AID Contributes to Accelerated Disease Progression in the TCL1 Mouse Transplant Model for CLL
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Sorting and DNA and RNA Preparation for Sequencing
2.3. B Cell Receptor (BCR) Analysis
2.4. Whole Exome Sequencing (WES)
2.5. Mutation Analysis
2.6. CNV Analysis
2.7. Mutational Signature Analysis
2.8. RNAseq
2.9. Sµ Region Sequencing
2.10. Sµ Region Mutation Analysis
2.11. Statistical Analysis and Visualization
3. Results
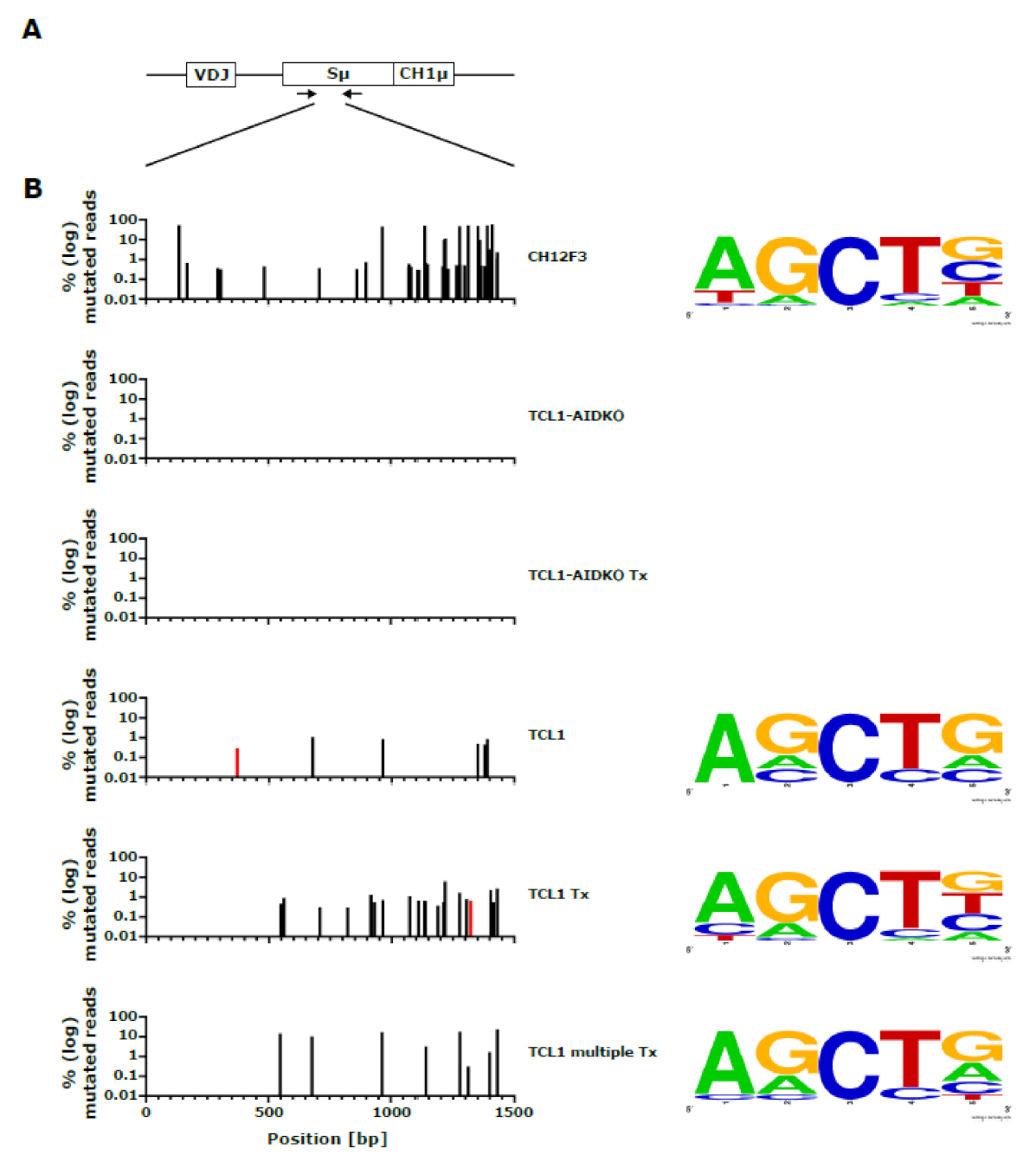
3.1. Mutation Analysis in TCL1 Mice
3.2. CLL Development in AID Deficient TCL1 Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, K.; Lieber, M.R. Current insights into the mechanism of mammalian immunoglobulin class switch recombination. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Noia, D.J.M.; Neuberger, M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007, 76, 1–22. [Google Scholar] [CrossRef]
- Rebhandl, S.; Huemer, M.; Greil, R.; Geisberger, R. AID/APOBEC deaminases and cancer. Oncoscience 2015, 2, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, T.J.; Orchard, J.A.; Gardiner, A.; Oscier, D.G.; Davis, Z.; Stevenson, F.K. Immunoglobulin V genes and CD38 expression in CLL. Blood 2000, 95, 2455–2457. [Google Scholar] [CrossRef] [PubMed]
- Patten, P.; Chu, C.C.; Albesiano, E.; Damle, R.N.; Yan, X.-J.; Kim, D.; Zhang, L.; Magli, A.R.; Barrientos, J.; Kolitz, J.E.; et al. IGHV-unmutated and IGHV-mutated chronic lymphocytic leukemia cells produce activation-induced deaminase protein with a full range of biologic functions. Blood 2012, 120, 4802–4811. [Google Scholar] [CrossRef]
- Kasar, S.; Kim, J.; Improgo, R.; Tiao, G.; Polak, P.; Haradhvala, N.; Lawrence, M.S.; Kiezun, A.; Fernandes, S.M.; Bahl, S.; et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat. Commun. 2015, 6, 8866. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.; Alsolami, R.; Becq, J.; Stamatopoulos, B.; Timbs, A.; Bruce, D.; Robbe, P.; Vavoulis, D.; Clifford, R.; Cabes, M.; et al. Whole-genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHV(mut) and IgHV(unmut) subgroups. Leukemia 2018, 32, 332–342. [Google Scholar] [CrossRef]
- Bichi, R.; Shinton, S.A.; Martin, E.S.; Koval, A.; Calin, G.A.; Cesari, R.; Russo, G.; Hardy, R.R.; Croce, C.M. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6955–6960. [Google Scholar] [CrossRef]
- Hofbauer, J.P.; Heyder, C.; Denk, U.; Kocher, T.; Holler, C.; Trapin, D.; Asslaber, D.; Tinhofer, I.; Greil, R.; Egle, A. Development of CLL in the TCL1 transgenic mouse model is associated with severe skewing of the T-cell compartment homologous to human CLL. Leukemia 2011, 25, 1452–1458. [Google Scholar] [CrossRef]
- Zaborsky, N.; Gassner, F.J.; Höpner, J.P.; Schubert, M.; Hebenstreit, D.; Stark, R.; Asslaber, D.; Steiner, M.; Geisberger, R.; Greil, R.; et al. Exome sequencing of the TCL1 mouse model for CLL reveals genetic heterogeneity and dynamics during disease development. Leukemia 2019, 33, 957–968. [Google Scholar] [CrossRef]
- Muramatsu, M.; Kinoshita, K.; Fagarasan, S.; Yamada, S.; Shinkai, Y.; Honjo, T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 2000, 102, 553–563. [Google Scholar] [CrossRef]
- Bolotin, D.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Brochet, X.; Lefranc, M.-P.; Giudicelli, V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008, 36, W503–W508. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Larson, D.E.; Wilson, R.K. Using VarScan 2 for germline variant calling and somatic mutation detection. Curr. Protoc. Bioinform. 2013, 44, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data, version 2010; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 May 2021).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Hu, E. enrichplot: Visualization of Functional Enrichment Result, R package version 1.10.1; 2020. Available online: https://yulab-smu.top/biomedical-knowledge-mining-book/ (accessed on 26 May 2021).
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kondo, S.; Sugai, M.; Nazarea, M.; Imamura, S.; Honjo, T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int. Immunol 1996, 8, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Rebhandl, S.; Zaborsky, N.; Gassner, F.J.; Hainzl, S.; Weiss, L.; Hebenstreit, D.; Greil, R.; Geisberger, R. AID induces intraclonal diversity and genomic damage in CD86 + chronic lymphocytic leukemia cells. Eur. J. Immunol. 2014, 44, 3747–3757. [Google Scholar] [CrossRef]
- Xue, K.; Rada, C.; Neuberger, M.S. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2-/- ung-/- mice. J. Exp. Med. 2006, 203, 2085–2094. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Calhoun, P. Exact: Unconditional Exact Test, R package version 2.0; 2019. Available online: https://cran.r-project.org/web/packages/Exact/Exact.pdf (accessed on 26 May 2021).
- Barnard, G.A. A new test for 2 × 2 tables. Nature 1945, 156, 177. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.-L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Lada, A.G.; Goncearenco, A.; Green, M.R.; De, S.; Nudelman, G.; Panchenko, A.R.; Koonin, E.V.; Pavlov, Y.I. Activation induced deaminase mutational signature overlaps with CpG methylation sites in follicular lymphoma and other cancers. Sci. Rep. 2016, 6, 38133. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Yan, X.-J.; Albesiano, E.; Zanesi, N.; Yancopoulos, S.; Sawyer, A.; Romano, E.; Petlickovski, A.; Efremov, D.G.; Croce, C.M.; Chiorazzi, N. B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2006, 103, 11713–11718. [Google Scholar] [CrossRef]
- Shimizu, T.; Marusawa, H.; Endo, Y.; Chiba, T. Inflammation-mediated genomic instability: Roles of activation-induced cytidine deaminase in carcinogenesis. Cancer Sci. 2012, 103, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, D.P.; Lee, E.L.; Takayama, S.; Coppé, J.-P.; Heo, S.-J.; Boffelli, D.; Di Noia, J.M.; Martin, D.I.K. Activation-induced cytidine deaminase (AID) is necessary for the epithelial-mesenchymal transition in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 2013, 110, E2977–E2986. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, P.M.; Teater, M.; Chambwe, N.; Kormaksson, M.; Redmond, D.; Ishii, J.; Vuong, B.; Chaudhuri, J.; Melnick, A.; VasanthaKumar, A.; et al. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Rep. 2015, 12, 2086–2098. [Google Scholar] [CrossRef]
- Morande, P.E.; Yan, X.-J.; Sepulveda-Yanez, J.H.; Seija, N.; Marquez, M.E.; Sotelo, N.S.; Abreu, C.; Crispo, M.; Fernández-Graña, G.; Rego, N.; et al. AID overexpression leads to aggressive murine CLL and non-Ig mutations that mirror human neoplasms. Blood 2021. [Google Scholar] [CrossRef]
- Gassner, F.J.; Zaborsky, N.; Buchumenski, I.; Levanon, E.Y.; Gatterbauer, M.; Schubert, M.; Rauscher, S.; Hebenstreit, D.; Nadeu, F.; Campo, E.; et al. RNA editing contributes to epitranscriptome diversity in chronic lymphocytic leukemia. Leukemia 2020, 35, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Hackl, H.; Gassner, F.J.; Greil, R.; Geisberger, R. Investigating epigenetic effects of activation-induced deaminase in chronic lymphocytic leukemia. PLoS ONE 2018, 13, e0208753. [Google Scholar] [CrossRef] [PubMed]
- Gassner, F.J.; Zaborsky, N.; Feldbacher, D.; Greil, R.; Geisberger, R. RNA editing alters mirna function in chronic lymphocytic leukemia. Cancers 2020, 12, 1159. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schubert, M.; Gassner, F.J.; Huemer, M.; Höpner, J.P.; Akimova, E.; Steiner, M.; Egle, A.; Greil, R.; Zaborsky, N.; Geisberger, R. AID Contributes to Accelerated Disease Progression in the TCL1 Mouse Transplant Model for CLL. Cancers 2021, 13, 2619. https://doi.org/10.3390/cancers13112619
Schubert M, Gassner FJ, Huemer M, Höpner JP, Akimova E, Steiner M, Egle A, Greil R, Zaborsky N, Geisberger R. AID Contributes to Accelerated Disease Progression in the TCL1 Mouse Transplant Model for CLL. Cancers. 2021; 13(11):2619. https://doi.org/10.3390/cancers13112619
Chicago/Turabian StyleSchubert, Maria, Franz Josef Gassner, Michael Huemer, Jan Philip Höpner, Ekaterina Akimova, Markus Steiner, Alexander Egle, Richard Greil, Nadja Zaborsky, and Roland Geisberger. 2021. "AID Contributes to Accelerated Disease Progression in the TCL1 Mouse Transplant Model for CLL" Cancers 13, no. 11: 2619. https://doi.org/10.3390/cancers13112619
APA StyleSchubert, M., Gassner, F. J., Huemer, M., Höpner, J. P., Akimova, E., Steiner, M., Egle, A., Greil, R., Zaborsky, N., & Geisberger, R. (2021). AID Contributes to Accelerated Disease Progression in the TCL1 Mouse Transplant Model for CLL. Cancers, 13(11), 2619. https://doi.org/10.3390/cancers13112619





