Decoding LncRNAs
Abstract
Simple Summary
Abstract
1. Introduction
2. LncRNAs Structure-Functions
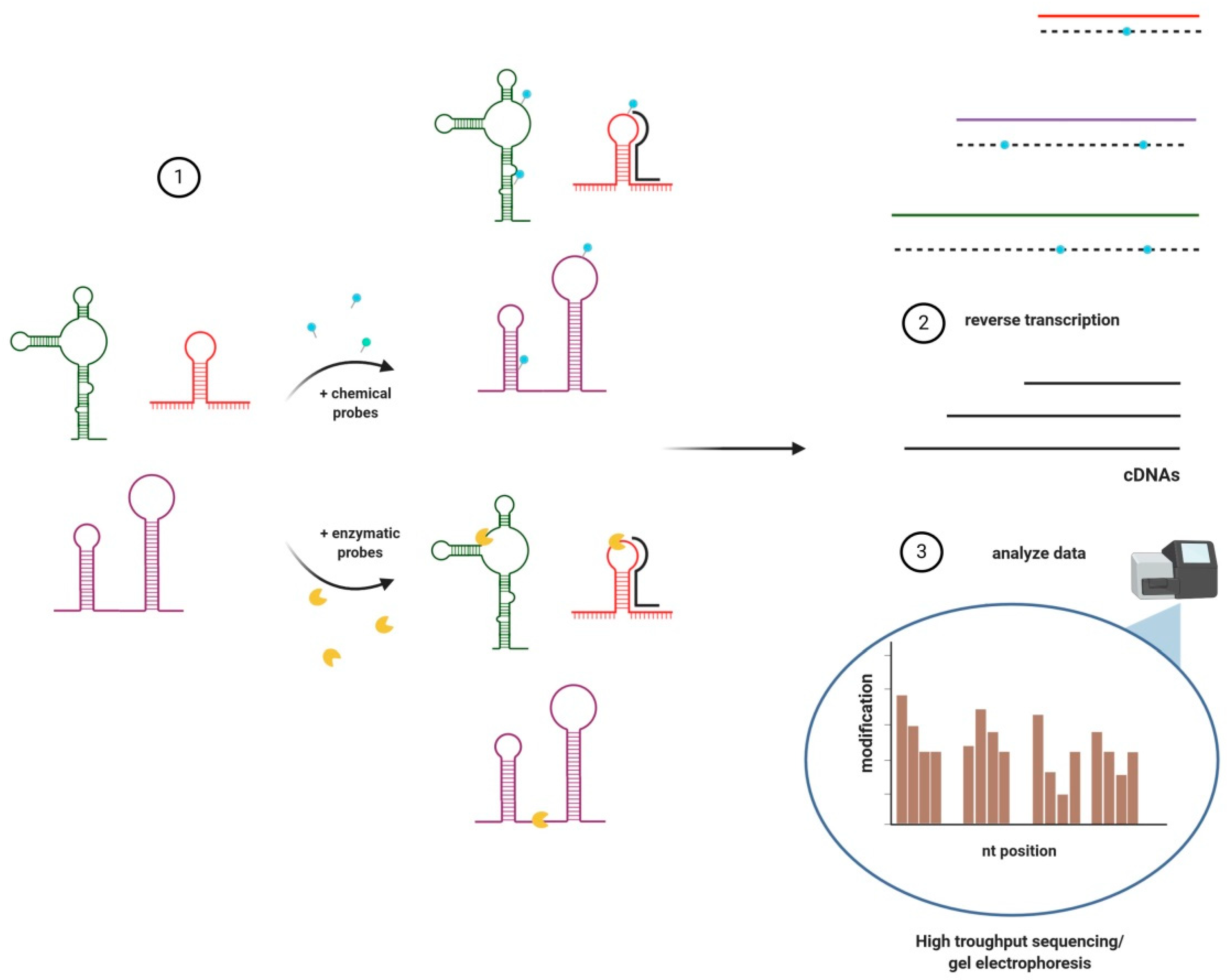
3. Structural Analyses of lncRNAs
4. Identification of lncRNAs’ Binding Partners
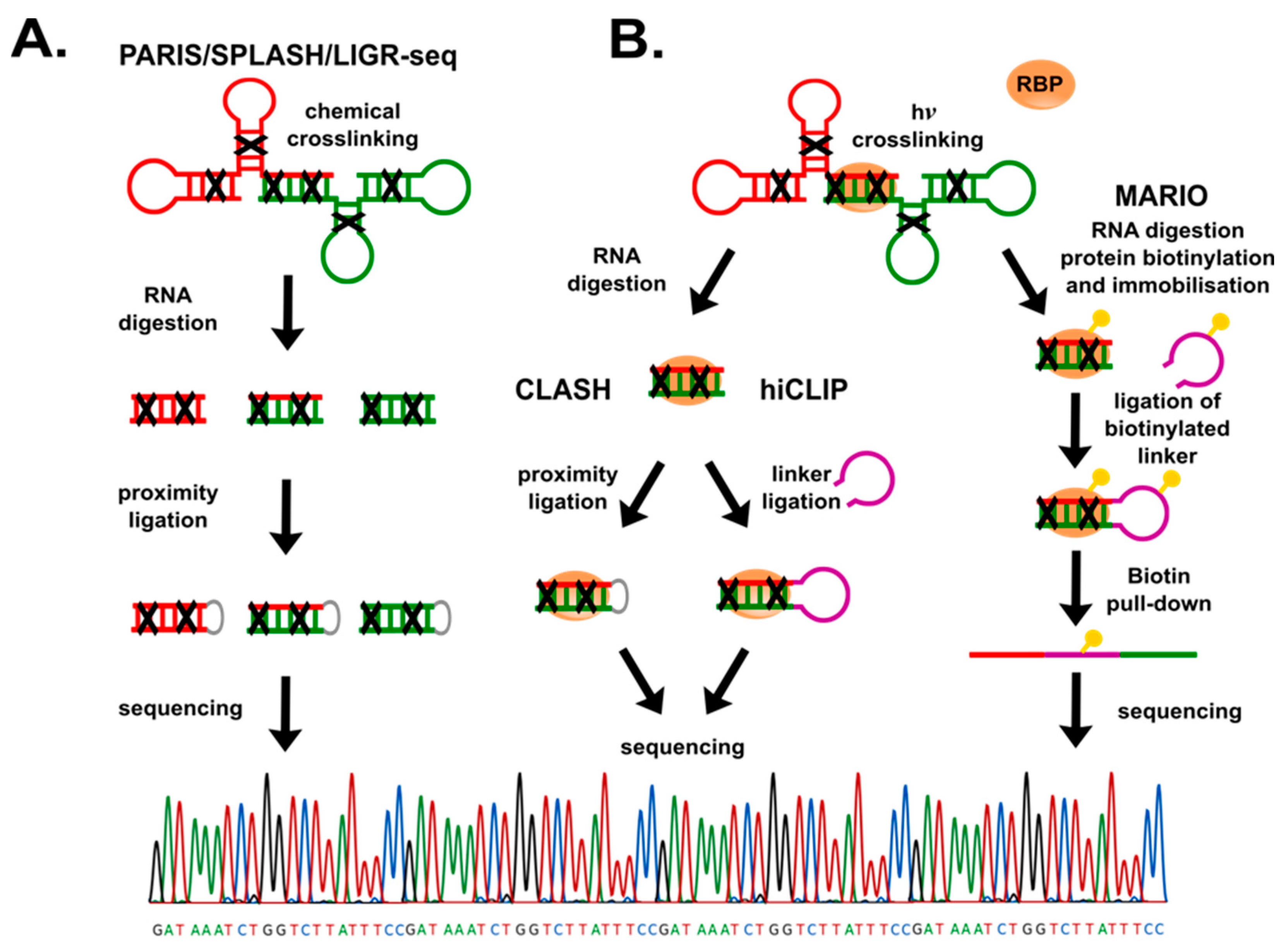
4.1. RNA-RNA Interactions
4.2. RNA-DNA Interactions
4.3. RNA-Protein Interactions
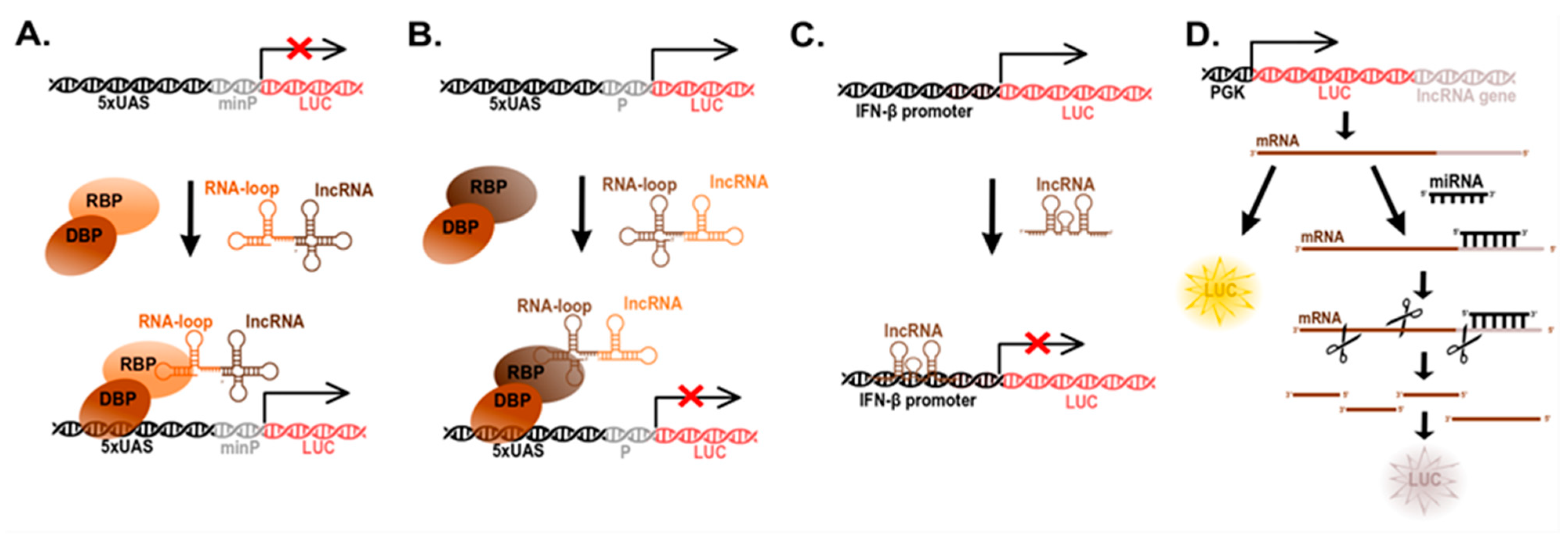
5. Determination of lncRNA Function (Reporter Systems)
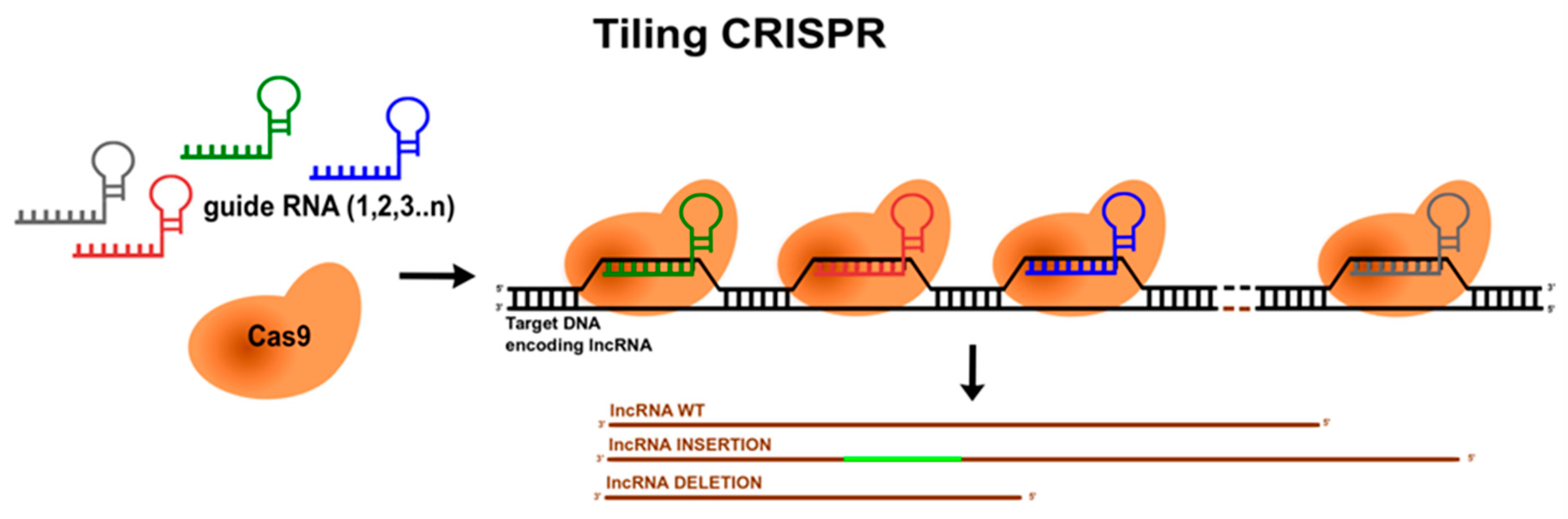
6. Mapping lncRNAs’ Functional Domains
7. LncRNAs Databases and Bioinformatic Tools
8. LncRNAs and Disease
9. Applications of lncRNAs
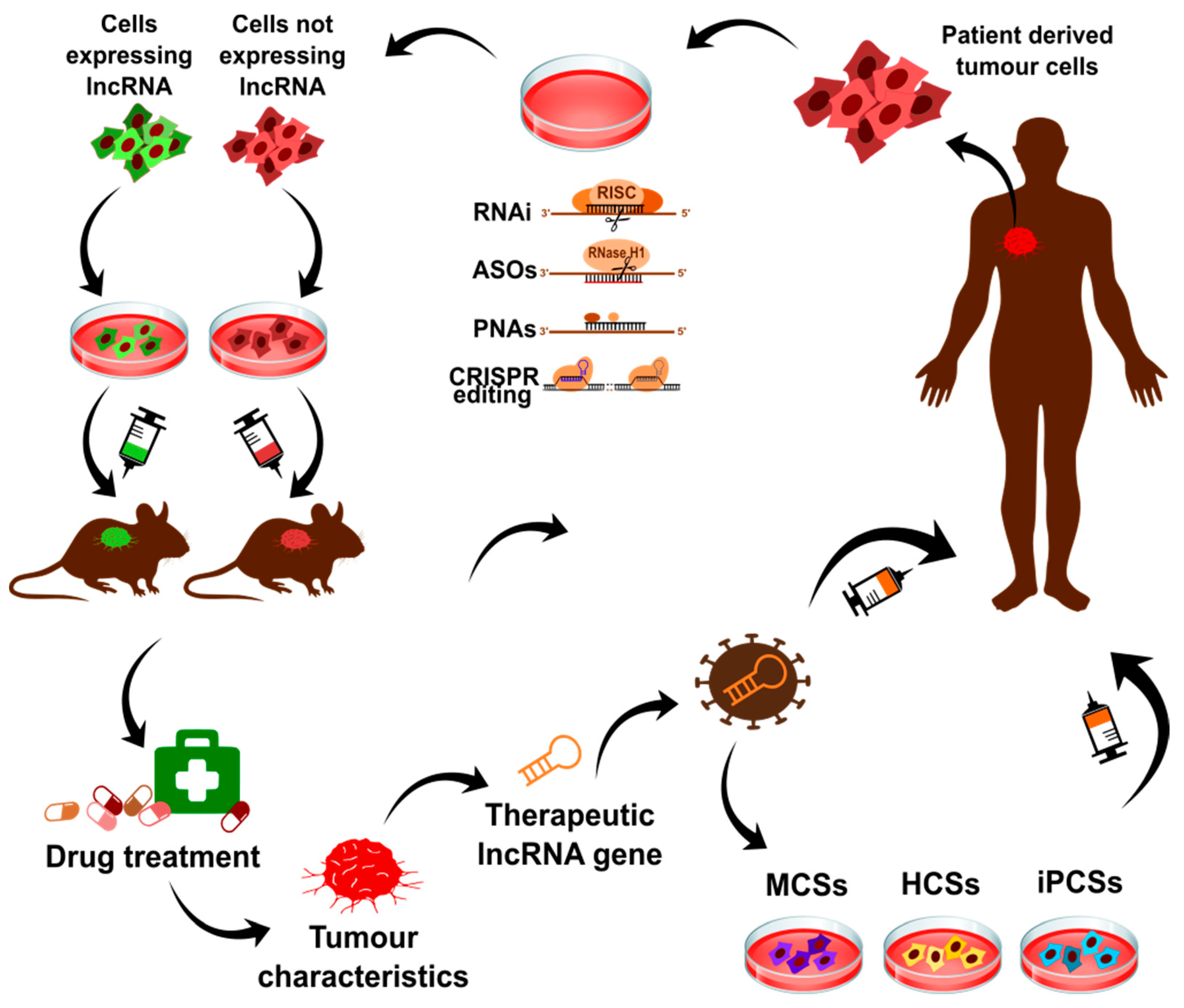
9.1. Diagnostic Biomarkers in Cancers and Targets for Therapy
9.2. Silencing of Single Chromosome/Trisomy Effects
9.3. Tissue/Muscle Regeneration
10. Discussion
11. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Saghafi, T.; Ali Taheri, R.; Parkkila, S.; Emameh, R.Z. Phytochemicals as modulators of long non-coding RNAs and inhibitors of cancer-related carbonic anhydrases. Int. J. Mol. Sci. 2019, 20, 2939. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long non-coding RNAs in the regulation of gene expression: Physiology and disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Niemczyk, M.; Ito, Y.; Huddleston, J.; Git, A.; Abu-Amero, S.; Caldas, C.; Moore, G.E.; Stojic, L.; Murrell, A. Imprinted chromatin around DIRAS3 regulates alternative splicing of GNG12-AS1, a long noncoding RNA. Am. J. Hum. Genet. 2013, 93, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Song, J.; Qiao, M.; Zhao, X.; Li, R.; Jiao, J.; Sun, Q. Long noncoding RNA expression profile and functional analysis in psoriasis. Mol. Med. Rep. 2019, 49, 3421–3430. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Mongelli, A.; Martelli, F.; Farsetti, A.; Gaetano, C. The dark that matters: Long noncoding RNAs as master regulators of cellular metabolism in noncommunicable diseases. Front. Physiol. 2019, 10, 369. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, J.; Guo, X.; She, J.; Liu, Y. Long non-coding RNA PVT1, A molecular sponge for miR-149, contributes aberrant metabolic dysfunction and inflammation in IL-1β-simulated osteoarthritic chondrocytes. Biosci. Rep. 2018, 38, BSR20180576. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, N.; Wang, C.; Zhao, H.; Gu, Y. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle 2018, 17, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, J.; Wu, C.; Chen, W.; Lin, R.; Zhou, Y.; Huang, X. The LINC01138 interacts with PRMT5 to promote SREBP1-mediated lipid desaturation and cell growth in clear cell renal cell carcinoma. Biochem. Biophys. Res. Commun. 2018, 507, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Polycarpou-Schwarz, M.; Groß, M.; Mestdagh, P.; Schott, J.; Grund, S.E.; Hildenbrand, C.; Rom, J.; Aulmann, S.; Sinn, H.P.; Vandesompele, J.; et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene 2018, 37, 4750–4768. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef]
- Li, J.; Chen, C.; Ma, X.; Geng, G.; Liu, B.; Zhang, Y.; Zhang, S.; Zhong, F.; Liu, C.; Yin, Y.; et al. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat. Commun. 2016, 7, 11730. [Google Scholar] [CrossRef]
- Legnini, I.; Morlando, M.; Mangiavacchi, A.; Fatica, A.; Bozzoni, I. A Feedforward Regulatory Loop between HuR and the Long Noncoding RNA linc-MD1 Controls Early Phases of Myogenesis. Mol. Cell 2014, 53, 506–514. [Google Scholar] [CrossRef]
- Kok, F.O.; Baker, A.H. The function of long non-coding RNAs in vascular biology and disease. Vasc. Pharmacol. 2019, 114, 23–30. [Google Scholar] [CrossRef]
- Pang, Y.; Mao, C.; Liu, S. Encoding activities of non-coding RNAs. Theranostics 2018, 8, 2496–2507. [Google Scholar] [CrossRef]
- Van Solingen, C.; Scacalossi, K.R.; Moore, K.J. Long noncoding RNAs in lipid metabolism. Curr. Opin. Lipidol. 2018, 29, 224–232. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.L.; Yang, L.; Chen, L.L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 2015, 4, e07540. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yang, B.; Chen, B.J.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Ragan, C.; Goodall, G.J.; Shirokikh, N.E.; Preiss, T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019, 9, 2048. [Google Scholar] [CrossRef] [PubMed]
- Sleutels, F.; Zwart, R.; Barlow, D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002, 415, 810–813. [Google Scholar] [CrossRef]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-Lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007, 2, e845. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, H.; Zhu, L.; Hao, B.; Zhang, W.; Hua, J.; Gu, H.; Jin, W.; Zhang, G. Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget 2016, 7, 82770–82782. [Google Scholar] [CrossRef] [PubMed]
- Imamura, K.; Takaya, A.; Ishida, Y.; Fukuoka, Y.; Taya, T.; Nakaki, R.; Kakeda, M.; Imamachi, N.; Sato, A.; Yamada, T.; et al. Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J. 2018, 37, e97723. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Ishida, E.; Matsumoto, S.; Shibusawa, N.; Okada, S.; Monden, T.; Satoh, T.; Yamada, M.; Mori, M. A liver X receptor (LXR)-β alternative splicing variant (LXRBSV) acts as an RNA co-activator of LXR-β. Biochem. Biophys. Res. Commun. 2009, 390, 1260–1265. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Girnita, L.; Varani, G.; Calin, G.A. Decrypting noncoding RNA interactions, structures, and functional networks. Genome Res. 2019, 29, 1377–1388. [Google Scholar] [CrossRef]
- Qian, X.; Zhao, J.; Yeung, P.Y.; Zhang, Q.C.; Kwok, C.K. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem. Sci. 2019, 44, 33–52. [Google Scholar] [CrossRef]
- Busan, S.; Weeks, K.M. Visualization of RNA structure models within the Integrative Genomics Viewer. RNA 2017, 23, 1012–1018. [Google Scholar] [CrossRef]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.M.; Pellequer, J.L.; Annibale, P.; Pessey, O.; Inga, A.; Chillón, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell 2019, 75, 982–995.e9. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Qu, K.; Ouyang, Z.; Kertesz, M.; Li, J.; Tibshirani, R.; Makino, D.L.; Nutter, R.C.; Segal, E.; Chang, H.Y. Genome-wide Measurement of RNA Folding Energies. Mol. Cell 2012, 48, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Rouskin, S.; Zubradt, M.; Washietl, S.; Kellis, M.; Weissman, J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 2014, 505, 701–705. [Google Scholar] [CrossRef]
- Tapsin, S.; Sun, M.; Shen, Y.; Zhang, H.; Lim, X.N.; Susanto, T.T.; Yang, S.L.; Zeng, G.S.; Lee, J.; Lezhava, A.; et al. Genome-wide identification of natural RNA aptamers in prokaryotes and eukaryotes. Nat. Commun. 2018, 9, 1289. [Google Scholar] [CrossRef]
- Zhou, K.I.; Parisien, M.; Dai, Q.; Liu, N.; Diatchenko, L.; Joseph, R.; Pan, T.; Program, S.T.; Division, B.S. Hairpin Predisposes Its Conformation To Protein Binding. J. Mol. Biol. 2017, 428, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Pintacuda, G.; Young, A.N.; Cerase, A. Function by structure: Spotlights on xist long non-coding RNA. Front. Mol. Biosci. 2017, 4, 90. [Google Scholar] [CrossRef]
- Amaral, P.P.; Clark, M.B.; Gascoigne, D.K.; Dinger, M.E.; Mattick, J.S. LncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res. 2011, 39 (Suppl. 1), 146–151. [Google Scholar] [CrossRef] [PubMed]
- Korostowski, L.; Raval, A.; Breuer, G.; Engel, N. Enhancer-driven chromatin interactions during development promote escape from silencing by a long non-coding RNA. Epigenetics Chromatin 2011, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Vallot, C.; Patrat, C.; Collier, A.J.; Huret, C.; Casanova, M.; Liyakat Ali, T.M.; Tosolini, M.; Frydman, N.; Heard, E.; Rugg-Gunn, P.J.; et al. XACT Noncoding RNA Competes with XIST in the Control of X Chromosome Activity during Human Early Development. Cell Stem Cell 2017, 20, 102–111. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. ALUstrious long ncRNAs and their roles in shortening mRNA half-lives. Cell Cycle 2011, 10, 1882–1883. [Google Scholar] [CrossRef]
- Filbin, M.E.; Kieft, J.S. Towards a structural understanding of IRES RNA function. Bone 2009, 19, 267–276. [Google Scholar] [CrossRef]
- Xia, X.; Holcik, M. Strong eukaryotic IRESs have weak secondary structure. PLoS ONE 2009, 4, e4136. [Google Scholar] [CrossRef]
- Fernández, N.; Fernandez-Miragall, O.; Ramajo, J.; García-Sacristán, A.; Bellora, N.; Eyras, E.; Briones, C.; Martínez-Salas, E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res. 2011, 39, 8572–8585. [Google Scholar] [CrossRef]
- May, J.; Johnson, P.; Saleem, H.; Simon, A.E. A Sequence-Independent, Unstructured Internal Ribosome Entry Site Is Responsible for Internal Expression of the Coat Protein of Turnip Crinkle Virus. J. Virol. 2017, 91, e02421-16. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins and their ‘Mysterious’ (meta)physics. Front. Phys. 2019, 7, 8–23. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012, 40, 5034–5051. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef]
- Wang, Y.; Zhong, Y.; Zhou, Y.; Tanaseichuk, O.; Li, Z.; Zhao, J.C. Identification of a Xist silencing domain by Tiling CRISPR. Sci. Rep. 2019, 9, 2408. [Google Scholar] [CrossRef]
- Smola, M.J.; Christy, T.W.; Inoue, K.; Nicholson, C.O.; Friedersdorf, M.; Keene, J.D.; Lee, D.M.; Calabrese, J.M.; Weeks, K.M. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA 2016, 113, 10322–10327. [Google Scholar] [CrossRef]
- Duszczyk, M.M.; Wutz, A.; Rybin, V.; Sattler, M. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 2011, 17, 1973–1982. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Guo, J.K.; Wei, Y.; Dou, D.R.; Zarnegar, B.; Ma, Q.; Li, R.; Zhao, Y.; Liu, F.; Choudhry, H.; et al. Structural modularity of the XIST ribonucleoprotein complex. Nat. Commun. 2020, 11, 6163. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, A.; Rose, D.; Fasold, M.; Reiche, K.; Stadler, P.F. Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 2015, 21, 801–812. [Google Scholar] [CrossRef]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of Long Noncoding RNA Evolution Derived from Direct Comparison of Transcriptomes in 17 Species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Orosz, F.; Ovádi, J. Proteins without 3D structure: Definition, detection and beyond. Bioinformatics 2011, 27, 1449–1454. [Google Scholar] [CrossRef]
- Vishwanath, S.; de Brevern, A.G.; Srinivasan, N. Same but not alike: Structure, flexibility and energetics of domains in multi-domain proteins are influenced by the presence of other domains. PLoS Comput. Biol. 2018, 14, e1006008. [Google Scholar] [CrossRef]
- Strobel, E.J.; Yu, A.M.; Lucks, J.B. High-throughput determination of RNA structures. Nat. Rev. Genet. 2018, 19, 615–634. [Google Scholar] [CrossRef]
- Ziehler, W.A.; Engelke, D.R. Probing RNA Structure with Chemical Reagents and Enzymes. Curr. Protoc. Nucleic Acid Chem. 2000, 6.1.1–6.1.21. [Google Scholar] [CrossRef]
- Weeks, K.M. Advances in RNA secondary and tertiary structure analysis by chemical probing. Curr. Opin. Struct. Biol. 2010, 20, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Martens, L.; Rühle, F.; Stoll, M. LncRNA secondary structure in the cardiovascular system. Non-Coding RNA Res. 2017, 2, 137–142. [Google Scholar] [CrossRef]
- Novikova, I.V.; Dharap, A.; Hennelly, S.P.; Sanbonmatsu, K.Y. 3S: Shotgun secondary structure determination of long non-coding RNAs. Methods 2013, 63, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Somarowthu, S.; Legiewicz, M.; Chilló, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, E.J.; Hennelly, S.P.; Novikova, I.V.; Irwin, J.A.; Dean, C.; Sanbonmatsu, K.Y. COOLAIR Antisense RNAs Form Evolutionarily Conserved Elaborate Secondary Structures. Cell Rep. 2016, 16, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Somarowthu, S.; Pyle, A.M. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017, 13, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Hennelly, S.; Doyle, B.; Gulati, A.A.; Novikova, I.V.; Sanbonmatsu, K.Y.; Boyer, L.A.; Xue, Z.; Hennelly, S.; Doyle, B.; et al. Article A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardiovascular Lineage Article A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardio. Mol. Cell 2016, 64, 37–50. [Google Scholar] [CrossRef]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018, 46, 3742–3752. [Google Scholar] [CrossRef] [PubMed]
- Underwood, J.G.; Uzilov, A.V.; Katzman, S.; Onodera, C.S.; Mainzer, J.E.; Mathews, D.H.; Lowe, T.M.; Salama, S.R.; Haussler, D. FragSeq: Transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods 2010, 7, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Ilik, I.A.; Quinn, J.J.; Georgiev, P.; Tavares-Cadete, F.; Maticzka, D.; Toscano, S.; Wan, Y.; Spitale, R.C.; Luscombe, N.; Backofen, R.; et al. Tandem stem-loops in roX RNAs act together to mediate X Chromosome dosage compensation in Drosophila. Mol. Cell 2013, 51, 156–173. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yang, Y.A.; Zhang, A.; Fong, K.-W.; Kim, J.; Song, B.; Li, S.; Zhao, J.C.; Yu, J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2015, 77, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Shi, H.; Xi, H.; Wu, X.; Cui, J.; Gao, Y.; Liang, W.; Hu, C.; Liu, Y.; Li, J.; et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncrnas for detection of gastric cancer. Theranostics 2017, 7, 213–227. [Google Scholar] [CrossRef]
- Ponti, R.D.; Armaos, A.; Marti, S.; Tartaglia, G.G. A method for RNA structure prediction shows evidence for structure in lncRNAs. Front. Mol. Biosci. 2018, 5, 111. [Google Scholar] [CrossRef]
- Brown, J.A.; Bulkley, D.; Wang, J.; Valenstein, M.L.; Yario, T.A.; Steitz, T.A.; Steitz, J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014, 21, 633–640. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef]
- Aw, J.G.A.; Shen, Y.; Wilm, A.; Sun, M.; Lim, X.N.; Boon, K.L.; Tapsin, S.; Chan, Y.S.; Tan, C.P.; Sim, A.Y.L.; et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 2016, 62, 603–617. [Google Scholar] [CrossRef]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef]
- Sastry, S.S.; Ross, B.M.; P’arraga, A. Cross-linking of DNA-binding proteins to DNA with psoralen and psoralen furan-side monoadducts: Comparison of action spectra with DNA-DNA cross- linking. J. Biol. Chem. 1997, 272, 3715–3723. [Google Scholar] [CrossRef]
- Simon, M.D. Insight into lncRNA biology using hybridization capture analyses. Biochim. Biophys. Acta-Gene Regul. Mech. 2016, 1859, 121–127. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef]
- Granneman, S.; Kudla, G.; Petfalski, E.; Tollervey, D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 9613–9618. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Sugimoto, Y.; Vigilante, A.; Darbo, E.; Zirra, A.; Militti, C.; D’Ambrogio, A.; Luscombe, N.M.; Ule, J. HiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015, 519, 491–494. [Google Scholar] [CrossRef]
- Kashi, K.; Henderson, L.; Bonetti, A.; Carninci, P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim. Biophys. Acta-Gene Regul. Mech. 2016, 1859, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.C.; Cao, X.; Yu, P.; Xiao, S.; Lu, J.; Biase, F.H.; Sridhar, B.; Huang, N.; Zhang, K.; Zhong, S. Mapping RNA-RNA interactome and RNA structure in vivo by MARIO. Nat. Commun. 2016, 7, 12023. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, P.; Lin, R.; Rong, L.; Xue, Y.; Fang, Y. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Sci. Rep. 2015, 5, 13749. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Tang, J.; Deng, L.; Xie, Y.; Jiang, R.; Li, G.; Sun, B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget 2015, 6, 42813–42824. [Google Scholar] [CrossRef] [PubMed]
- Portoso, M.; Ragazzini, R.; Brenčič, Ž.; Moiani, A.; Michaud, A.; Vassilev, I.; Wassef, M.; Servant, N.; Sargueil, B.; Margueron, R. PRC 2 is dispensable for HOTAIR—Mediated transcriptional repression. EMBO J. 2017, 36, 981–994. [Google Scholar] [CrossRef]
- Pachnis, V.; Belayew, A.; Tilghman, S.M. Locus unlinked to α-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. USA 1984, 81, 5523–5527. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Borsani, G.; Tonlorenzi, R.S.M. Characterization of a murine gene expressed from the inactive X chromosome. Nature 1991, 354, 56–58. [Google Scholar] [CrossRef] [PubMed]
- Koziol, M.J.; Rinn, J.L. RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 2010, 20, 142–148. [Google Scholar] [CrossRef]
- Long, Y.; Hwang, T.; Gooding, A.R.; Goodrich, K.J.; Rinn, J.L.; Cech, T.R. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 2020, 52, 931–938. [Google Scholar] [CrossRef]
- Simon, M.D.; Wang, C.I.; Kharchenko, P.V.; West, J.A.; Chapman, B.A.; Alekseyenko, A.A.; Borowsky, M.L.; Kuroda, M.I.; Kingston, R.E. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 20497–20502. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, B.; Rivas-Astroza, M.; Nguyen, T.C.; Chen, W.; Yan, Z.; Cao, X.; Hebert, L.; Zhong, S. Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr. Biol. 2017, 27, 602–609. [Google Scholar] [CrossRef]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.T.; Li, H.; Fu, X.D. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef]
- Bell, J.C.; Jukam, D.; Teran, N.A.; Risca, V.I.; Smith, O.K.; Johnson, W.L.; Skotheim, J.M.; Greenleaf, W.J.; Straight, A.F. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. eLife 2018, 7, e27024. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, M.; Porter, D.F.; Khavari, P.A. Methods to study RNA–protein interactions. Nat. Methods 2019, 16, 225–234. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; Da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Tagwerker, C.; Flick, K.; Cui, M.; Guerrero, C.; Dou, Y.; Auer, B.; Baldi, P.; Huang, L.; Kaiser, P. A tandem affinity tag for two-step purification under fully denaturing conditions: Application in ubiquitin profiling complex identification combined with in vivo cross-linking. Mol. Cell. Proteom. 2006, 5, 737–748. [Google Scholar] [CrossRef]
- Tsai, B.P.; Wang, X.; Huang, L.; Waterman, M.L. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteom. 2011, 10, M110.007385. [Google Scholar] [CrossRef]
- Smith, K.C.; Aplin, R.T. A Mixed Photoproduct of Uracil and Cysteine (5-S-Cysteine-6-hydrouracil). A Possible Model for the in Vivo Cross-Linking of Deoxyribonucleic Acid and Protein by Ultraviolet Light. Biochemistry 1966, 5, 2125–2130. [Google Scholar] [CrossRef]
- Goddard, J.; Streeter, D.; Weber, C.; Gordon, M.P. Studies on the Inactivation of Tobacco Mosaic Virus By Ultraviolet Light. Photochem. Photobiol. 1966, 5, 213–222. [Google Scholar] [CrossRef]
- Ule, J.; Jensen, K.B.; Ruggiu, M.; Mele, A.; Ule, A.; Darnell, R.B. CLIP Identifies Nova-Regulated RNA Networks in the Brain. Science 2003, 302, 1212–1215. [Google Scholar] [CrossRef]
- Licatalosi, D.D.; Mele, A.; Fak, J.J.; Ule, J.; Kayikci, M.; Chi, S.W.; Clark, T.A.; Schweitzer, A.C.; Blume, J.E.; Wang, X.; et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008, 456, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, E.L.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 2016, 13, 508–514. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Trimarchi, T.; Bilal, E.; Ntziachristos, P.; Fabbri, G.; Dalla-Favera, R.; Tsirigos, A.; Aifantis, I. Genome-wide mapping and characterization of notch-regulated long noncoding RNAs in acute leukemia. Cell 2014, 158, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Wang, W.; Gu, C.; Chen, C.; Zeng, B.; Yang, Y.; Ji, P.; Sun, J.; Wu, J.; Lu, W.; et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. J. Exp. Clin. Cancer Res. 2019, 38, 411. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; Mcclanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological Organization of Multi-chromosomal Regions by Firre HHS Public Access. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef]
- Kelsey, A.D.; Yang, C.; Leung, D.; Minks, J.; Dixon-McDougall, T.; Baldry, S.E.L.; Bogutz, A.B.; Lefebvre, L.; Brown, C.J. Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genome Biol. 2015, 16, 208. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef]
- Fan, S.; Tian, T.; Lv, X.; Lei, X.; Yang, Z.; Liu, M.; Liang, F.; Li, S.; Lin, X.; Lin, Z.; et al. lncRNA CISAL Inhibits BRCA1 Transcription by Forming a Tertiary Structure at Its Promoter. iScience 2020, 23, 100835. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, G.; Lu, M.; Chai, W.; Li, Y.; Tong, X.; Li, J.; Jia, X.; Liu, W.; Qi, D.; et al. Long Noncoding RNA Lnc-MxA Inhibits Beta Interferon Transcription by Forming RNA-DNA Triplexes at Its Promoter. J. Virol. 2019, 93, e00786-19. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- López-Urrutia, E.; Bustamante Montes, L.P.; Ladrón de Guevara Cervantes, D.; Pérez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef]
- Cui, P.; Su, J.; Li, Q.; Xu, G.; Zhu, N. LncRNA RHPN1-AS1 targeting miR-625/REG3A promotes cell proliferation and invasion of glioma cells. OncoTargets Ther. 2019, 12, 7911–7921. [Google Scholar] [CrossRef]
- Noviello, T.M.R.; Di Liddo, A.; Ventola, G.M.; Spagnuolo, A.; D’Aniello, S.; Ceccarelli, M.; Cerulo, L. Detection of long non-coding RNA homology, a comparative study on alignment and alignment-free metrics. BMC Bioinform. 2018, 19, 407. [Google Scholar] [CrossRef]
- Seemann, S.E.; Mirza, A.H.; Hansen, C.; Bang-berthelsen, C.H. The identification and functional annotation of RNA. Genome Res. 2017, 27, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Thiel, B.C.; Ochsenreiter, R.; Gadekar, V.P.; Tanzer, A.; Hofacker, I.L. RNA structure elements conserved between mouse and 59 other vertebrates. Genes 2018, 9, 392. [Google Scholar] [CrossRef]
- Sprague, D.; Waters, S.A.; Kirk, J.M.; Wang, J.R.; Samollow, P.B.; Waters, P.D.; Calabrese, J.M. Nonlinear sequence similarity between the Xist and Rsx long noncoding RNAs suggests shared functions of tandem repeat domains. RNA 2019, 25, 1004–1019. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, D.; Kappei, D.; Theis, M.; Nitzsche, A.; Ding, L.; Paszkowski-Rogacz, M.; Surendranath, V.; Berger, N.; Schulz, H.; Saar, K.; et al. Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat. Methods 2012, 9, 360–362. [Google Scholar] [CrossRef] [PubMed]
- Kittler, R.; Pelletier, L.; Heninger, A.K.; Slabicki, M.; Theis, M.; Miroslaw, L.; Poser, I.; Lawo, S.; Grabner, H.; Kozak, K.; et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 2007, 9, 1401–1412. [Google Scholar] [CrossRef]
- Huang, J.; Zhou, N.; Watabe, K.; Lu, Z.; Wu, F.; Xu, M.; Mo, Y.-Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014, 5, e1008. [Google Scholar] [CrossRef]
- Goff, L.A.; Rinn, J.L. Linking RNA biology to lncRNAs. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef]
- Chiang, M.Y.; Chan, H.; Zounes, M.A.; Freier, S.M.; Lima, W.F.; Bennett, C.F. Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J. Biol. Chem. 1991, 266, 18162–18171. [Google Scholar] [CrossRef]
- Bennett, C.F. Therapeutic Antisense Oligonucleotides Are Coming of Age. Annu. Rev. Med. 2019, 70, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Behlke, M. Mini-review on current strategies to knockdown long non-coding RNAs. J. Rare Dis. Res. Treat. 2016, 1, 66–70. [Google Scholar] [CrossRef]
- Beletskii, A.; Hong, Y.K.; Pehrson, J.; Egholm, M.; Strauss, W.M. PNA interference mapping demonstrates functional domains in the noncoding RNA Xist. Proc. Natl. Acad. Sci. USA 2001, 98, 9215–9220. [Google Scholar] [CrossRef]
- Pellestor, F.; Paulasova, P. The peptide nucleic acids (PNAs), powerful tools for molecular genetics and cytogenetics. Eur. J. Hum. Genet. 2004, 12, 694–700. [Google Scholar] [CrossRef]
- Sarma, K.; Levasseur, P.; Aristarkhov, A.; Lee, J.T. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc. Natl. Acad. Sci. USA 2010, 107, 22196–22201. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Kaini, P.; Sander, J.D.; Joung, J.K.; Peterson, R.T.; Yeh, J.R.J. Heritable and Precise Zebrafish Genome Editing Using a CRISPR-Cas System. PLoS ONE 2013, 8, e68708. [Google Scholar] [CrossRef]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef]
- Hirose, T.; Yamazaki, T.; Nakagawa, S. Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip. Rev. RNA 2019, 10, e1545. [Google Scholar] [CrossRef]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. Lncipedia 5: Towards a reference set of human long non-coding rnas. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Cao, J.; Liu, L.; Du, Q.; Li, Z.; Zou, D.; Bajic, V.B.; Zhang, Z. Lncbook: A curated knowledgebase of human long non-coding rnas. Nucleic Acids Res. 2019, 47, D128–D134. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human microRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L. RNA Coding Potential Prediction Using Alignment-Free Logistic Regression Model. Methods Mol. Biol. 2021, 2254, 27–39. [Google Scholar] [CrossRef]
- Aguet, F.; Barbeira, A.N.; Bonazzola, R.; Brown, A.; Castel, S.E.; Jo, B.; Kasela, S.; Kim-Hellmuth, S.; Liang, Y.; Oliva, M.; et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Szcześniak, M.W.; Bryzghalov, O.; Ciomborowska-Basheer, J.; Makałowska, I. CANTATAdb 2.0: Expanding the Collection of Plant Long Noncoding RNAs. Methods Mol. Biol. 2019, 1933, 415–429. [Google Scholar] [CrossRef]
- Wen, X.; Gao, L.; Guo, X.; Li, X.; Huang, X.; Wang, Y.; Xu, H.; He, R.; Jia, C.; Liang, F. LncSLdb: A resource for long non-coding RNA subcellular localization. Database 2018, 2018, 1–6. [Google Scholar] [CrossRef]
- Mas-Ponte, D.; Carlevaro-Fita, J.; Palumbo, E.; Pulido, T.H.; Guigo, R.; Johnson, R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 2017, 23, 1080–1087. [Google Scholar] [CrossRef]
- Lin, Y.; Pan, X.; Shen, H.-B. lncLocator 2.0: A cell-line-specific subcellular localization predictor for long non-coding RNAs with interpretable deep learning. Bioinformatics 2021, btab127. [Google Scholar] [CrossRef]
- Tong, X.; Liu, S. CPPred: Coding potential prediction based on the global description of RNA sequence. Nucleic Acids Res. 2019, 47, e43. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Jungreis, I.; Kellis, M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xu, Z.; Hu, B.; Lu, Z.J. COME: A robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Res. 2017, 45, e2. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.C.; Fang, S.S.; Wu, Y.; Zhang, J.H.; Chen, Y.; Liu, J.; Wu, B.; Wu, J.R.; Li, E.M.; Xu, L.Y.; et al. CNIT: A fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res. 2019, 47, W516–W522. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Yesselman, J.D.; Tian, S.; Liu, X.; Shi, L.; Li, J.B.; Das, R. Updates to the RNA mapping database (RMDB), version 2. Nucleic Acids Res. 2018, 46, D375–D379. [Google Scholar] [CrossRef]
- Pinkney, H.R.; Wright, B.M.; Diermeier, S.D. The lncrna toolkit: Databases and in silico tools for lncrna analysis. Non-Coding RNA 2020, 6, 49. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Y.; Zhong, X.; Liu, H.; Lu, C.; Li, C.; Zhang, H. DMFold: A novel method to predict RNA secondary structure with pseudoknots based on deep learning and improved base pair maximization principle. Front. Genet. 2019, 10, 143. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Stadler, P.F. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 2006, 22, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Pyfrom, S.C.; Luo, H.; Payton, J.E. PLAIDOH: A novel method for functional prediction of long non-coding RNAs identifies cancer-specific LncRNA activities. BMC Genom. 2019, 20, 137. [Google Scholar] [CrossRef]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, T.; Iwakiri, J.; Ono, Y.; Hamada, M. Lncrrisearch: A web server for lncRNA-RNA interaction prediction integrated with tissue-specific expression and subcellular localization data. Front. Genet. 2019, 10, 462. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, G.; Shi, A.; Hu, J.; Li, F.; Zhang, X.; Zhang, Y.; Huang, J.; Xiao, Y.; Li, X.; et al. LnChrom: A resource of experimentally validated lncRNA-chromatin interactions in human and mouse. Database 2018, 2018, bay039. [Google Scholar] [CrossRef]
- Buske, F.A.; Bauer, D.C.; Mattick, J.S.; Bailey, T.L. Triplexator: Detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012, 22, 1372–1381. [Google Scholar] [CrossRef]
- Hao, Y.; Wu, W.; Li, H.; Yuan, J.; Luo, J.; Zhao, Y.; Chen, R. NPInter v3.0: An upgraded database of noncoding RNA-associated interactions. Database 2016, 2016, baw057. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Chen, X.; Xue, H.; Tang, Y.; Zhang, P.; Kang, Q.; Hao, Y.; Chen, R.; Zhao, Y.; He, S. NPInter v4.0: An integrated database of ncRNA interactions. Nucleic Acids Res. 2020, 48, D160–D165. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Peng, L.; Liu, F.; Yang, J.; Liu, X.; Meng, Y.; Deng, X.; Peng, C.; Tian, G.; Zhou, L. Probing lncRNA–Protein Interactions: Data Repositories, Models, and Algorithms. Front. Genet. 2020, 10, 1346. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, T.; Cui, T.; Wang, Z.; Zhang, Y.; Tan, P.; Huang, Y.; Yu, J.; Wang, D. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020, 48, D189–D197. [Google Scholar] [CrossRef] [PubMed]
- Kirk, J.M.; Kim, S.O.; Inoue, K.; Smola, M.J.; Lee, D.M.; Schertzer, M.D.; Wooten, J.S.; Baker, A.R.; Sprague, D.; Collins, D.W.; et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 2018, 50, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Mills, J.D.; Kavanagh, T.; Kim, W.S.; Chen, B.J.; Waters, P.D.; Halliday, G.M.; Janitz, M. High expression of long intervening non-coding RNA OLMALINC in the human cortical white matter is associated with regulation of oligodendrocyte maturation. Mol. Brain 2015, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Chang, K.; Li, Z.; Gates, K.; Rana, Z.A.; Zhang, D.; Han, T.; Yang, C.; Cunningham, T.J.; Head, R.; et al. An Evolutionarily Conserved Long Noncoding RNA TUNA Controls Pluripotency and Neural Lineage Commitment. Mol. Cell 2015, 53, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Scheuermann, J.C.; Surface, L.E.; Robert, K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a long non-coding RNA required for cardiovascular lineage commitment. Cell 2014, 152, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, M.; Tavallaie, M.; Soltani, B.M.; Hajipoor, S.; Hosseini, S.M. LncRNA HSPC324 plays role in lung development and tumorigenesis. Genomics 2020, 112, 2615–2622. [Google Scholar] [CrossRef]
- Tano, K.; Mizuno, R.; Okada, T.; Rakwal, R.; Shibato, J.; Masuo, Y.; Ijiri, K.; Akimitsu, N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010, 584, 4575–4580. [Google Scholar] [CrossRef]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef]
- Tontonoz, P.; Wu, X.; Jones, M.; Zhang, Z.; Salisbury, D.; Sallam, T. Long noncoding RNA facilitated gene therapy reduces atherosclerosis in a murine model of familial hypercholesterolemia. Circulation 2017, 136, 776–778. [Google Scholar] [CrossRef]
- Hosono, Y.; Niknafs, Y.S.; Prensner, J.R.; Iyer, M.K.; Dhanasekaran, S.M.; Mehra, R.; Pitchiaya, S.; Tien, J.; Escara-Wilke, J.; Poliakov, A.; et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 2017, 171, 1559–1572.e20. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Kretz, M.; Webster, D.E.; Flockhart, R.J.; Lee, C.S.; Zehnder, A.; Lopez-Pajares, V.; Qu, K.; Zheng, G.X.Y.; Chow, J.; Kim, G.E.; et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26, 338–343. [Google Scholar] [CrossRef]
- Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; Kim, G.E.; Spitale, R.C.; Flynn, R.A.; Zheng, G.X.Y.; Aiyer, S.; Raj, A.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Cipolla, G.A.; de Oliveira, J.C.; Salviano-Silva, A.; Lobo-Alves, S.C.; Lemos, D.S.; Oliveira, L.C.; Jucoski, T.S.; Mathias, C.; Pedroso, G.A.; Zambalde, E.P.; et al. Long non-coding RNAs in multifactorial diseases: Another layer of complexity. Non-Coding RNA 2018, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Chen, Y. Long noncoding RNAs and Alzheimer’s disease. Clin. Int. Aging 2016, 11, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Long Noncoding RNAs as Biomarkers in Cancer. Dis. Markers 2017, 2017, 7243968. [Google Scholar] [CrossRef]
- Magistri, M.; Velmeshev, D.; Makhmutova, M.; Faghihi, M.A. Transcriptomics Profiling of Alzheimer’s Disease Reveal Neurovascular Defects, Altered Amyloid-β Homeostasis, and Deregulated Expression of Long Noncoding RNAs. J. Alzheimer’s Dis. 2015, 48, 647–665. [Google Scholar] [CrossRef]
- Ni, Y.; Huang, H.; Chen, Y.; Cao, M.; Zhou, H.; Zhang, Y. Investigation of Long Non-coding RNA Expression Profiles in the Substantia Nigra of Parkinson’s Disease. Cell. Mol. Neurobiol. 2017, 37, 329–338. [Google Scholar] [CrossRef]
- Hosseini, E.; Bagheri-Hosseinabadi, Z.; De Toma, I.; Jafarisani, M.; Sadeghi, I. The importance of long non-coding RNAs in neuropsychiatric disorders. Mol. Asp. Med. 2019, 70, 127–140. [Google Scholar] [CrossRef]
- Fallah, H.; Sayad, A.; Ranjbaran, F.; Talebian, F.; Ghafouri-Fard, S.; Taheri, M. IFNG/IFNG-AS1 expression level balance: Implications for autism spectrum disorder. Metab. Brain Dis. 2020, 35, 327–333. [Google Scholar] [CrossRef]
- Chen, S.; Zhu, X.; Niu, W.; Yao, G.; Kong, L.; He, M.; Chen, C.; Lu, Z.; Cui, X.; Zhang, L. OverexpressionRegulatory Role of lncRNA NONHSAT089447 in the Dopamine Signaling Pathway in Schizophrenic Patients. Med. Sci. Monit. 2019, 25, 4322–4332. [Google Scholar] [CrossRef]
- Safari, M.R.; Komaki, A.; Arsang-Jang, S.; Taheri, M.; Ghafouri-Fard, S. Expression Pattern of Long Non-coding RNAs in Schizophrenic Patients. Cell. Mol. Neurobiol. 2019, 39, 211–221. [Google Scholar] [CrossRef]
- Santer, L.; López, B.; Ravassa, S.; Baer, C.; Riedel, I.; Chatterjee, S.; Moreno, M.U.; González, A.; Querejeta, R.; Pinet, F.; et al. Circulating long noncoding RNA LIPCAR Predicts heart failure outcomes in patients without chronic kidney disease. Hypertension 2019, 73, 820–828. [Google Scholar] [CrossRef]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; De Groote, P.; Pinet, F.; Thum, T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef]
- Skuratovskaia, D.; Vulf, M.; Komar, A.; Kirienkova, E.; Litvinova, L. Promising directions in atherosclerosis treatment based on epigenetic regulation using microRNAs and long noncoding RNAs. Biomolecules 2019, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Trenner, M.; Boon, R.A.; Spin, J.M.; Maegdefessel, L. Long noncoding RNAs in key cellular processes involved in aortic aneurysms. Atherosclerosis 2020, 292, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Liu, Y.; Guo, S.; Yao, R.; Wu, L.; Xiao, L.; Wang, Z.; Liu, Y.; Zhang, Y. Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction. Cell. Physiol. Biochem. 2017, 44, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, W.; Jin, M.; Chen, J.; Xu, W.; Kong, X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017, 8, e2929. [Google Scholar] [CrossRef]
- Morán, I.; Akerman, İ.; Van De Bunt, M.; Xie, R.; Benazra, M.; Nammo, T.; Arnes, L.; Naki, N.; García-hurtado, J.; Pasquali, L.; et al. Human β Cell Transcriptome Analysis Uncovers lncRNAs That Are Tissue-Specific, Dynamically Regulated, and Abnormally Expressed in Type 2 Diabetes. Cell Metab. 2013, 16, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wu, J.; Li, D.; Zhou, J.; Yu, H.; Ma, L. Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction through miR-320-Pten axis. Biomed. Pharmacother. 2018, 106, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nurnberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Cheng, M.; Chi, X.; Liu, X.; Yang, W. A Human Long Non-coding RNA LncATV Promotes Virus Replication Through Restricting RIG-I-Mediated Innate Immunity. Front. Immunol. 2019, 10, 1711. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.; Xu, J.; Yang, W.; You, X.; Zhong, Q.; Wang, X. The research progress of LncRNA involved in the regulation of inflammatory diseases. Mol. Immunol. 2018, 101, 182–188. [Google Scholar] [CrossRef]
- Mathy, N.W.; Chen, X.M. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J. Biol. Chem. 2017, 292, 12375–12382. [Google Scholar] [CrossRef]
- Zemmour, D.; Pratama, A.; Loughhead, S.M.; Mathis Di Benoist, C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. USA 2017, 114, E3472–E3480. [Google Scholar] [CrossRef]
- Bi, X.; Guo, X.H.; Mo, B.Y.; Wang, M.L.; Luo, X.Q.; Chen, Y.X.; Liu, F.; Olsen, N.; Pan, Y.F.; Zheng, S.G. LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine 2019, 50, 408–420. [Google Scholar] [CrossRef]
- Zhang, H.J.; Wei, Q.F.; Wang, S.J.; Zhang, H.J.; Zhang, X.Y.; Geng, Q.; Cui, Y.H.; Wang, X.H. LncRNA HOTAIR alleviates rheumatoid arthritis by targeting miR-138 and inactivating NF-κB pathway. Int. Immunopharmacol. 2017, 50, 283–290. [Google Scholar] [CrossRef]
- Haberman, Y.; Benshoshan, M.; Di Segni, A.; Dexheimer, P.J.; Braun, T.; Weiss, B.; Walters, T.D.; Baldassano, R.N.; Noe, J.D.; Markowitz, J.; et al. Long ncRNA Landscape in the Ileum of Treatment-Naive Early-Onset Crohn Disease. Inflamm. Bowel Dis. 2018, 24, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Munschauer, M.; Vogel, J. Nuclear lnc RNA stabilization in the host response to bacterial infection. EMBO J. 2018, 37, 2–4. [Google Scholar] [CrossRef] [PubMed]
- zur Bruegge, J.; Einspanier, R.; Sharbati, S. A long journey Ahead: Long non-coding RNAs in bacterial infections. Front. Cell. Infect. Microbiol. 2017, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Duval, M.; Cossart, P.; Lebreton, A. Mammalian microRNAs and long noncoding RNAs in the host-bacterial pathogen crosstalk. Semin. Cell Dev. Biol. 2017, 65, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Bao, J.; Wang, Y.; Chen, W.; Wu, T.; Wang, L.; Lv, X.; Gao, W.; Wang, B.; Zhu, G.; et al. Changes in long non-coding RNA expression profiles related to the antagonistic effects of Escherichia coli F17 on lamb spleens. Sci. Rep. 2018, 8, 16514. [Google Scholar] [CrossRef]
- Westermann, A.J.; Förstner, K.U.; Amman, F.; Barquist, L.; Chao, Y.; Schulte, L.N.; Müller, L.; Reinhardt, R.; Stadler, P.F.; Vogel, J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 2016, 529, 496–501. [Google Scholar] [CrossRef]
- He, J.; Ou, Q.; Liu, C.; Shi, L.; Zhao, C.; Xu, Y.; Kong, S.K.; Loo, J.F.C.; Li, B.; Gu, D. Differential expression of long non-coding RNAs in patients with tuberculosis infection. Tuberculosis 2017, 107, 73–79. [Google Scholar] [CrossRef]
- Roy, S.; Schmeier, S.; Kaczkowski, B.; Arner, E.; Alam, T.; Ozturk, M.; Tamgue, O.; Parihar, S.P.; Kawaji, H.; Itoh, M.; et al. Transcriptional landscape of Mycobacterium tuberculosis infection in macrophages. Sci. Rep. 2018, 8, 6758. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yang, B.; Lindahl, A.J.; Damaschke, N.; Boersma, M.D.; Huang, W.; Corey, E.; Jarrard, D.F.; Denu, J.M. Identifying Dysregulated Epigenetic Enzyme Activity in Castrate-Resistant Prostate Cancer Development. ACS Chem. Biol. 2017, 12, 2804–2814. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, J.; Wang, J.; Wen, Q.; Wang, H.; He, J.; Hu, S.; He, W.; Du, X.; Liu, S.; et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 38963. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Deshpande, N.P.; Man, S.M.; Burgos-Portugal, J.A.; Khattak, F.A.; Raftery, M.J.; Wilkins, M.R.; Mitchell, H.M. Transcriptomic and proteomic analyses reveal key innate immune signatures in the host response to the gastrointestinal pathogen Campylobacter concisus. Infect. Immun. 2015, 83, 832–845. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, Q.; Yao, Y.; Fang, J.; Sun, F.; Ni, Y.; Shen, Y.; Wang, H.; Shao, S. Microarray analysis of Long non-coding RNA expression profiles in human gastric cells and tissues with Helicobacter pylori infection. BMC Med. Genom. 2015, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Long, Y.; Li, C.; Cao, L.; Gan, H.; Huang, K.; Jia, Y. Genome-wide analysis of long noncoding RNA profile in human gastric epithelial cell response to Helicobacter pylori. Jpn. J. Infect. Dis. 2015, 68, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, C.Y.; Yedavalli, V.S.R.K.; Jeang, K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 2013, 4, e00596-12. [Google Scholar] [CrossRef]
- Morenikeji, O.B.; Bernard, K.; Strutton, E.; Wallace, M.; Thomas, B.N. Evolutionarily Conserved Long Non-coding RNA Regulates Gene Expression in Cytokine Storm During COVID-19. Front. Bioeng. Biotechnol. 2021, 8, 582953. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Huang, L.; Wei, Q.; Zhang, Y.; Zhang, S.; Zhang, W.; Cai, L.; Liang, S. Microarray analysis of long non-coding RNA expression profiles uncovers a Toxoplasma-induced negative regulation of host immune signaling. Parasites Vectors 2018, 11, 174. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G.S.; Wahlestedt, C.; Kapranov, P. The Landscape of long non-coding RNA classification Georges. Trends Genet. 2016, 31, 239–251. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Rafiee, A.; Riazi-Rad, F.; Havaskary, M.; Nuri, F. Long noncoding RNAs: Regulation, function and cancer. Biotechnol. Genet. Eng. Rev. 2018, 34, 153–180. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Xia, Z.; Yan, R.; Duan, F.; Song, C.; Wang, P.; Wang, K. Genetic polymorphisms in long noncoding RNA H19 are associated with susceptibility to breast cancer in Chinese population. Medicine 2016, 95, e2771. [Google Scholar] [CrossRef]
- Richard, J.L.C.; Eichhorn, P.J.A. Deciphering the roles of lncRNAs in breast development and disease. Oncotarget 2018, 9, 20179–20212. [Google Scholar] [CrossRef]
- Qian, K.; Liu, G.; Tang, Z.; Hu, Y.; Fang, Y.; Chen, Z.; Xu, X. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch. Biochem. Biophys. 2017, 615, 1–9. [Google Scholar] [CrossRef]
- Xu, T.; Hu, X.X.; Liu, X.X.; Wang, H.J.; Lin, K.; Pan, Y.Q.; Sun, H.L.; Peng, H.X.; Chen, X.X.; Wang, S.K.; et al. Association between SNPs in long non-coding RNAs and the risk of female breast cancer in a chinese population. J. Cancer 2017, 8, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Pan, C.F.; He, Z.C.; Wang, J.; Wang, P.L.; Ma, T.; Xia, Y.; Chen, Y.J. Long Noncoding RNA-LET Suppresses Tumor Growth and EMT in Lung Adenocarcinoma. BioMed Res. Int. 2016, 2016, 4693471. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, H.; Liu, Z.; Owzar, K.; Han, Y.; Su, L.; Wei, Y.; Hung, R.J.; McLaughlin, J.; Brhane, Y.; et al. A Novel Genetic Variant in Long Non-coding RNA Gene NEXN-AS1 is Associated with Risk of Lung Cancer. Sci. Rep. 2016, 6, 34234. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Yang, F.; Zhang, Y.L.; Liu, B.; Wang, M.; Hong, X.; Yu, Y.; Zhou, Y.H.; Zeng, H. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017, 18, 95–104. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Stitkovcova, K.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Kala, Z.; Svoboda, M.; Kiss, I.; Vyzula, R.; et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ahmed, M.; Zhang, F.; Yao, C.Q.; Li, S.; Liang, Y.; Hua, J.; Soares, F.; Sun, Y.; Langstein, J.; et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016, 48, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Xu, C.; Xie, H.; Xu, H.; Zhan, P.; Yu, L.; Fang, X. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS ONE 2014, 9, e108133. [Google Scholar] [CrossRef]
- Zhang, C.L.; Zhu, K.P.; Shen, G.Q.; Zhu, Z.S. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumor Biol. 2016, 37, 2737–2748. [Google Scholar] [CrossRef]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.J.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.F.; et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef]
- Matullo, G.; Naccarati, A.; Pardini, B. MicroRNA expression profiling in bladder cancer: The challenge of next-generation sequencing in tissues and biofluids. Int. J. Cancer 2016, 138, 2334–2345. [Google Scholar] [CrossRef] [PubMed]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding rnas in extracellular fluids as cancer biomarkers: The new frontier of liquid biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef] [PubMed]
- Sarfi, M.; Abbastabar, M.; Khalili, E. Long noncoding RNAs biomarker-based cancer assessment. J. Cell. Physiol. 2019, 234, 16971–16986. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Long noncoding RNAs: From genomic junk to rising stars in the early detection of cancer. Anal. Bioanal. Chem. 2019, 411, 4265–4275. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yu, F.; Wang, J.; Li, Y.; Li, Y.; Li, Z.; Zhou, Q. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed. Rep. 2015, 3, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Donovan, M.J.; Noerholm, M.; Bentink, S.; Belzer, S.; Skog, J.; O’Neill, V.; Cochran, J.S.; Brown, G.A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015, 18, 370–375. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef]
- Duan, W.; Du, L.; Jiang, X.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; Wang, L.; Zhang, X.; et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 2016, 7, 78850–78858. [Google Scholar] [CrossRef]
- Arantes, L.M.R.B.; De Carvalho, A.C.; Melendez, M.E.; Carvalho, A.L. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev. Mol. Diagn. 2017, 18, 85–112. [Google Scholar] [CrossRef]
- Tang, H.; Wu, Z.; Zhang, J.; Su, B. Salivary lncRNA as a potential marker for Oral squamous cell carcinoma diagnosis. Mol. Med. Rep. 2013, 7, 761–766. [Google Scholar] [CrossRef]
- Shao, Y.; Ye, M.; Jiang, X.; Sun, W.; Ding, X.; Liu, Z.; Ye, G.; Zhang, X.; Xiao, B.; Guo, J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 2014, 120, 3320–3328. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Chen, Z.; He, A.; Xie, H.; Zhang, Q.; Cai, Z.; Liu, Y.; Huang, W. SPRY4-IT1: A novel oncogenic long non-coding rna in human cancers. Tumor Biol. 2017, 39, 1010428317711406. [Google Scholar] [CrossRef]
- Yue, B.; Qiu, S.; Zhao, S.; Liu, C.; Zhang, D.; Yu, F.; Peng, Z.; Yan, D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J. Gastroenterol. Hepatol. 2016, 31, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Qian, J.; Chen, F.; Fan, Y.; Long, J. LINC00461 promotes cell migration and invasion in breast cancer through miR-30a-5p/integrin β3 axis. J. Cell. Biochem. 2019, 120, 4851–4862. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Li, Z.; Zheng, H.; Chan, M.T.V.; Ka Kei Wu, W. CCAT2: A novel oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017, 50, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Esmaeili, M.; Taheri, M. H19 lncRNA: Roles in tumorigenesis. Biomed. Pharmacother. 2020, 123, 109774. [Google Scholar] [CrossRef]
- Fang, Z.; Chen, W.; Yuan, Z.; Liu, X.; Jiang, H. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed. Pharmacother. 2018, 101, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Lee, S.H.; Jones, A.; Fiegl, H.; Kalwa, M.; Wagner, W.; Chindera, K.; Evans, I.; Dubeau, L.; Orjalo, A.; et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015, 7, 108. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.; Zhang, C.; Duan, C. Multiple Roles of Exosomal Long Noncoding RNAs in Cancers. BioMed Res. Int. 2019, 2019, 1460572. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, L.; Yu, F.; Zhang, Y.; Li, P.; Wang, K. The functional roles of exosomal long non-coding RNAs in cancer. Cell. Mol. Life Sci. 2019, 76, 2059–2076. [Google Scholar] [CrossRef]
- Bullock, M.D.; Silva, A.M.; Kanlikilicer-Unaldi, P.; Filant, J.; Rashed, M.H.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. Exosomal non-coding RNAs: Diagnostic, prognostic and therapeutic applications in cancer. Non-Coding RNA 2015, 1, 53–68. [Google Scholar] [CrossRef]
- Silva, A.; Bullock, M.; Calin, G. The clinical relevance of long non-coding RNAs in cancer. Cancers 2015, 7, 2169–2182. [Google Scholar] [CrossRef]
- Askarian-Amiri, M.E.; Leung, E.; Finlay, G.; Baguley, B.C. The regulatory role of long noncoding RNAs in cancer drug resistance. Methods Mol. Biol. 2016, 1395, 207–227. [Google Scholar] [CrossRef]
- Pan, J.J.; Xie, X.J.; Li, X.; Chen, W. Long Non-coding RNAs and Drug Resistance. Asian Pac. J. Cancer Prev. 2016, 16, 8067–8073. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Ji, J.; Shen, J.; Lu, X. When Long Noncoding RNAs Meet Genome Editing in Pluripotent Stem Cells. Stem Cells Int. 2017, 2017, 3250624. [Google Scholar] [CrossRef]
- Majidinia, M.; Yousefi, B. Long non-coding RNAs in cancer drug resistance development. DNA Repair 2016, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, Y.; Zhang, K.; Liu, X.; Dai, Y.; Jiao, X. Targeted inhibition of long non-coding RNA H19 blocks anaplastic thyroid carcinoma growth and metastasis. Bioengineered 2019, 10, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Ballister, E.R.; Rodgers, J.; Martial, F.; Lucas, R.J. A live cell assay of GPCR coupling allows identification of optogenetic tools for controlling Go and Gi signaling. BMC Biol. 2018, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. P53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Saeinasab, M.; Bahrami, A.R.; González, J.; Marchese, F.P.; Martinez, D.; Mowla, S.J.; Matin, M.M.; Huarte, M. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J. Exp. Clin. Cancer Res. 2019, 38, 172. [Google Scholar] [CrossRef]
- Xing, Z.; Zhang, Y.; Liang, K.; Yan, L.; Xiang, Y.; Li, C.; Hu, Q.; Jin, F.; Putluri, V.; Putluri, N.; et al. Expression of Long Noncoding RNA YIYA Promotes Glycolysis in Breast Cancer. Cancer Res. 2018, 78, 4524–4532. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Gong, N.; Teng, X.; Li, J.; Liang, X.-J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Barwal, T.S.; Acharya, V.; Tamang, S.; Vasquez, K.M.; Jain, A. Cancer Susceptibility Candidate 9 (CASC9): A Novel Targetable Long Noncoding RNA in Cancer Treatment. Transl. Oncol. 2020, 13, 100774. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, G.; Qiu, H.; Yang, J.; Bu, X.; Zhu, S.; Zheng, J.; Dang, C.; Wang, W.; Chu, D. The Novel Notch-induced Long Noncoding RNA LUNAR1 Determines the Proliferation and Prognosis of Colorectal Cancer. Sci. Rep. 2019, 9, 19915. [Google Scholar] [CrossRef]
- Zare, K.; Shademan, M.; Ghahramani Seno, M.M.; Dehghani, H. CRISPR/Cas9 Knockout Strategies to Ablate CCAT1 lncRNA Gene in Cancer Cells 06 Biological Sciences 0604 Genetics. Biol. Proced. Online 2018, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Jing, Y.; Cost, G.J.; Chiang, J.C.; Kolpa, H.J.; Cotton, A.M.; Carone, D.M.; Carone, B.R.; Shivak, D.A.; Guschin, D.Y.; et al. Translating dosage compensation to trisomy 21. Nature 2013, 500, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.C.; Jiang, J.; Newburger, P.E.; Lawrence, J.B. Trisomy silencing by XIST normalizes Down syndrome cell pathogenesis demonstrated for hematopoietic defects in vitro. Nat. Commun. 2018, 9, 5180. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qian, T.; Wang, X.; Liu, J.; Gu, X. Noncoding RNAs and Their Potential Therapeutic Applications in Tissue Engineering. Engineering 2017, 3, 3–15. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Y.; Bao, X.; Zhu, X.; Kwok, Y.K.Y.; Sun, K.; Chen, X.; Huang, Y.; Jauch, R.; Esteban, M.A.; et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015, 25, 335–350. [Google Scholar] [CrossRef] [PubMed]

| Type of Experiment | Limitations | Methods | Structure | lncRNA | References | |
|---|---|---|---|---|---|---|
| In vitro |
| Enzymatic probing | ||||
| PARS/nuclease S1 and RNaseV1 | double- and single- stranded regions of RNA | Rox1 Rox2 | [73] | ||
| Chemical probing | ||||||
| SHAPE-seq | 2′-OH acylation | Braveheart RepA Rox1 Rox2 SRA HOTAIR COOLAIR MALAT1 NEAT1 | [66,67,68,69,71,73,74,75] | ||
| DMS-seq (DMS) | unpaired adenine and cytosine residues | BraveheartRepA SRA HOTAIR MALAT1 | [66,67,69,74,75] | ||
| In vivo |
| Chemical Probing: | ||||
| SHAPE-MaP (1M7,1M6,NMI1) | 2′-OH acylation | Xist | [55] | |||
| In silico |
| CROSS (Computational Recognition of Secondary Structure) | RepA, D2 domain | Xist HOTAIR | [76] | |
| Biophysical |
| X-ray | A-rich 3′-UTR | MALAT1 | [77] | |
| NMR spectroscopy | AUCG tetraloop | Xist | [56] | |||
| RBP | RNA Sequence/ Motif | Interacting RNA | Method | References |
|---|---|---|---|---|
| λN | Box B loop | any fused with Box B loop | Gal4-λN/BoxB reporter system | [90,91] |
| MCP | MS2 loop | any fused with MS2 loop | RNA-tethering | [92] |
| IGF2BP1,2,3 | CAUH | mostly exons, i.e., eEF2 | PAR-CLIP | [84] |
| PUM2 | UGUANAUA | 3′ untranslated region (UTR) | PAR-CLIP | [84] |
| QKI | ACUAAY | mostly introns | PAR-CLIP | [84] |
| AGO (most enriched 7 nucleotide- mers) | AUGCUGC | miR-103,-107 | PAR-CLIP | [84] |
| GCUGCUA | miR-15a/b,-16,196a | PAR-CLIP | [84] | |
| UUUGCAC | miR-19a/b | PAR-CLIP | [84] | |
| UGCACUU | miR-130a/b,-148a/b,-301a/b | PAR-CLIP | [84] | |
| CACUUUA | miR-106a/b,-20a/b | PAR-CLIP | [84] | |
| UUGCUGC | miR-424 | PAR-CLIP | [84] | |
| UUGCACU | miR-130a/b,181a,-301a/b,-454 | PAR-CLIP | [84] | |
| GCACUUU | miR-17,-20a/b,-93,-106a/b | PAR-CLIP | [84] | |
| UGCUGCU | miR-15a/b,-16,196a,-103,107,-424 | PAR-CLIP | [84] | |
| STAU1 | 3′ UTRs (Alu, 858 nt duplex) | XBP1 | hiCLIP | [87] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borkiewicz, L.; Kalafut, J.; Dudziak, K.; Przybyszewska-Podstawka, A.; Telejko, I. Decoding LncRNAs. Cancers 2021, 13, 2643. https://doi.org/10.3390/cancers13112643
Borkiewicz L, Kalafut J, Dudziak K, Przybyszewska-Podstawka A, Telejko I. Decoding LncRNAs. Cancers. 2021; 13(11):2643. https://doi.org/10.3390/cancers13112643
Chicago/Turabian StyleBorkiewicz, Lidia, Joanna Kalafut, Karolina Dudziak, Alicja Przybyszewska-Podstawka, and Ilona Telejko. 2021. "Decoding LncRNAs" Cancers 13, no. 11: 2643. https://doi.org/10.3390/cancers13112643
APA StyleBorkiewicz, L., Kalafut, J., Dudziak, K., Przybyszewska-Podstawka, A., & Telejko, I. (2021). Decoding LncRNAs. Cancers, 13(11), 2643. https://doi.org/10.3390/cancers13112643






