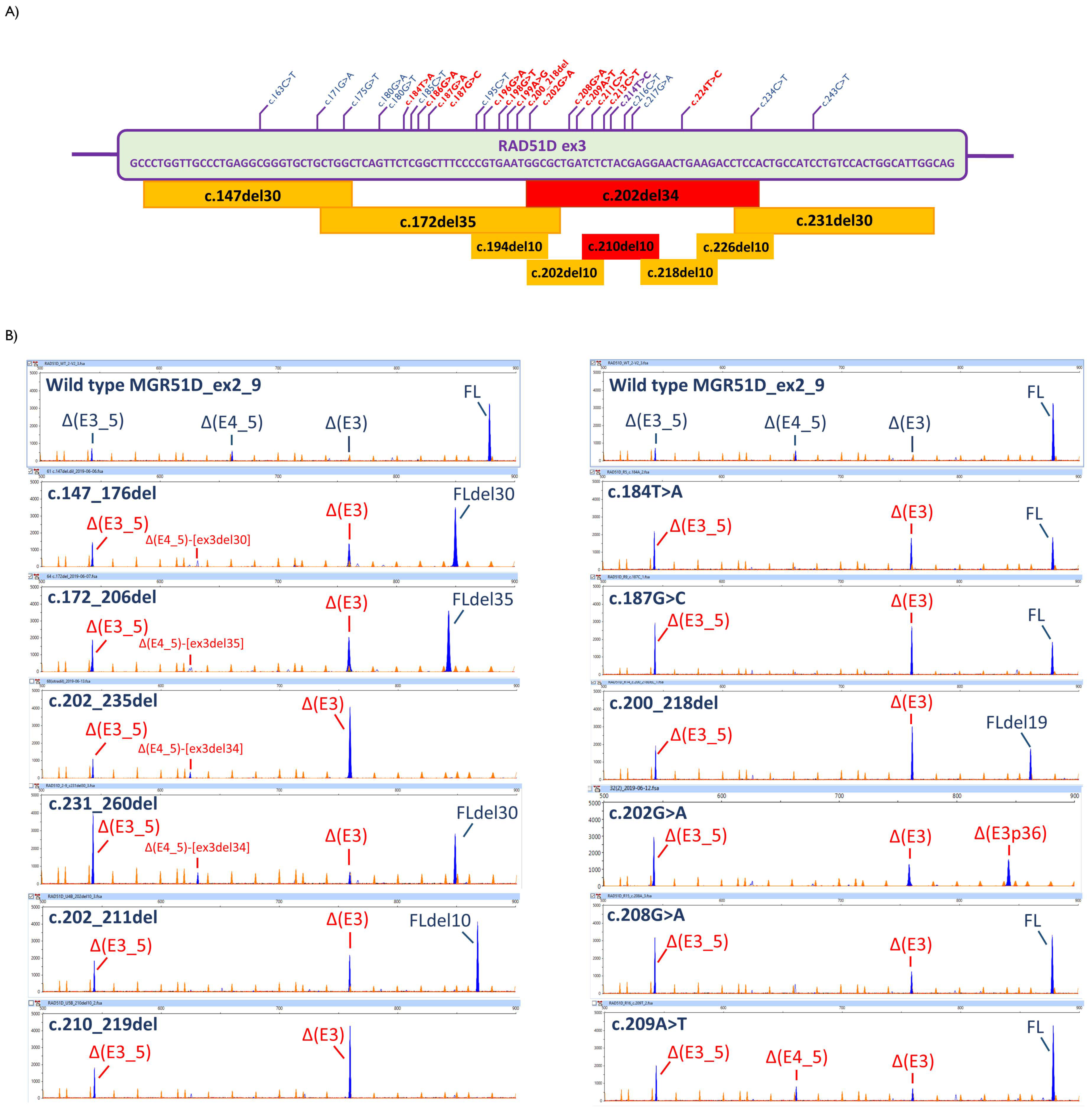
| Variant 1 | Transcripts 2 | In Silico Tools 3 | |||||
|---|---|---|---|---|---|---|---|
| Canonical | PTC | In-Frame | HSF | HEXplorer | Hot-Skip | ΔtESRseq | |
| Wild type | 73.1% ± 5.6 | ∆(E4_5) (9.4% ± 3) | ∆(E3_5) (17.5% ± 5.2) | - | |||
| BRIDGES variants | |||||||
| c.171G>A | 76.0% ± 1.4 | ∆(E4_5) (8.4% ± 0.4) | ∆(E3_5) (18.6% ± 1.1) | - | −40.3 | 0 | −1.03 |
| c.175G>T | 52.9% ± 2.6 | ∆(E4_5) (9.7% ± 1.1) ∆(E3) (9.6% ± 0.4) | ∆(E3_5) (27.7% ± 1.2) | - | −27.0 | 2 | 1.30 |
| c.180G>A | 67.5% ± 2.6 | ∆(E4_5) (14.4% ± 1.1) | ∆(E3_5) (18.1% ± 1.5) | - | 2.5 | 0 | 0.25 |
| c.180G>T | 69.4% ± 0.5 | ∆(E4_5) (13.7% ± 0.1) | ∆(E3_5) (16.9% ± 0.4) | - | −65.1 | 5 | 0.25 |
| c.184T>A | 37.2% ± 0.6 | ∆(E3) (31.1% ± 1.5) | ∆(E3_5) (31.7% ± 1.5) | + | 23.3 | 0 | −0.67 |
| c.185C>T | 66.3% ± 1.2 | ∆(E4_5) (9.9% ± 0.5) | ∆(E3_5) (23.8% ± 1.5) | - | −53.9 | 7 | −1.85 |
| c.186G>A | 64.3% ± 0.5 | ∆(E4_5) (7.4% ± 0.1) ∆(E3) (8.5% ± 0.0) | ∆(E3_5) (19.7% ± 0.4) | + | −23.2 | 0 | −1.88 |
| c.187G>A | 59.8% ± 2.9 | ∆(E3) (13.9% ± 0.9) | ∆(E3_5) (26.3% ± 2.3) | + | 13.4 | 0.3 | 0.65 |
| c.187G>C | 27.4% ± 0.4 | ∆(E3) (38.3% ± 0.4) | ∆(E3_5) (34.2% ± 0.3) | - | −12.4 | 1 | −0.05 |
| c.195C>T | 69.7% ± 0.3 | ∆(E4_5) (10.2% ± 0.1) | ∆(E3_5) (20.1% ± 0.3) | - | −22.8 | 0.4 | −0.26 |
| c.196G>A | 58.8% ± 1.6 | ∆(E4_5) (5.4% ± 0.3) ∆(E3) (10.6% ± 0.8) | ∆(E3_5) (33.3% ± 2.1) | - | −17.2 | 0 | −1.72 |
| c.198G>T | 48.7% ± 1.8 | ∆(E4_5) (5.9% ± 0.3) ∆(E3) (17.1% ± 0.7) | ∆(E3_5) (28.3% ± 0.9) | - | −47.3 | 5 | −2.69 |
| c.199A>G | 65.1% ± 1.0 | ∆(E3_5) (34.9% ± 1.0) | - | 37.8 | 0.7 | 1.31 | |
| c.200_218del | 28.7% ± 0.5 | ∆(E3) (46.9% ± 0.0) | ∆(E3_5) (24.4% ± 0.5) | - | −119.8 | 0.26 | −4.14 |
| c.202G>A | - | ∆(E3) (26.7% ± 0.9) | ∆(E3_5) (41.1% ± 3.1); ∆(E3p36) (32.2% ± 2.9) | - | −38.5 | 0 | −1.58 |
| c.208G>A | 46.8% ± 1.1 | ∆(E3) (16.8% ± 0.2) | ∆(E3_5) (36.4% ± 1.2) | + | −52.1 | 0 | −1.37 |
| c.209A>T | 58.2% ± 1.0 | ∆(E4_5) (10.0% ± 0.3) ∆(E3) (9.2% ± 0.2) | ∆(E3_5) (22.6% ± 0.9) | - | −58.0 | 5 | −0.30 |
| c.211C>T | 60.6% ± 0.7 | ∆(E4_5) (8.6% ± 0.0) ∆(E3) (7.9% ± 0.3) | ∆(E3_5) (23.0% ± 0.5) | - | −63.2 | 2.5 | 0.19 |
| c.213C>T | 57.2% ± 0.4 | ∆(E3) (19.5% ± 0.0) | ∆(E3_5) (23.3% ± 0.3) | - | −53.1 | 4 | −1.55 |
| c.214T>C | 100.0% ± 0.0 | + | 6.7 | 0 | −0.30 | ||
| c.216C>T | 68.4% ± 0.2 | ∆(E3) (14.0% ± 0.0) | ∆(E3_5) (17.6% ± 0.4) | - | −34.1 | 0 | −2.38 |
| c.217G>A | 66.5% ± 2.0 | ∆(E4_5) (12.5% ± 0.3) | ∆(E3_5) (21.0% ± 1.7) | + | −7.8 | 0 | −0.94 |
| c.224T>C | 59.9% ± 0.9 | ∆(E4_5) (13.4% ± 0.1) | ∆(E3_5) (26.7% ± 0.8) | - | −9.3 | 0 | 0.27 |
| c.234C>T | 70.7% ± 0.5 | ∆(E4_5) (11.5% ± 0.2) | ∆(E3_5) (17.8% ± 0.4) | + | −0.3 | 0 | 0.10 |
| c.243C>T | 50.9% ± 1.2 | ∆(E3) (16.8% ± 0.1) | ∆(E3_5) (32.3% ± 1.1) | - | −59.7 | 4 | −0.58 |
| Artificial Variants c.212_217 | |||||||
| c.212T>A | 63.6% ± 2.3 | ∆(E3) (5.5% ± 0.3) ∆(E4_5) (5.5% ± 0.0) | ∆(E3_5) (25.4% ± 2.6) | + | 16.1 | 0 | 0.51 |
| c.212T>C | 62.7% ± 1.0 | ∆(E3) (5.5% ± 0.1) ∆(E4_5) (7.1% ± 0.0) | ∆(E3_5) (24.7% ± 0.9) | + | −0.4 | 0 | −0.01 |
| c.212T>G | 63.4% ± 2.1 | ∆(E3) (5.9% ± 0.2) ∆(E4_5) (4.9% ± 0.2) | ∆(E3_5) (25.8% ± 1.9) | + | 41.8 | 0 | 1.58 |
| c.213C>A | 41.4% ± 0.3 | ∆(E3) (11.1% ± 0.2) ∆(E4_5) (6.4% ± 0.0) | ∆(E3_5) (41.1% ± 0.5) | - | −25.6 | 0.5 | −0.81 |
| c.213C>G | 49.6% ± 0.6 | ∆(E3) (6.7% ± 0.1) ∆(E4_5) (6.1% ± 0.0) | ∆(E3_5) (37.6% ± 0.7) | - | −20.1 | 0.5 | 0.57 |
| c.214T>A | 64.3% ± 1.3 | ∆(E4_5) (10.9% ± 0.1) | ∆(E3_5) (24.8% ± 1.3) | + | 19.1 | 0 | 0.87 |
| c.214T>G | 65.8% ± 2.4 | ∆(E4_5) (11.3% ± 0.3) | ∆(E3_5) (18.9% ± 0.4) | - | 18.3 | 0 | 3.41 |
| c.215A>C | 28.7% ± 0.2 | ∆(E4_5) (9.6% ± 0.3) | ∆(E3_5) (61.7% ± 0.4) | - | −40.5 | 0 | 1.82 |
| c.215A>G | 69.9% ± 1.6 | ∆(E4_5) (9.4% ± 0.2) | ∆(E3_5) (20.7% ± 1.5) | - | −27.9 | 0.33 | 1.78 |
| c.215A>T | 70.6% ± 1.7 | ∆(E4_5) (8.7% ± 0.1) | ∆(E3_5) (20.7% ± 1.6) | - | −21.2 | 0.67 | 1.90 |
| c.216C>A | 54.7% ± 1.0 | ∆(E3) (12.8% ± 0.4) | ∆(E3_5) (32.5% ± 1.4) | - | −73.7 | 0.13 | −0.56 |
| c.216C>G | 36.4% ± 0.6 | ∆(E3) (22.1% ± 1.3) | ∆(E3_5) (41.6% ± 1.9) | - | −91 | 0.33 | −1.15 |
| c.217G>C | 62.7% ± 0.5 | ∆(E3) (8.3% ± 0.1) | ∆(E3_5) (29.0% ± 0.4) | - | −21.2 | 0 | −0.27 |
| c.217G>T | 63.6% ± 0.4 | ∆(E3) (14.3% ± 0.3) | ∆(E3_5) (22.1% ± 0.5) | - | −114.2 | 1.67 | −1.66 |
| Hot-Skip Variants | |||||||
| c.163C>G | 38.7% ± 3.0 | ∆(E3) (26.9% ± 0.7) | ∆(E3_5) (34.4% ± 2.5) | - | −92.7 | 18 | −2.81 |
| c.163C>T (BRIDGES) | 62.0% ± 0.5 | ∆(E4_5) (12.3% ± 0.3) ∆(E3) (5.6% ± 0.1) | ∆(E3_5) (20.1% ± 0.6) | - MES: new 5′SS (7.07) | −52.0 | 16 | −1.978 |
| c.178C>T | 54.3% ± 1.0 | ∆(E3) (16.0% ± 0.1); ∆(E4_5) (4.8% ± 0.1) | ∆(E3_5) (24.9% ± 0.8) | - | −44.7 | 12 | −2.08 |
| c.HGVS 1 | p.HGVS 1 | Clinvar 2 | PS3/BS3 3 | PM2/BS1/BA1 4 | Proxy for Allele Counts 5 | Variant Classification 6 |
|---|---|---|---|---|---|---|
| c.83-4_-3delinsAG | p.? | VUS (**) | PS3_VS | (0/251433) PM2 | rs780590372 (=) | Likely Pathogenic (PS3_VS + PM2) |
| c.83-2A>G | p.? | LP (*) | PS3_VS (91%PS3_VS + 9%N/A) 7 | (0/251433) PM2 | rs780590372 (−1) | Likely Pathogenic (PS3_VS + PM2) |
| c.145-2A>G | p.? | not reported | PS3_VS | (0/251102) PM2 | rs201974522 (−1) | Likely Pathogenic (PS3_VS + PM2) |
| c.163C>T | p.(Arg55Trp) | VUS (**) | N/A (62%N/A + 38%PS3_VS) | (2/251433) PM2 | - | Uncertain Significance (PM2 only) 8 |
| c.171G>A | p.(Leu57=) | LB (**) | BS3 (76%BS3 + 24%PS3_VS) | (0/249480) PM2 | rs745307359 (−4) | Uncertain Significance (BS3 + PM2) |
| c.175G>T | p.(Ala59Ser) | not reported | N/A (53%N/A + 47%PS·_VS) | (0/251298) PM2 | rs780689600 (+4) | Uncertain Significance (PM2 only) 8 |
| c.180G>A | p.(Gln60=) | LB (*) | BS3 (68%BS3 + 32%PS3_VS) | (0/251298) PM2 | rs780689600 (−1) | Uncertain Significance (BS3 + PM2) |
| c.180G>T | p.(Gln60His) | VUS (**) | N/A (69%N/A + 31%PS3_VS) | (0/251298) PM2 | Uncertain Significance (PM2 only) 8 | |
| c.184T>A | p.(Ser62Thr) | not reported | N/A (37%N/A + 63%PS3_VS) | (0/251382) PM2 | rs374357106 (+3) | Uncertain Significance (PM2 only) 8 |
| c.185C>T | p.(Ser62Leu) | VUS (**) | N/A (66%N/A + 34%PS3_VS) | (5/251382) N/A | - | Uncertain Significance (no codes) 8 |
| c.186G>A | p.(Ser62=) | LB (**) | N/A (64%BS3 + 36%PS3_VS) | (3/251380) N/A | - | Uncertain Significance (no codes) 8 |
| c.187G>A | p.(Ala63Thr) | VUS (**) | N/A (60%N/A + 40%PS3_VS) | (1/251408) PM2 | - | Uncertain Significance (PM2 only) 8 |
| c.187G>C | p.(Ala63Pro) | not reported | N/A (27%N/A + 73%PS3_VS) | (0/251408) PM2 | c.187G>A | Uncertain Significance (PM2 only) 8 |
| c.195C>T | p.(Pro65=) | LB (**) | BS3 (70%BS3 + 30%PS3_VS) | (5/251420) N/A | - | Uncertain Significance (BS3 only) |
| c.196G>A | p.(Val66Met) | B (2), LB (4), VUS (4) | N/A (59%N/A + 41%PS3_VS) | (80/251384) BS1 | - | Uncertain Significance (BS1) 8 |
| c.198G>T | p.(Val66=) | LB (**) | N/A (49%BS3 + 51%PS3_VS) | (9/251448) N/A | - | Uncertain Significance (no codes) 8 |
| c.199A>G | p.(Asn67Asp) | not reported | N/A (65%N/A + 35%PS3_VS) | (0/251448) PM2 | rs546461804 (+1) | Uncertain Significance (PM2 only) 8 |
| c.200_218del | p.(Asn67Argfs*7) | not reported | PS3_VS | (0/251448) PM2 | rs54661804 (=) | Likely Pathogenic (PS3_VS + PM2) |
| c.202G>A | p.(Gly68Ser) | VUS (**) | PS3 (68%PS3_VS + 32%PS3) | (9/251454) N/A | - | Uncertain Significance (PS3 only) |
| c.208G>A | p.(Asp70Asn) | VUS (**) | N/A (47%N/A + 53%PS3_VS) | (8/251458) N/A | - | Uncertain Significance (no codes) 8 |
| c.209A>T | p.(Asp70Val) | VUS (**) | N/A (58%N/A + 42%PS3_VS) | (1/251450) PM2 | - | Uncertain Significance (PM2 only) 8 |
| c.211C>T | p.(Leu71Phe) | VUS (*) | N/A (61%N/A + 39%PS3_VS) | (0/251476) PM2 | rs559850711 (+1) | Uncertain Significance (PM2 only) 8 |
| c.213C>T | p.(Leu71=) | LB (**) | N/A (57%BS3 + 43%PS3_VS) | (2/251466) PM2 | - | Uncertain Significance (PM2 only) 8 |
| c.214T>C | p.(Tyr72His) | not reported | N/A | (0/251466) PM2 | rs745546403 (−1) | Uncertain Significance (PM2 only) |
| c.216C>T | p.(Tyr72=) | B/LB (**) | BS3 (67%BS3 + 33%PS3_VS) | (27/251474) N/A | - | Uncertain Significance (BS3) |
| c.217G>A | p.(Glu73Lys) | VUS (**) | N/A (67%N/A + 33%PS3_VS) | (2/251462) PM2 | - | Uncertain Significance (PM2 only) 8 |
| c.224T>C | p.(Leu75Pro) | not reported | N/A (60%N/A + 40%PS3_VS) | (0/251466) PM2 | rs746929682 (−1) | Uncertain Significance (PM2 only) 8 |
| c.234C>T | p.(Ser78=) | B (**) | BS3 (71%BS3 + 29%PS3_VS) | (29273/251448) BA1 | - | Benign (BS3 + BA1) |
| c.243C>T | p.(Ile81=) | LB (**) | N/A (60%BS3 + 40%PS3_VS) | (1/251472) PM2 | - | Uncertain Significance (PM2 only) 8 |
| c.263 + 6T>C | p.? | not reported | N/A (49%BS3 + 51%PS3_VS) | (0/251424) PM2 | rs56218020 (+1) | Uncertain Significance (PM2) 8 |
| c.343C>T | p.(Gln115Ter) | P (**) | PS3_VS | (0/141295) PM2 | rs786202507 (−4)rs878854562(+7) | Likely Pathogenic (PS3_VS + PM2) |
| c.345+2T>C | p.? | LP(1); VUS(1) | PS3_VS | (0/251220) PM2 | rs878854562 (+3) | Likely Pathogenic (PS3_VS + PM2) |
| c.476_480+1dup | p.? | not reported | N/A (40%PS3_VS + 60%N/A) | (0/251474) PM2 | rs1057521922 (=) | Uncertain Significance (PM2 only) 8 |
| c.480+1G>A | p.? | (-) | PS3 (70%PS3_VS + 30%PS3) | (0/251474) PM2 | rs1057521922 (−3) | Likely Pathogenic (PS3 + PM2) |
| c.481-8C>A | p.? | not reported | N/A (30%PS3_VS + 70%N/A) | (0/241990) PM2 | rs762247126 (=) | Uncertain Significance (PM2 only) 8 |
| c.577-2A>G | p.? | P/LP (**) | PS3 (80%PS3_VS + 20%PS3) | (0/250980) PM2 | rs1210749655 (−4) | Likely Pathogenic (PS3 + PM2) |
| c.738+1G>A | p.? | LP (**) | PS3_VS (70%PS3_VS + 10%N/A) 7 | (0/240992) PM2 | rs1210620444 (−1) | Likely Pathogenic (PS3 + PM2) 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bueno-Martínez, E.; Sanoguera-Miralles, L.; Valenzuela-Palomo, A.; Lorca, V.; Gómez-Sanz, A.; Carvalho, S.; Allen, J.; Infante, M.; Pérez-Segura, P.; Lázaro, C.; et al. RAD51D Aberrant Splicing in Breast Cancer: Identification of Splicing Regulatory Elements and Minigene-Based Evaluation of 53 DNA Variants. Cancers 2021, 13, 2845. https://doi.org/10.3390/cancers13112845
Bueno-Martínez E, Sanoguera-Miralles L, Valenzuela-Palomo A, Lorca V, Gómez-Sanz A, Carvalho S, Allen J, Infante M, Pérez-Segura P, Lázaro C, et al. RAD51D Aberrant Splicing in Breast Cancer: Identification of Splicing Regulatory Elements and Minigene-Based Evaluation of 53 DNA Variants. Cancers. 2021; 13(11):2845. https://doi.org/10.3390/cancers13112845
Chicago/Turabian StyleBueno-Martínez, Elena, Lara Sanoguera-Miralles, Alberto Valenzuela-Palomo, Víctor Lorca, Alicia Gómez-Sanz, Sara Carvalho, Jamie Allen, Mar Infante, Pedro Pérez-Segura, Conxi Lázaro, and et al. 2021. "RAD51D Aberrant Splicing in Breast Cancer: Identification of Splicing Regulatory Elements and Minigene-Based Evaluation of 53 DNA Variants" Cancers 13, no. 11: 2845. https://doi.org/10.3390/cancers13112845
APA StyleBueno-Martínez, E., Sanoguera-Miralles, L., Valenzuela-Palomo, A., Lorca, V., Gómez-Sanz, A., Carvalho, S., Allen, J., Infante, M., Pérez-Segura, P., Lázaro, C., Easton, D. F., Devilee, P., Vreeswijk, M. P. G., de la Hoya, M., & Velasco, E. A. (2021). RAD51D Aberrant Splicing in Breast Cancer: Identification of Splicing Regulatory Elements and Minigene-Based Evaluation of 53 DNA Variants. Cancers, 13(11), 2845. https://doi.org/10.3390/cancers13112845










