Simple Summary
Quantification of liver metastases on imaging is of utmost importance in therapy response assessment, wherein gadoxetic acid (Gd-EOB)-enhanced magnetic resonance imaging (MRI) shows the highest accuracy. Common criteria for assessing therapy response simplify measuring liver metastasis, as full 3D quantification is very time-consuming. Therefore, we trained a deep-learning model using manual 3D segmentation of liver metastases and hepatic parenchyma in 278 Gd-EOB MRI scans of 149 patients with neuroendocrine neoplasms (NEN). The clinical relevance of the model was evaluated in 33 additional consecutive patients with NEN and liver metastases, comparing the model’s segmentation of baseline and follow-up examinations with the therapy response evaluation of an expert multidisciplinary cancer conference (MCC). The model showed high accuracy in quantifying liver metastases and hepatic tumor load, and its measurements matched the response evaluation of an MCC so that its use to support treatment decision-making would be possible.
Abstract
Background: Rapid quantification of liver metastasis for diagnosis and follow-up is an unmet medical need in patients with secondary liver malignancies. We present a 3D-quantification model of neuroendocrine liver metastases (NELM) using gadoxetic-acid (Gd-EOB)-enhanced MRI as a useful tool for multidisciplinary cancer conferences (MCC). Methods: Manual 3D-segmentations of NELM and livers (149 patients in 278 Gd-EOB MRI scans) were used to train a neural network (U-Net architecture). Clinical usefulness was evaluated in another 33 patients who were discussed in our MCC and received a Gd-EOB MRI both at baseline and follow-up examination (n = 66) over 12 months. Model measurements (NELM volume; hepatic tumor load (HTL)) with corresponding absolute (ΔabsNELM; ΔabsHTL) and relative changes (ΔrelNELM; ΔrelHTL) between baseline and follow-up were compared to MCC decisions (therapy success/failure). Results: Internal validation of the model’s accuracy showed a high overlap for NELM and livers (Matthew’s correlation coefficient (φ): 0.76/0.95, respectively) with higher φ in larger NELM volume (φ = 0.80 vs. 0.71; p = 0.003). External validation confirmed the high accuracy for NELM (φ = 0.86) and livers (φ = 0.96). MCC decisions were significantly differentiated by all response variables (ΔabsNELM; ΔabsHTL; ΔrelNELM; ΔrelHTL) (p < 0.001). ΔrelNELM and ΔrelHTL showed optimal discrimination between therapy success or failure (AUC: 1.000; p < 0.001). Conclusion: The model shows high accuracy in 3D-quantification of NELM and HTL in Gd-EOB-MRI. The model’s measurements correlated well with MCC’s evaluation of therapeutic response.
1. Introduction
The incidence of neuroendocrine neoplasms (NEN) has increased in the past 30 years considerably, while at the same time, multiple treatment options are available for this disease [1]. The radiological workload for follow-up of patients with NENs has, therefore, increased accordingly. However, not only because of the increasing incidence but also because of the lower aggressiveness of NENs compared to liver metastases of other entities (e.g., colorectal carcinoma), the number of follow-up examinations is increasing [2,3,4,5,6,7,8,9]. Based on the indolent clinical course of NENs, patients often present at an advanced stage for first diagnosis [4,6,9,10]. The liver represents the predominant site for metastases, and accurate calculation of the hepatic metastatic tumor burden is mandatory for therapeutic follow-up [11]. The measurement of diffuse liver lesions, which occur in 60–70% of patients, can be challenging and is–at present-time-consuming. A further challenge is that common therapeutic response criteria intended to characterize how metastases develop over time are not always suitable for each patient [9]. Response criteria in solid tumors (RECIST, Version 1.1) are based on changes in diameters of a few lesions, which are considered representative [12]. However, hepatic tumor load (HTL), which is neglected if only measuring the diameter of metastases, is an important prognostic marker in hepatically metastasized NEN [4,13,14,15,16]. The quantitative evaluation of the metastatic volume can potentially provide a practical method for assessing the disease’s course and may show improved prognostic value.
Magnetic resonance imaging (MRI) is the most sensitive technique to detect and quantify neuroendocrine liver metastases (NELMs) compared to conventional computed tomography (CT), ultrasound (US), and somatostatin receptor imaging [16,17,18]. Gadoxetic acid-enhanced (Gd-EOB) MRI is even more sensitive than conventional extracellular gadolinium chelate-enhanced MRI [17,19,20]. In addition to the use of contrast-enhanced MRI, the use of diffusion-weighted imaging (DWI) sequences increases the sensitivity in the detection of NELM [21,22,23]. Thus, the combination of DWI and hepatobiliary phase (HBP) sequences with Gd-EOB is now the imaging modality with the highest sensitivity for NELM [24]. Hepatic metastases of NELM typically demonstrate a hypervascularization pattern in dynamic contrast phases (arterial, portal-venous and transitional phase), which aids in the differentiation of NELM from other liver lesions [19,25,26]. Despite the value of dynamic contrast phases in differential diagnoses, lesion measurement, and thus response evaluation, is preferably performed in the hepatobiliary phase (HBP) when hepatocyte-specific contrast agents are used [27].
Advances in artificial intelligence (AI) technology have led to generating image recognition algorithms poised to aid and improve medical imaging procedures. AI has already demonstrated strong performance in various medical applications, especially in image-based diagnoses [28]. Although several studies suggest that the performance of AI in imaging diagnosis is superior to human experts, the consensus is that AI should play a supporting role to radiologists and that AI tools could especially be used to save time in clinical routine [28,29,30,31,32,33]. The various fields of AI support in liver imaging include segmentation, lesion detection and classification of diffuse or focal liver diseases [34,35].
Here we provide the first data using a high-precision AI algorithm for the 3D quantification of the hepatic tumor burden of NELM and provide a useful tool for clinical decision-making, for example, in multidisciplinary cancer conferences (MCC).
2. Material and Methods
2.1. Patient Cohorts
2.1.1. AI Development (AI dev) Cohort
398 MRI scans in 149 patients with NEN, who underwent Gd-EOB enhanced MRI (MAGNETOM Aera (1.5T), Siemens Healthcare, Erlangen, Germany) between January 2015 and August 2018 at our institution were retrospectively identified from our radiology database. 120 of these scans were not suitable for the model’s training because of missing evidence of NELM (n = 112) or due to non-standard scan protocols (n = 8), resulting in a total inclusion of 278 Gd-EOB MRI datasets. Pretreatments (e.g., partial liver resection, transarterial or local ablative therapies), which may influence the morphology of the liver, were not an exclusion criterion.
2.1.2. MCC Cohort
In a second institutional database search, we consecutively identified 33 patients discussed in our MCC between January 2019 and January 2020 and received a Gd-EOB MRI both as a baseline and as a follow-up examination (n = 66). All 33 patients had liver metastases and were selected independently of the hepatic tumor volume or their disease history. In these patients, all MCC decisions were based on the course of the metastatic liver disease. Patients in whom the MCC decision was based on extrahepatic tumor manifestations were excluded.
2.2. Magnetic Resonance Imaging
AI dev cohort: MRI was obtained with 1.5 T using phased-array body coils in all patients (MAGNETOM Aera, Siemens Healthcare, Erlangen, Germany) at the same institution. All patients received Gd-EOB (Primovist, Bayer, Berlin, Germany) as an intravenous contrast agent (0.025 mmol/kg body weight; automatic injection at 2 mL/s flow rate, 40 mL saline flush). All MRI examination protocols comprised a 3D T1-weighted (T1w) gradient echo (GRE) sequence with fat saturation (FS) during hepatobiliary contrast phase (HBP) (VIBE: “volumetric interpolated breath-hold examination”; repetition time (TR): 4.58 ms; echo time (TE): 2.21 ms; slice thickness 3 mm, flip angle (FA): 25°; acquisition matrix: 320\0\0\165). The HBP sequence was acquired 20 mins after contrast administration.
MCC cohort: Gd-EOB MRI scans were performed on five different institutional MRI scanners and included both 1.5 T and 3 T examinations. All examinations contained a 3D T1w GRE FS sequence during HBP. Due to the different scanners, the scan parameters (TR, TE, FA and matrix) varied between the examinations. The HBP sequence was acquired between 10 and 20 mins after contrast administration. Among others, diffusion-weighted imaging (DWI) sequences were acquired in the time between Gd-EOB injection and the HBP sequence. All DWI sequences contained at least two b values (b = 0 and b = 800) [36].
All examination protocols corresponded to the ENETS consensus guidelines for the standard of care in neuroendocrine tumors [15].
2.3. Manual Segmentation
All HBP sequences of the MRI scans (AI dev and MCC cohort) were anonymized and segmented using the Medical Imaging Interaction Toolkit (MITK) [37]. Volumetry (3D segmentation) of the liver and all liver metastases was performed in the HBP 3D T1w-GRE FS sequence. There was no limit on the number of metastases segmented per patient. Segmentation was performed manually using the polygonal region of interest (ROI) tool and is based on the planimetry method. Margins of the liver metastases were defined by the signal difference between hypointense liver metastases and the contrast-enhanced liver parenchyma. Adjacent vessels and biliary ducts were excluded if reasonably possible. All segmentations were refined by a radiologist with >5 years of experience in abdominal MRI. Distribution patterns of NELM were scored according to the number: singular, multiple (≤10 metastases) and diffuse (>10 metastases) and distribution: unilobar (left or right) and bilobar.
For subanalysis, all NELM and livers were manually segmented in DWI sequences in the MCC cohort. The segmentation process was equivalent to that previously described in the HBP sequences.
2.4. Model Training and Validation
The model was trained using the MIC-DKFZ nnU-Net (Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany) deep-learning framework. nnU-Net is an open-source tool. The source code and comprehensive documentation are publicly available on GitHub [38]. nnU-Net enables 3D semantic segmentation in many biomedical imaging applications without requiring designing respective specialized solutions [39]. Out of the 278 MRI scans, 222 (80%) HBP sequences were randomly chosen for the model training.
The HBP sequences of the remaining 56 scans (20%) were used to test the model’s accuracy (internal validation). External validation of the model’s accuracy was performed in the MCC cohort (different scanners (1.5 T and 3 T) and sequence parameters were used compared to the model’s training).
2.5. Clinical Correlation
Our model analyzed the NELM volume and liver volume of the 33 patients with MCC decisions in the baseline (BL) scan and the follow-up (FU) scan on which the MCC decisions were based. The MCC is part of our European Neuroendocrine Tumor Society (ENETS) center of excellence and consists of specialized gastroenterologists, endocrine surgeons, pathologists, nuclear medicine specialists, radiotherapists and radiologists. Absolute and relative changes in NELM volume and HTL calculated by the model were analyzed and compared to the MCC decisions. MCC decisions were classified as therapy success (stable disease (SD) or partial regression (PR)) or therapy failure (progressive disease (PD)) based on the presented images. The evaluation within the board was guided by the response criteria in solid tumors (RECIST, Version 1.1).
2.6. Statistics
Statistical analysis was performed using SPSS Statistics (IBM, Version 25, Armonk, NY, USA). The Kolmogorov–Smirnov test showed a non-normal distribution of the data. Therefore, nonparametric testing was performed.
Descriptive data were accordingly presented as the median and interquartile range (IQR). Relative size differences in segmentations were calculated by the following formula: (model’s volume–radiologists’ volume)/radiologists’ volume. Matthew’s correlation coefficients (φ) were calculated to measure the model’s segmentation accuracy as previously published [40]. MCC decisions were compared to the automatized volume evaluation of the model. HTL was calculated by the formula: (NELM volume/(liver volume–NELM volume)) × 100. Absolute volume changes were calculated by the difference: VolumeFollow-up–VolumeBaseline. Relative volume changes were calculated by the formula: ((VolumeFollow-up–VolumeBaseline)/VolumeBaseline) × 100. Mann–Whitney U test was used as a dominance test comparing two independent groups of quantitative data. A sign test was used to compare two related samples. Spearman’s rank test was used for correlation analysis in continuous variables, and the corresponding correlation coefficients (rs) were calculated. ROC analysis was performed, and Youden indexes were calculated to determine optimal cutoff values.
3. Results
3.1. Patient Cohorts
3.1.1. AI dev Cohort
Characteristics of the 149 patients with NEN are summarized in Table 1. The most common primary tumor sites were the ileum (51.0%) and the pancreas (43.0%). Confirmed (histologically or with the aid of SR imaging) liver metastases were present in 118 patients (79.2%), which were used for the model training. Out of these 118 patients, 4 patients (3.4%) had singular liver metastasis, 59 patients (50%) had multiple metastases (≤10 metastases), and 55 patients (46.6%) had a diffuse metastatic pattern (>10 metastases). Both liver lobes were involved in 91 patients (77.1%). Unilobar disease limited to a single liver lobe was found in 27 patients (22.9%) (right liver: 24 patients, left liver: 3 patients).

Table 1.
Patient and disease characteristics of the AI dev cohort and MCC cohort.
3.1.2. MCC Cohort
Characteristics of the 33 patients with MCC decisions are summarized in Table 1. Comparably to the training cohort, the most common primary tumor sites were the pancreas and ileum (36.4% each). Therapeutic response was classified by the MCC as therapeutic success in 16 (48%) patients (SD: n = 14; PR: n = 2) and therapeutic failure in 17 (PD, 52%) patients.
3.2. Validation of the Model
The median NELM volume in the 56 patients (internal validation) of the AI dev group was 17.25 cm3 (IQR: 4.48–60.93 cm3) as determined by the nnU-Net model and 16.17 cm3 (IQR: 4.87–58.16 cm3) in the radiologists’ manual segmentation. The median relative volume difference between the model’s and the radiologist’s segmentation of NELM was −3.7% (IQR: −24.54−+11.83%). The model showed a median φ of 0.76 (IQR: 0.68–0.83) for the segmentation of metastases (Figure 1, left side). Analysis of the data in a case-wise fashion identified three out of 56 patients (5.4%), whereby the model’s segmentation only achieved a weak overlap (φ < 0.2). Two out of these patients had very low NELM volume (0.1 and 0.2 cm3). The third patient showed atypical, hyperintense signal intensities of the metastases in HBP; NELM were subsequently missed by the model. Dividing the cohort by the median NELM volume (16.17 cm3) into high and low NELM volume, the model showed significant higher φin patients with higher NELM volume (median φ: 0.80; IQR: 0.73–0.84) compared to low NELM volume (median φ: 0.71; IQR: 0.64–0.78; p = 0.003). For liver segmentation, the median volume was 1639.9 cm3 (IQR: 1366.1–1960.7 cm3) in the manual segmentation and 1659.0 cm3 (IQR: 1404.2–1966.9 cm3) in the model’s segmentation. The median relative volume difference between the model’s and the manual segmentations of livers was +0.9% (IQR: −0.7−+4.2%). The model showed a median φ of 0.95 (IQR: 0.95–0.96) in liver segmentation (Figure 1, right side). The external validation (MCC cohort) confirmed the high accuracy of the model. The model achieved a median φof 0.86 (IQR: 0.81–0.91) in the segmentation of NELM and of 0.96 (IQR: 0.95–0.96) in liver segmentation.
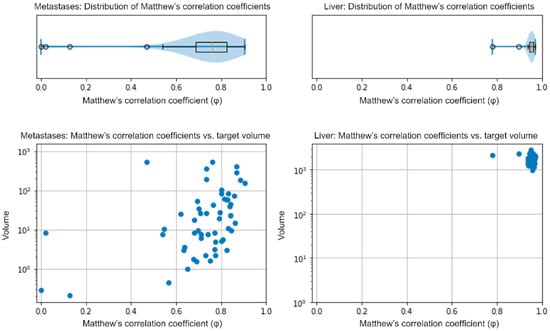
Figure 1.
Internal validation–distribution of φ in NELM and liver segmentations (upper row) and its distribution in correlation to the target volume in cm3 (lower row).
3.3. Automatized NELM Volume Analysis and Clinical Correlation (MCC Cohort)
The model’s measurements of the MCC cohort are summarized in Table 2 and exemplarily visualized in Figure 2.

Table 2.
Summary of the model’s segmentation results for the MCC cohort and their absolute and relative changes between baseline and follow-up MRI examinations.
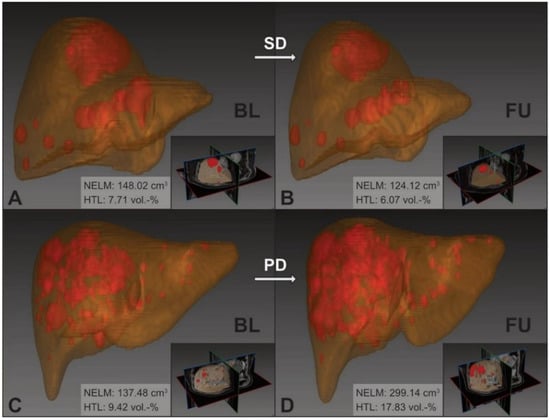
Figure 2.
nnU-Net 3D segmentation of NELM and livers in the MCC cohort. Upper row: example images of therapy success (patient ID: 11) with stable disease between baseline (A) and follow-up (B); ΔrelNELM: −16.14% and ΔrelHTL: −21.23%. Lower row: example images of therapy failure (patient ID: 08) with progressive disease between baseline (C) and follow-up (D); ΔrelNELM: +117.58% and ΔrelHTL: +89.32%. BL: baseline; FU: follow-up; SD: stable disease; PD: progressive disease; NELM: neuroendocrine liver metastasis; HTL: hepatic tumor load.
The comparison between patients with therapy success (n = 16) and therapy failure (n = 17) showed significant differences for all absolute and relative volume changes (p < 0.001). Patients classified as therapy success by the MCC showed significant lower values in median absolute NELM volume change (ΔabsNELM), median absolute HTL change (ΔabsHTL), median relative NELM volume change (ΔrelNELM) and median relative HTL change (ΔrelHTL) than patients with therapy failure (p < 0.001) (Figure 3).
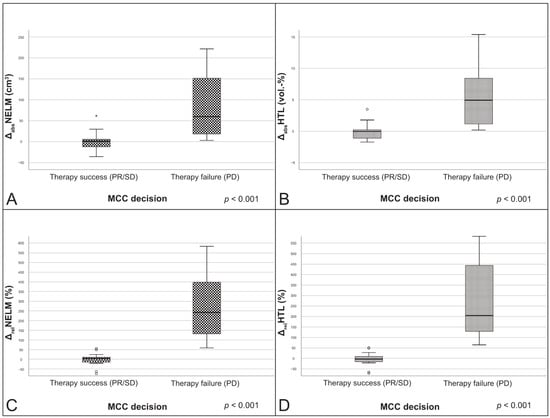
Figure 3.
Boxplots of the change variables in correlation to the MCC decisions. (A) ΔabsNELM; (B) ΔabsHTL; (C) ΔrelNELM; (D) ΔrelHTL. PR: partial response; ΔabsNELM: absolute NELM volume change; ΔabsHTL: absolute HTL change; ΔrelNELM: relative NELM volume change; ΔrelHTL: relative HTL change.
The case-wise analysis of the 33 MCC patients is summarized in Table 3. The case-wise analysis showed that the model correctly detected increased NELM volume in all of the 17 patients with therapeutic failure (100%). The ΔabsNELM increase in these 17 patients ranged from +3.02 cm3 to +864.45 cm3 and ΔabsHTL ranging from +0.18 vol.-% to +36.41 vol.-%. The relative increase of ΔrelNELM ranged from +58.52% to +4513.64% and in ΔrelHTL from +64.97% to +2497.20%. In patients with therapeutic success (n = 16), the ΔabsNELM ranged from −394.57 cm3 to −34.75 cm3 (in PR) and −35.70 cm3 to +61.56 cm3 (in SD) and the ΔabsHTL from −16.96 vol.-% to −1.73 vol.-% (in PR) and −1.64 vol.-% to +3.48 vol.-% (in SD). The relative change variables of ΔrelNELM ranged from −74.68% to −63.68% in PR and from −20.19% to 55.25% in SD, and the ΔrelHTL ranged from −71.13% to −65.03% in PR and from −21.23% to +50.51% in SD (Figure 4).

Table 3.
Case-wise summary of the model’s measurements and response variables in the MCC cohort.
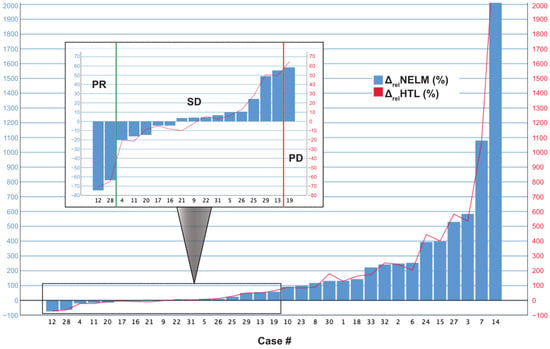
Figure 4.
Case-wise illustration of the relative volume changes (ΔrelNELM and ΔrelHTL) between baseline and follow-up examination in the MCC cohort. The box within the figure shows an optimized scaling of cases with less change to illustrate the significance thresholds for partial response (PR) and progressive disease (PD).
In the total cohort, ROC analysis of MCC decision and the relative changes (ΔrelNELM and ΔrelHTL) showed an area under the curve (AUC) of 1.000 (p < 0.001) for both variables. The absolute changes showed an AUC of 0.908 for ΔabsNELM and of 0.926 for ΔabsHTL (p < 0.001). To determine the best cutoff values for progressive disease, a Youden index was calculated. For ΔrelNELM, the highest Youden index (1.000; 100% sensitivity and 100% specificity) was found at the cutoff value +56.88%. For ΔrelHTL, the highest Youden index (1.000; 100% sensitivity and 100% specificity) was found at a cutoff value of +57.73% (Figure 5).
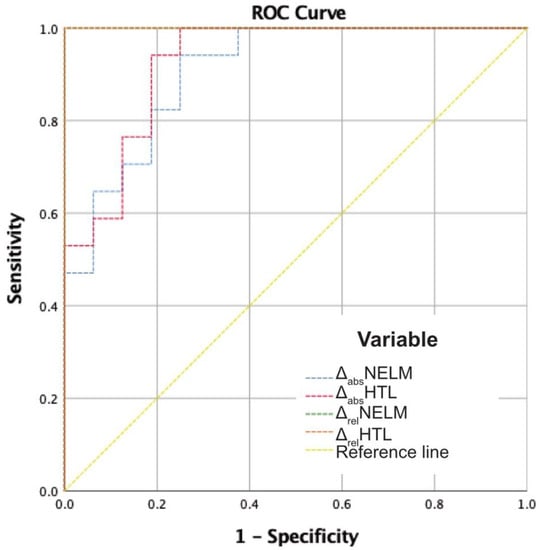
Figure 5.
ROC analysis of the absolute and relative change variables in relation to the MCC decisions. AUC ΔabsNELM: 0.908; AUC ΔabsHTL: 0.926; AUC ΔrelNELM: 1.000; AUC ΔrelHTL: 1.000; p < 0.001.
3.4. Comparison of 3D Quantification between HBP and DWI Sequences
In the MCC cohort, manual segmentations of NELM (rs: 0.981; p < 0.001), livers (rs: 0.966; p < 0.001) and HTL (rs: 0.956, p < 0.001) showed a high correlation between HBP and DWI sequences. However, direct comparison of the measured values for NELM and livers showed significant differences between HBP and DWI (p < 0.001; Table 4). When looking at the changes between BL and FU, a high correlation between DWI and HBP sequences was also shown for ΔabsNELM (rs: 0.919; p < 0.001), ΔrelNELM (rs: 0.960; p < 0.001), ΔabsHTL (rs: 0.883, p < 0.001) and ΔrelHTL (rs: 0.952; p < 0.001). There were no significant difference of ΔabsNELM, ΔrelNELM, ΔabsHTL and ΔrelHTL between DWI and HBP-based measurements (p = 0.072 to 0.719; Table 4).

Table 4.
Comparison of 3D quantification between HBP and DWI sequences.
4. Discussion
This is the most extensive study presenting AI data quantifying the total volume of hepatic tumor burden in NEN using a deep-learning model combined with Gd-EOB MRI. The model achieved high accuracy, especially in patients with higher NELM volume and delivered results corresponding to the MCC consensus decision-making regarding therapeutic success or failure.
The presented deep-learning model differs from previous studies in several aspects. First, the training data set of 278 Gd-EOB MRI examinations is the largest published to date in the automated assessment of focal liver lesions [41]. The high proportion of patients with more than ten metastases resulted in more than 2000 segmented metastases. Second, various hepatic conditions were included in the model’s training. Previous liver resection, excessive pretreatment, ablation therapies or preceding intraarterial treatments (e.g., transarterial chemoembolization (TACE) or selective internal radiation therapy (SIRT)) were no exclusion criteria for training. The combination of high case numbers and various pretreatments should improve the robustness of the model in preparation for everyday clinical usage [42,43]. Due to the broad training, it is possible to quantify patients under different therapies with the model. However, individual pitfalls must be considered. Therapy-induced hemorrhage of NELM affects the visualization of lesions in HBP sequences. Our study identified one case in which the model had achieved low accuracy for this reason. In addition, in two cases with very low tumor burden, our model showed only unsatisfactory accuracy. Though, these cases are also of less interest for an automated volume analysis since a conventional, manual evaluation could easily be performed. The aim of our study was not to replace manual evaluation but to complement and improve it.
Our results demonstrate that accurate, automated 3D segmentation of NELM is feasible in HBP from Gd-EOB MRI examinations. Due to the comparatively lower growth dynamics of NELM compared to metastases from other primary tumors, we believe automated quantification is particularly valuable based on the numerous follow-up studies in patients with NEN. Even if NELM is characterized by marked arterial hypervascularization or cystic components on imaging, these features do not affect the HBP sequence [25]. Liver metastases from a wide variety of primary tumors show the same typical imaging characteristics in the HBP sequences with marked hypointensity of the lesion compared to the surrounding liver parenchyma [44]. Therefore, by using HBP in Gd-EOB MRI, our model is not limited to the segmentation of NELM, and its use should also be investigated for liver metastases of other primary tumors.
The high value of Gd-EOB HBP sequences in the determination of NELM size has already been shown and corroborates our approach to using this sequence for 3D segmentation [45]. The high lesion to liver contrast also provides optimum conditions for automated segmentation [46,47]. However, besides its excellent imaging characteristics, Gd-EOB MRI has some disadvantages. These include the comparatively high costs due to the contrast agent itself and the resulting examination time, the general side effects and possible deposition of gadolinium [48]. As a non-contrast alternative with high sensitivity, DWI sequences can also be effectively used to measure NELM without the disadvantages of Gd-EOB MRI [49,50]. Currently, however, DWI sequences are used for detection rather than measurements of liver lesions. In particular, a 3D measurement may be limited by the lower axial resolution of commonly used DWI sequences. In our subanalysis, we could show that DWI-based measurements correlate strongly with those in HBP. However, the absolute measurements of NELM and livers showed significant differences between the two sequences, so that an exact 3D quantification using DWI was not possible. Nevertheless, this inaccuracy was relativized when the measurements were compared in evaluating treatment response. The relative and absolute changes of NELM volume and HTL between baseline and follow-up examination showed no significant difference between HBP and DWI so that evaluation of treatment response using 3D measurements in DWI seems feasible. Therefore, the results of our study encourage developing similar automation for non-contrast DWI MRI as well.
The limitation to lesion diameters versus volume in clinical routine can be best explained by the time required for full 3D volumetry. Up to now, 3D volumetry of liver lesions has only been carried out within the framework of studies [51,52]. Besides the volumetric assessment of tumor burden, the 3D segmentations generated by the model presented in this study could be used for further lesion analysis, such as texture analysis, radiomics or contrast-uptake used in Choi criteria [53,54]. To date, most studies concerning artificial intelligence and liver imaging focus on diffuse liver disease or the classification of liver tumors [55,56,57]. With the help of the presented model and the associated time saving by the automatized segmentation, not only 3D quantification of HTL but also more sophisticated tumor analyses could find their way into clinical routine.
Assessment of therapeutic response in liver metastases, independently from primary tumor origin, is most commonly based on the Response Evaluation Criteria in Solid Tumors (RECIST, Version 1.1). RECIST1.1 is suitable for study cohorts and facilitates response evaluation by defining a limited number (maximum two per organ) of target lesions [58]. From a practical point, response criteria vary regarding increasing versus decreasing tumors. Partial response (PR) is defined as a decrease of at least 30% in the sum of the largest diameter of target lesions. By contrast, progressive disease is defined as increasing at least 20% of target lesions or the appearance of one or more new lesions in a 2D measurement [59]. Considering this somehow simplified approach, the pure volumetric determination of growth behavior should allow a more precise measuring method for therapeutic decision-making in the individual patient. The simplification of RECIST1.1 can lead to patients being interpreted incorrectly or inconsistently during their illness. The limitations of RECIST1.1 become even more evident when evaluating the effects of targeted molecular agents, especially in slow-growing tumors, such as NENs [60,61]. RECIST1.1 treatment response strongly depends on which target lesions were chosen at the baseline scan. Heterogeneous treatment response, which can be seen in different types of primary cancers and systemic treatments, is not represented by RECIST1.1 [62]. Additionally, volumetric measurement methods show a higher intra-observer reproducibility compared to RECIST1.1 [63]. Quantification of total HTL in clinical routine is not routinely performed, and in most cases, tumor load is visually estimated by the radiologist. However, several studies have shown that hepatic tumor burden is an important prognostic imaging marker [13,64,65]. Volumetric evaluation of the HTL, as performed by our model, provides useful information on lesion distribution and allows a more realistic quantification of hepatic tumor extent than the (2D) diameter measurements, which are commonly used [66]. In addition, the model considers all lesions, which would also allow capturing of heterogeneous treatment responses.
The new challenge in volumetric tumor mass determination will be developing new cutoff values. If metastasis is seen as a sphere mathematically, an increase of the diameter of the lesion of 20%, which defines a lesion to be classified as a progressive disease in RECIST1.1, would result in a volume increase of approximately 73%. In our cohort, the MCC stated progressive disease and therapy failure when the tumor volume, as determined by the model, increased by 57%. Furthermore, a NELM volume decrease of −57% correctly identified the two patients with partial response. Our results show that 3D assessment of NELM could be useful, but further studies are needed to evaluate its superiority over 2D methods regarding clinical endpoints [67].
MCCs are designed to optimize patient outcomes by elaborating the best treatment plans or changes in cases of therapy failure in a multidisciplinary approach [68,69]. The number of cases discussed in each MCC is steadily rising. This can be explained by the increasing acceptance of the multidisciplinary approach and the rising incidence of cancers due to improved diagnostics [70]. Our study shows that deep-learning models can assist the MCC’s decisions by automatized the quantification of HTL. Besides the time-saving aspect, the model could also provide decision support to physicians who have no access to a regularly held MCC.
Our study has some limitations. As mentioned above, the 3D assessment approach needs to be further evaluated on larger clinical collectives with direct comparison to 2D measurements and the impact on clinical endpoints. Another limitation of the study is that the ground truth of accuracy is based on manual segmentation of liver metastasis. Due to the sometimes pronounced, even small foci of liver metastases, manual segmentation is not perfect. To minimize this limitation, all segmentations were checked multiple times to capture all metastases (no limit on the number of lesions per patient) and to train the model as realistically as possible.
5. Conclusions
In conclusion, the deep-learning model presented shows high accuracy in 3D volumetry of NELM and determination of HTL in Gd-EOB MRI and paves the way for fully automated 3D assessment of hepatic disease. The model also provides useful (potentially prognostic) information about HTL and NELM volume and can be used to assist physicians in response evaluation and the decision-making about therapeutic success or failure comparable to the decisions of an expert multidisciplinary cancer conference.
Author Contributions
U.F.: designed research, performed the study, collected data, analyzed data, wrote the manuscript; S.X.: collected data, analyzed data, wrote the manuscript; A.H.: analyzed data, contributed important data; T.A.A.: collected data, analyzed data; F.D.: collected data, analyzed data; K.F.: collected data, analyzed data; H.J.: contributed important data; M.M.: contributed important data; H.A.: contributed important data; D.G.: contributed important data; T.D.: designed research; B.W.: designed research, analyzed data, wrote the manuscript; T.P.: designed research, performed the study, analyzed data, wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge APC funding from the German Research Foundation (DFG) and the Open Access Publication Fund of Charité-Universitätsmedizin Berlin.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Institutional Review Board of Charité Berlin (protocol code EA2/033/18, 10 April 2018).
Informed Consent Statement
Patient consent was waived due to the retrospective study design.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
Fehrenbach reports honoraria and travel expenses for scientific meetings (outside of submitted work): Bayer, Siemens, GE; Geisel reports honoraria and travel expenses for scientific meetings (outside of submitted work): Bayer, Siemens, GE; Amthauer reports lecture and travel expenses (outside of submitted work): Pfizer, Sirtex, Novartis, GE, Norgine, Terumo; Denecke reports honoraria and travel expenses for scientific meetings (outside of submitted work): Siemens, Bayer, Canon, IPSEN, Novartis, Astra Zeneca, MSD, Roche, Merck; Penzkofer was supported by Berlin Institute of Health (Clinician Scientist Grant, Platform Grant) and reports research agreements (no personal payments, outside of submitted work) with AGO, Aprea AB, ARCAGY-GINECO, Astellas Pharma Global Inc. (APGD), Astra Zeneca, Clovis Oncology, Inc., Dohme Corp, Holaira, Incyte Corporation, Karyopharm, Lion Biotechnologies, Inc., MedImmune, Merck Sharp, Millennium Pharmaceuticals, Inc., Morphotec Inc., NovoCure Ltd., PharmaMar S.A. and PharmaMar USA, Inc., Roche, Siemens Healthineers, and TESARO Inc.
References
- Maggio, I.; Manuzzi, L.; Lamberti, G.; Ricci, A.D.; Tober, N.; Campana, D. Landscape and Future Perspectives of Immunotherapy in Neuroendocrine Neoplasia. Cancers 2020, 12, 832. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.-H.; Zhang, Y.-Q.; Shi, S.-S.; Chen, Y.-J.; Yuan, X.-H.; Jiang, L.-M.; Wang, S.-M.; Ma, L.; Su-Sheng, S.; Feng, C.-Y.; et al. A nation-wide retrospective epidemiological study of gastroenteropancreatic neuroendocrine neoplasms in china. Oncotarget 2017, 8, 71699–71708. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Igarashi, H.; Nakamura, K.; Sasano, H.; Okusaka, T.; Takano, K.; Komoto, I.; Tanaka, M.; Imamura, M.; Jensen, R.T.; et al. Epidemiological trends of pancreatic and gastrointestinal neuroendocrine tumors in Japan: A nationwide survey analysis. J. Gastroenterol. 2015, 50, 58–64. [Google Scholar] [CrossRef]
- Cives, M.; Strosberg, J.R. Gastroenteropancreatic Neuroendocrine Tumors. Cancer J. Clin. 2018, 68, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.C.; Hassan, M.M.; Phan, A.T.; Dagohoy, C.G.; Leary, C.C.; Mares, J.E.; Abdalla, E.K.; Fleming, J.B.; Vauthey, J.-N.; Rashid, A.; et al. One Hundred Years after “Carcinoid”: Epidemiology of and Prognostic Factors for Neuroendocrine Tumors in 35,825 Cases in the United States. J. Clin. Oncol. 2008, 26, 3063–3072. [Google Scholar] [CrossRef] [PubMed]
- Man, D.; Wu, J.; Shen, Z.; Zhu, X. Prognosis of patients with neuroendocrine tumor: A SEER database analysis. Cancer Manag. Res. 2018, 10, 5629–5638. [Google Scholar] [CrossRef]
- Cetinkaya, R.B.; Aagnes, B.; Thiis-Evensen, E.; Tretli, S.; Bergestuen, D.S.; Hansen, S.M. Trends in Incidence of Neuroendocrine Neoplasms in Norway: A Report of 16,075 Cases from 1993 through 2010. Neuroendocrinology 2015, 104, 1–10. [Google Scholar] [CrossRef]
- Dasari, A.; Shen, C.; Halperin, D.; Zhao, B.; Zhou, S.; Xu, Y.; Shih, T.; Yao, J.C. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients with Neuroendocrine Tumors in the United States. JAMA Oncol. 2017, 3, 1335–1342. [Google Scholar] [CrossRef]
- Pavel, M.; Baudin, E.; Couvelard, A.; Krenning, E.; Öberg, K.; Steinmüller, T.; Anlauf, M.; Wiedenmann, B.; Salazar, R. ENETS Consensus Guidelines for the Management of Patients with Liver and Other Distant Metastases from Neuroendocrine Neoplasms of Foregut, Midgut, Hindgut, and Unknown Primary. Neuroendocrinology 2012, 95, 157–176. [Google Scholar] [CrossRef]
- Rindi, G.; Wiedenmann, B. Neuroendocrine neoplasia of the gastrointestinal tract revisited: Towards precision medicine. Nat. Rev. Endocrinol. 2020, 16, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.; Buell, J.F.; Kandil, E. Surgical treatment of liver metastases in patients with neuroendocrine tumors. Ann. Transl. Med. 2013, 1, 6. [Google Scholar]
- Schwartz, L.H.; Litière, S.; De Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Update and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C.N.; Nagasaka, T.; Goel, A.; Scharf, I.; Grabowski, P.; Sosnowski, A.; Schmitt-Gräff, A.; Boland, C.R.; Arnold, R.; Blum, H.E. Molecular characteristics and predictors of survival in patients with malignant neuroendocrine tumors. Int. J. Cancer 2008, 123, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Dromain, C.; De Baere, T.; Lumbroso, J.; Caillet, H.; Laplanche, A.; Boige, V.; Ducreux, M.; Duvillard, P.; Elias, D.; Schlumberger, M.; et al. Detection of Liver Metastases from Endocrine Tumors: A Prospective Comparison of Somatostatin Receptor Scintigraphy, Computed Tomography, and Magnetic Resonance Imaging. J. Clin. Oncol. 2005, 23, 70–78. [Google Scholar] [CrossRef]
- Sundin, A.; Arnold, R.; Baudin, E.; Cwikla, J.B.; Eriksson, B.; Fanti, S.; Fazio, N.; Giammarile, F.; Hicks, R.J.; Kjaer, A.; et al. ENETS Consensus Guidelines for the Standards of Care in Neuroendocrine Tumors: Radiological, Nuclear Medicine and Hybrid Imaging. Neuroendocrinology 2017, 105, 212–244. [Google Scholar] [CrossRef]
- Ronot, M.; Clift, A.K.; Baum, R.P.; Singh, A.; Kulkarni, H.R.; Frilling, A.; Vilgrain, V. Morphological and Functional Imaging for Detecting and Assessing the Resectability of Neuroendocrine Liver Metastases. Neuroendocrinology 2018, 106, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Vreugdenburg, T.D.; Ma, N.; Duncan, J.K.; Riitano, D.; Cameron, A.L.; Maddern, G.J. Comparative diagnostic accuracy of hepatocyte-specific gadoxetic acid (Gd-EOB-DTPA) enhanced MR imaging and contrast enhanced CT for the detection of liver metastases: A systematic review and meta-analysis. Int. J. Colorectal Dis. 2016, 31, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Giesel, F.; Kratochwil, C.; Mehndiratta, A.; Wulfert, S.; Moltz, J.; Zechmann, C.; Kauczor, H.; Haberkorn, U.; Ley, S. Comparison of neuroendocrine tumor detection and characterization using DOTATOC-PET in correlation with contrast enhanced CT and delayed contrast enhanced MRI. Eur. J. Radiol. 2012, 81, 2820–2825. [Google Scholar] [CrossRef]
- Karaosmanoglu, A.D.; Onur, M.R.; Ozmen, M.N.; Akata, D.; Karcaaltincaba, M. Magnetic Resonance Imaging of Liver Metastasis. Semin. Ultrasound CT MRI 2016, 37, 533–548. [Google Scholar] [CrossRef]
- Feuerlein, S.; Gupta, R.T.; Boll, D.T.; Merkle, E.M. Hepatocellular MR contrast agents: Enhancement characteristics of liver parenchyma and portal vein after administration of gadoxetic acid in comparison to gadobenate dimeglumine. Eur. J. Radiol. 2012, 81, 2037–2041. [Google Scholar] [CrossRef]
- D’Assignies, G.; Fina, P.; Bruno, O.; Vullierme, M.-P.; Tubach, F.; Paradis, V.; Sauvanet, A.; Ruszniewski, P.; Vilgrain, V. High Sensitivity of Diffusion-weighted MR Imaging for the Detection of Liver Metastases from Neuroendocrine Tumors: Comparison with T2-weighted and Dynamic Gadolinium-enhanced MR Imaging. Radiology 2013, 268, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Sankowski, A.J.; Ćwikla, J.B.; Nowicki, M.L.; Chaberek, S.; Pech, M.; Lewczuk, A.; Walecki, J. The clinical value of MRI using single-shot echoplanar DWI to identify liver involvement in patients with advanced gastroenteropancreatic-neuroendocrine tumors (GEP-NETs), compared to FSE T2 and FFE T1 weighted image after i.v. Gd-EOB-DTPA contrast enhancement. Med. Sci. Monit. 2012, 18, MT33–MT40. [Google Scholar] [CrossRef][Green Version]
- Minon, M.; Soriano, C.; Morland, D.; Walter, T.; Lepage, C.; Tabarin, A.; DeBlock, M.; Rousset, P.; Barbe, C.; Hoeffel, C.; et al. Prospective comparison of whole-body MRI with diffusion-weighted and conventional imaging for the follow-up of neuroendocrine tumors. Endocrine 2019, 67, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Hayoz, R.; Vietti-Violi, N.; Duran, R.; Knebel, J.-F.; LeDoux, J.-B.; Dromain, C. The combination of hepatobiliary phase with Gd-EOB-DTPA and DWI is highly accurate for the detection and characterization of liver metastases from neuroendocrine tumor. Eur. Radiol. 2020, 30, 6593–6602. [Google Scholar] [CrossRef]
- Danet, I.-M.; Semelka, R.C.; Leonardou, P.; Braga, L.; Vaidean, G.; Woosley, J.T.; Kanematsu, M. Spectrum of MRI Appearances of Untreated Metastases of the Liver. Am. J. Roentgenol. 2003, 181, 809–817. [Google Scholar] [CrossRef]
- Khosa, F.; Khan, A.N.; Eisenberg, R.L. Hypervascular Liver Lesions on MRI. Am. J. Roentgenol. 2011, 197, W204–W220. [Google Scholar] [CrossRef]
- Luersen, G.F.; Wei, W.; Tamm, E.P.; Bhosale, P.R.; Szklaruk, J. Evaluation of Magnetic Resonance (MR) Biomarkers for Assessment of Response with Response Evaluation Criteria in Solid Tumors: Comparison of the Measurements of Neuroendocrine Tumor Liver Metastases (NETLM) with Various MR Sequences and at Multiple Phases of Contrast Administration. J. Comput. Assist. Tomogr. 2016, 40, 717–722. [Google Scholar] [CrossRef]
- Liang, H.; Tsui, B.Y.; Ni, H.; Valentim, C.C.S.; Baxter, S.L.; Liu, G.; Cai, W.; Kermany, D.S.; Sun, X.; Chen, J.; et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med. 2019, 25, 433–438. [Google Scholar] [CrossRef]
- Rauschecker, A.M.; Rudie, J.D.; Xie, L.; Wang, J.; Duong, M.T.; Botzolakis, E.J.; Kovalovich, A.M.; Egan, J.; Cook, T.C.; Bryan, R.N.; et al. Artificial Intelligence System Approaching Neuroradiologist-level Differential Diagnosis Accuracy at Brain MRI. Radiology 2020, 295, 626–637. [Google Scholar] [CrossRef]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: An open, web-based, international, diagnostic study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, C.J.P.; Jiang, B.; Chen, J.; Song, J.; Liu, Z.; He, Z.; Krittanawong, C.; Fang, P.-H.; Ming, W.-K. Artificial Intelligence Versus Clinicians in Disease Diagnosis: Systematic Review. JMIR Med. Inform. 2019, 7, e10010. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Sumkin, J.H.; Berg, W.A.; Carter, G.J.; Bandos, A.I.; Chough, D.M.; Ganott, M.A.; Hakim, C.M.; Kelly, A.E.; Zuley, M.L.; Houshmand, G.; et al. Diagnostic Performance of MRI, Molecular Breast Imaging, and Contrast-enhanced Mammography in Women with Newly Diagnosed Breast Cancer. Radiology 2019, 293, 531–540. [Google Scholar] [CrossRef]
- Azer, S.A. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: A systematic review. World J. Gastrointest. Oncol. 2019, 11, 1218–1230. [Google Scholar] [CrossRef]
- Zhou, L.-Q.; Wang, J.-Y.; Yu, S.-Y.; Wu, G.-G.; Wei, Q.; Deng, Y.-B.; Wu, X.-L.; Cui, X.-W.; Dietrich, C.F. Artificial intelligence in medical imaging of the liver. World J. Gastroenterol. 2019, 25, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Kaya, B.; Koc, Z. Diffusion-weighted MRI and optimal b-value for characterization of liver lesions. Acta Radiol. 2014, 55, 532–542. [Google Scholar] [CrossRef]
- Nolden, M.; Zelzer, S.; Seitel, A.; Wald, D.; Müller, M.; Franz, A.M.; Maleike, D.; Fangerau, M.; Baumhauer, M.; Maier-Hein, L.; et al. The Medical Imaging Interaction Toolkit: Challenges and advances. Int. J. Comput. Assist. Radiol. Surg. 2013, 8, 607–620. [Google Scholar] [CrossRef]
- MIC-DKFZ nnUNet. Available online: https://github.com/MIC-DKFZ/nnUNet (accessed on 1 October 2020).
- Isensee, F.; Jäger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. Automated Design of Deep Learning Methods for Biomedical Image Segmentation. arXiv 2019, arXiv:1904.08128v2. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Goehler, A.; Hsu, T.-M.H.; Lacson, R.; Gujrathi, I.; Hashemi, R.; Chlebus, G.; Szolovits, P.; Khorasani, R. Three-Dimensional Neural Network to Automatically Assess Liver Tumor Burden Change on Consecutive Liver MRIs. J. Am. Coll. Radiol. 2020, 17, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Larouche, V.; Akirov, A.; AlShehri, S.; Ezzat, S. Management of Small Bowel Neuroendocrine Tumors. Cancers 2019, 11, 1395. [Google Scholar] [CrossRef]
- Akirov, A.; Larouche, V.; AlShehri, S.; Asa, S.L.; Ezzat, S. Treatment Options for Pancreatic Neuroendocrine Tumors. Cancers 2019, 11, 828. [Google Scholar] [CrossRef]
- Tsurusaki, M.; Sofue, K.; Murakami, T. Current evidence for the diagnostic value of gadoxetic acid-enhanced magnetic resonance imaging for liver metastasis. Hepatol. Res. 2016, 46, 853–861. [Google Scholar] [CrossRef]
- Morse, B.; Jeong, D.; Thomas, K.; Diallo, D.; Strosberg, J.R. Magnetic Resonance Imaging of Neuroendocrine Tumor Hepatic Metastases. Pancreas 2017, 46, 1219–1224. [Google Scholar] [CrossRef]
- Tirumani, S.H.; Jagannathan, J.P.; Braschi-Amirfarzan, M.; Qin, L.; Balthazar, P.; Ramaiya, N.H.; Shinagare, A.B. Value of hepatocellular phase imaging after intravenous gadoxetate disodium for assessing hepatic metastases from gastroenteropancreatic neuroendocrine tumors: Comparison with other MRI pulse sequences and with extracellular agent. Abdom. Radiol. 2018, 43, 2329–2339. [Google Scholar] [CrossRef]
- Grieser, C.; Denecke, T.; Rothe, J.-H.; Geisel, D.; Stelter, L.; Walter, T.C.; Seehofer, D.; Steffen, I.G. Gd-EOB enhanced MRI T1-weighted 3D-GRE with and without elevated flip angle modulation for threshold-based liver volume segmentation. Acta Radiol. 2014, 56, 1419–1427. [Google Scholar] [CrossRef]
- Kahn, J.; Posch, H.; Steffen, I.G.; Geisel, D.; Bauknecht, C.; Liebig, T.; Denecke, T. Is There Long-term Signal Intensity Increase in the Central Nervous System on T1-weighted Images after MR Imaging with the Hepatospecific Contrast Agent Gadoxetic Acid? A Cross-sectional Study in 91 Patients. Radiology 2017, 282, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Lestra, T.; Kanagaratnam, L.; Mulé, S.; Janvier, A.; Brixi, H.; Cadiot, G.; Dohan, A.; Hoeffel, C. Measurement variability of liver metastases from neuroendocrine tumors on different magnetic resonance imaging sequences. Diagn. Interv. Imaging 2018, 99, 73–81. [Google Scholar] [CrossRef]
- Lavelle, L.; O’Neill, A.; McMahon, C.; Cantwell, C.; Heffernan, E.; Malone, D.; Daly, L.; Skehan, S. Is diffusion-weighted MRI sufficient for follow-up of neuroendocrine tumour liver metastases? Clin. Radiol. 2016, 71, 863–868. [Google Scholar] [CrossRef]
- Kaye, E.A.; Cornelis, F.H.; Petre, E.N.; Tyagi, N.; Shady, W.; Shi, W.; Zhang, Z.; Solomon, S.B.; Sofocleous, C.T.; Durack, J.C. Volumetric 3D assessment of ablation zones after thermal ablation of colorectal liver metastases to improve prediction of local tumor progression. Eur. Radiol. 2019, 29, 2698–2705. [Google Scholar] [CrossRef]
- Sakakibara, M.; Ohkawa, K.; Katayama, K.; Imanaka, K.; Ishihara, A.; Hasegawa, N.; Kimura, H. Three-Dimensional Registration of Images Obtained before and after Radiofrequency Ablation of Hepatocellular Carcinoma to Assess Treatment Adequacy. Am. J. Roentgenol. 2014, 202, W487–W495. [Google Scholar] [CrossRef]
- Wang, L.; Tan, J.; Ge, Y.; Tao, X.; Cui, Z.; Fei, Z.; Lu, J.; Zhang, H.; Pan, Z. Assessment of liver metastases radiomic feature reproducibility with deep-learning-based semi-automatic segmentation software. Acta Radiol. 2021, 62, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Solis-Hernandez, M.P.; Del Valle, A.F.; Carmona-Bayonas, A.; Garcia-Carbonero, R.; Custodio, A.; Benavent, M.; Gordoa, T.A.; Nuñez-Valdovino, B.; Canovas, M.S.; Matos, I.; et al. Evaluating radiological response in pancreatic neuroendocrine tumours treated with sunitinib: Comparison of Choi versus RECIST criteria (CRIPNET_ GETNE1504 study). Br. J. Cancer 2019, 121, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Hamm, C.A.; Wang, C.J.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Duncan, J.S.; Weinreb, J.C.; Chapiro, J.; et al. Deep learning for liver tumor diagnosis part I: Development of a convolutional neural network classifier for multi-phasic MRI. Eur. Radiol. 2019, 29, 3338–3347. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.J.; Hamm, C.A.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Weinreb, J.C.; Duncan, J.S.; Chapiro, J.; et al. Deep learning for liver tumor diagnosis part II: Convolutional neural network interpretation using radiologic imaging features. Eur. Radiol. 2019, 29, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, J.; Ng, C.W.; Ma, Y.; Mo, S.; Fong, E.L.S.; Xing, J.; Song, Z.; Xie, Y.; Si, K.; et al. Deep learning enables automated scoring of liver fibrosis stages. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Gehan, E.A.; Tefft, M.C. Will There Be Resistance to the RECIST (Response Evaluation Criteria in Solid Tumors)? J. Natl. Cancer Inst. 2000, 92, 179–181. [Google Scholar] [CrossRef]
- Lamarca, A.; Barriuso, J.; Kulke, M.; Borbath, I.; Lenz, H.-J.; Raoul, J.L.; Meropol, N.J.; Lombard-Bohas, C.; Posey, J.; Faivre, S.; et al. Determination of an optimal response cut-off able to predict progression-free survival in patients with well-differentiated advanced pancreatic neuroendocrine tumours treated with sunitinib: An alternative to the current RECIST-defined response. Br. J. Cancer 2017, 118, 181–188. [Google Scholar] [CrossRef]
- Lamarca, A.; Crona, J.; Ronot, M.; Opalinska, M.; Lopez, C.L.; Pezzutti, D.; Najran, P.; Carvhalo, L.; Bezerra, R.O.F.; Borg, P.; et al. Value of Tumor Growth Rate (TGR) as an Early Biomarker Predictor of Patients’ Outcome in Neuroendocrine Tumors (NET)—The GREPONET Study. Oncology 2019, 24, e1082. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Kuhl, C.K.; Engelke, H.; Bettermann, G.; Keil, S. Factors That Drive Heterogeneity of Response-to-Treatment of Different Metastatic Deposits Within the Same Patients as Measured by RECIST 1.1 Analyses. Acad. Radiol. 2020. [Google Scholar] [CrossRef]
- Rothe, J.H.; Steffen, I.G.; Lehmkuhl, L.; Grieser, C.; Mußler, A.; Schnapauff, D.; Stelter, L.; Denecke, T. Volume Measurement of Liver Metastases Using Multidetector Computed Tomography: Comparison of Lesion Diameter and Volume segmentation—A Phantom Study. In RöFo—Fortschritte auf dem Gebiet der Röntgenstrahlen und der bildgebenden Verfahren; Thieme: Stuttgart, Germany, 2010; Volume 182, pp. 1082–1090. [Google Scholar] [CrossRef]
- Palazzo, M.; Lombard-Bohas, C.; Cadiot, G.; Matysiak-Budnik, T.; Rebours, V.; Vullierme, M.-P.; Couvelard, A.; Hentic, O.; Ruszniewski, P. Ki67 proliferation index, hepatic tumor load, and pretreatment tumor growth predict the antitumoral efficacy of lanreotide in patients with malignant digestive neuroendocrine tumors. Eur. J. Gastroenterol. Hepatol. 2013, 25, 232–238. [Google Scholar] [CrossRef]
- Beleù, A.; Rizzo, G.; De Robertis, R.; Drudi, A.; Aluffi, G.; Longo, C.; Sarno, A.; Cingarlini, S.; Capelli, P.; Landoni, L.; et al. Liver Tumor Burden in Pancreatic Neuroendocrine Tumors: CT Features and Texture Analysis in the Prediction of Tumor Grade and 18F-FDG Uptake. Cancers 2020, 12, 1486. [Google Scholar] [CrossRef] [PubMed]
- Cieciera, M.; Kratochwil, C.; Moltz, J.; Kauczor, H.-U.; Holland-Letz, T.; Choyke, P.; Mier, W.; Haberkorn, U.; Giesel, F.L. Semi-automatic 3D-volumetry of liver metastases from neuroendocrine tumors to improve combination therapy with 177Lu-DOTATOC and 90Y-DOTATOC. Diagn. Interv. Radiol. 2016, 22, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.K. RECIST Needs Revision: A Wake-up Call for Radiologists. Radiology 2019, 292, 110–111. [Google Scholar] [CrossRef]
- Keating, N.L.; Landrum, M.B.; Lamont, E.B.; Bozeman, S.R.; Shulman, L.N.; McNeil, B.J. Tumor Boards and the Quality of Cancer Care. J. Natl. Cancer Inst. 2012, 105, 113–121. [Google Scholar] [CrossRef]
- Croke, J.M.; El-Sayed, S. Multidisciplinary Management of Cancer Patients: Chasing a Shadow or Real Value? An Overview of the Literature. Curr. Oncol. 2012, 19, 232–238. [Google Scholar] [CrossRef]
- Hofland, J.; Kaltsas, G.; De Herder, W.W. Advances in the Diagnosis and Management of Well-Differentiated Neuroendocrine Neoplasms. Endocr. Rev. 2020, 41, 371–403. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).