Expression of NK Cell Receptor Ligands on Leukemic Cells Is Associated with the Outcome of Childhood Acute Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
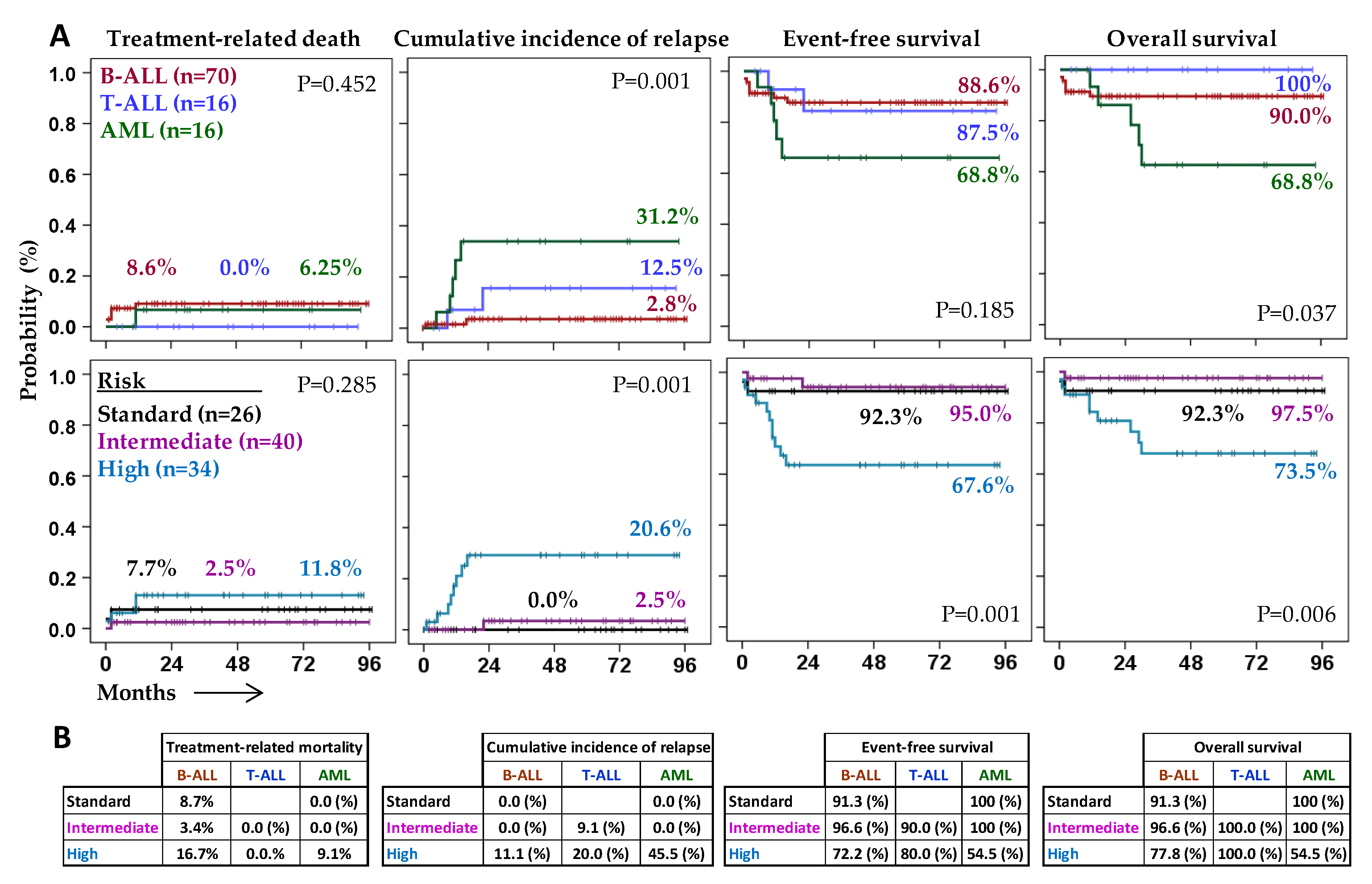
2.1. Characteristics and Outcome of Patients According to the Type of Acute Leukemia
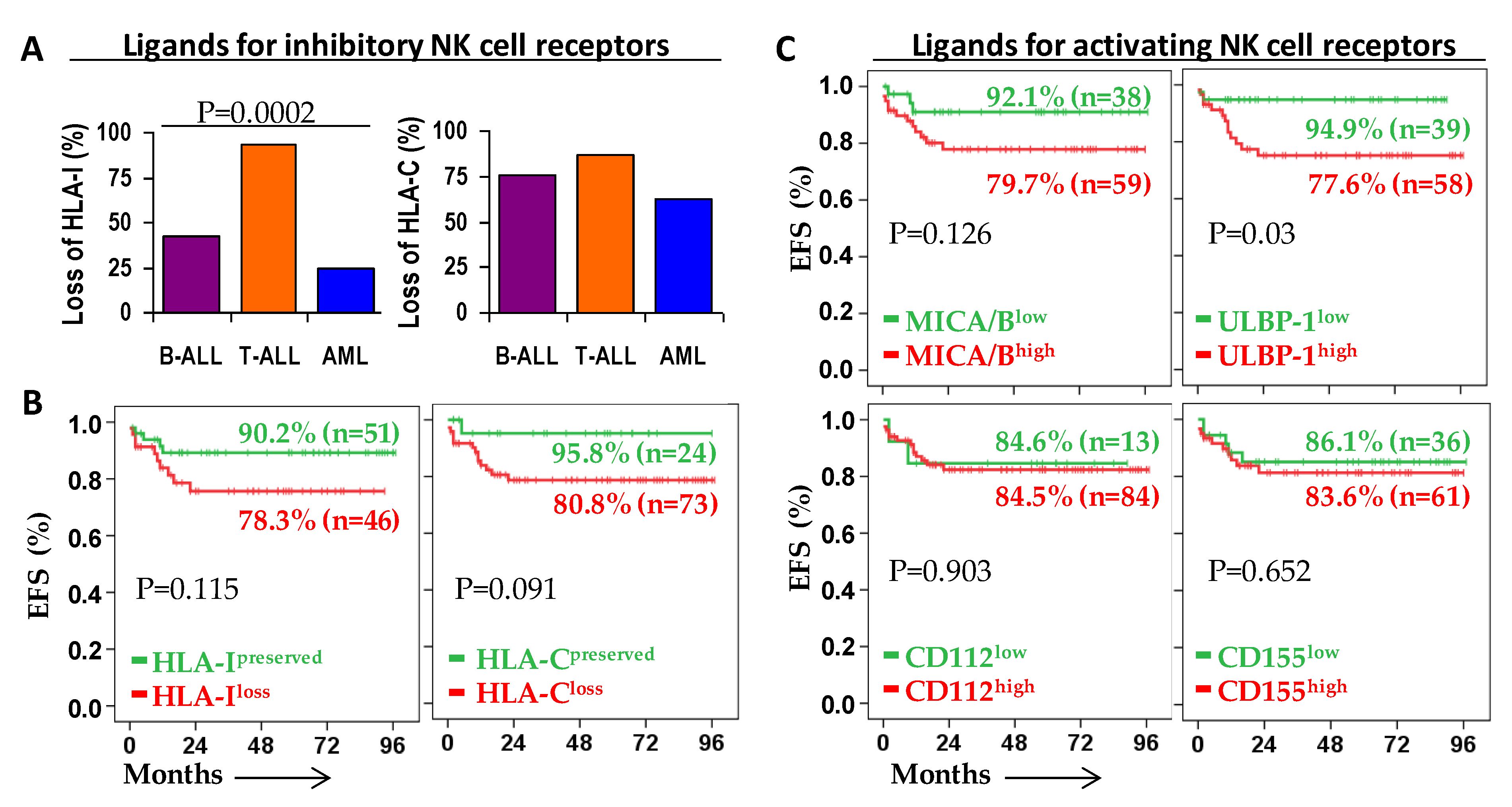
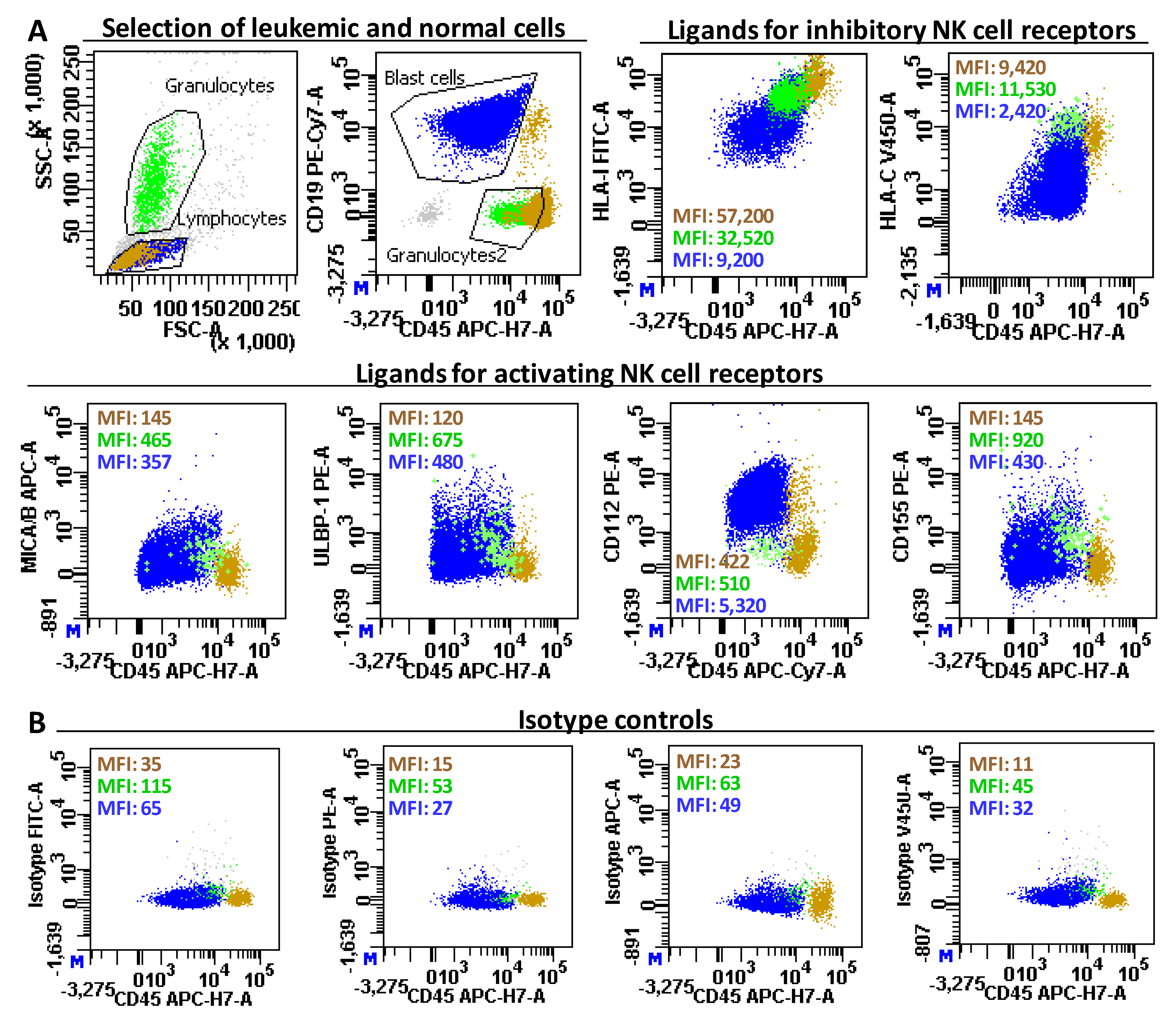
2.2. Patient Outcome According to the Expression of Ligands for NK Cell Receptors on Leukemic Blast Cells
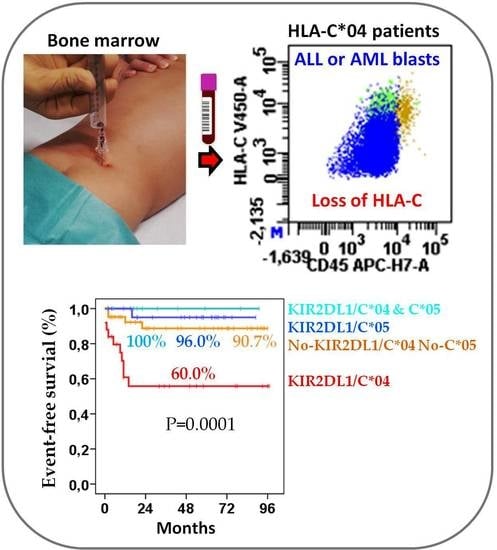
2.3. Role of KIR/HLA-Ligand Interactions in the Outcome of Acute Leukemia
2.4. KIR2DL1/C*04 Interaction Is an Independent Prognostic Biomarker That Complements Risk Status at Enrollment
2.5. Models to Predict NK Cell Alloreactivity in Allogeneic HSCT in Our Series
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. KIR and HLA Genotyping
4.3. Immunophenotyping of Normal Cells and Leukemic Blasts
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lonetti, A.; Pession, A.; Masetti, R. Targeted Therapies for Pediatric AML: Gaps and Perspective. Front. Pediatr. 2019, 7, 463. [Google Scholar] [CrossRef]
- Teachey, D.T.; Hunger, S.P.; Loh, M.L. Optimizing therapy in the modern age: Differences in length of maintenance therapy in acute lymphoblastic leukemia. Blood 2021, 137, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Bhojwani, D.; Pui, C.-H. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013, 14, e205-17. [Google Scholar] [CrossRef]
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef]
- von Stackelberg, A.; Locatelli, F.; Zugmaier, G.; Handgretinger, R.; Trippett, T.M.; Rizzari, C.; Bader, P.; O’Brien, M.M.; Brethon, B.; Bhojwani, D.; et al. Phase I/Phase II Study of Blinatumomab in Pediatric Patients with Relapsed/Refractory Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2016, 34, 4381–4389. [Google Scholar]
- Bhojwani, D.; Sposto, R.; Shah, N.N.; Rodriguez, V.; Yuan, C.; Stetler-Stevenson, M.; O’Brien, M.M.; McNeer, J.L.; Quereshi, A.; Cabannes, A.; et al. Inotuzumab ozogamicin in pediatric patients with relapsed/refractory acute lymphoblastic leukemia. Leukemia 2019, 33, 884–892. [Google Scholar] [CrossRef]
- Fuster, J.L.; Molinos-Quintana, A.; Fuentes, C.; Fernández, J.M.; Velasco, P.; Pascual, T.; Rives, S.; Dapena, J.L.; Sisinni, L.; López-Godino, O.; et al. Blinatumomab and inotuzumab for B cell precursor acute lymphoblastic leukaemia in children: A retrospective study from the Leukemia Working Group of the Spanish Society of Pediatric Hematology and Oncology (SEHOP). Br. J. Haematol. 2020, 190, 764–771. [Google Scholar] [PubMed]
- Davies, S.M.; Iannone, R.; Alonzo, T.A.; Wang, Y.-C.; Gerbing, R.; Soni, S.; Kolb, E.A.; Meshinchi, S.; Orchard, P.J.; Burns, L.J.; et al. A Phase 2 Trial of KIR-Mismatched Unrelated Donor Transplantation Using in Vivo T Cell Depletion with Antithymocyte Globulin in Acute Myelogenous Leukemia: Children’s Oncology Group AAML05P1 Study. Biol. Blood Marrow Transplant. 2020, 26, 712–717. [Google Scholar] [PubMed]
- Mamcarz, E.; Madden, R.; Qudeimat, A.; Srinivasan, A.; Talleur, A.; Sharma, A.; Suliman, A.; Maron, G.; Sunkara, A.; Kang, G.; et al. Improved survival rate in T-cell depleted haploidentical hematopoietic cell transplantation over the last 15 years at a single institution. Bone Marrow Transplant. 2020, 55, 929–938. [Google Scholar]
- Rubnitz, J.E.; Inaba, H.; Kang, G.; Gan, K.; Hartford, C.; Triplett, B.M.; Dallas, M.; Shook, D.; Gruber, T.; Pui, C.-H.; et al. Natural killer cell therapy in children with relapsed leukemia. Pediatr. Blood Cancer 2015, 62, 1468–1472. [Google Scholar] [CrossRef]
- Fernández, A.; Navarro-Zapata, A.; Escudero, A.; Matamala, N.; Ruz-Caracuel, B.; Mirones, I.; Pernas, A.; Cobo, M.; Casado, G.; Lanzarot, D.; et al. Optimizing the Procedure to Manufacture Clinical-Grade NK Cells for Adoptive Immunotherapy. Cancers 2021, 13, 577. [Google Scholar] [CrossRef] [PubMed]
- Yanir, A.D.; Martinez, C.A.; Sasa, G.; Leung, K.; Gottschalk, S.; Omer, B.; Ahmed, N.; Hegde, M.; Eunji, J.; Liu, H.; et al. Current Allogeneic Hematopoietic Stem Cell Transplantation for Pediatric Acute Lymphocytic Leukemia: Success, Failure and Future Perspectives-A Single-Center Experience, 2008 to 2016. Biol. Blood Marrow Transplant. 2018, 24, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Moretta, L.; Bottino, C.; Pende, D.; Castriconi, R.; Mingari, M.C.; Moretta, A. Surface NK receptors and their ligands on tumor cells. Semin. Immunol. 2006, 18, 151–158. [Google Scholar] [PubMed]
- Kärre, K. Natural killer cell recognition of missing self. Nat. Immunol. 2008, 9, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Parham, P. MHC class I molecules and KIRS in human history, health and survival. Nat. Rev. Immunol. 2005, 5, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Middleton, D.; Vilchez, J.R.; Cabrera, T.; Meenagh, A.; Williams, F.; Halfpenny, I.; Maleno, I.; Ruiz-Cabello, F.; Lopez-Nevot, M.A.; Garrido, F. Analysis of KIR gene frequencies in HLA class I characterised bladder, colorectal and laryngeal tumours. Tissue Antigens 2007, 69, 220–226. [Google Scholar] [CrossRef]
- Ruggeri, L.; Mancusi, A.; Perruccio, K.; Burchielli, E.; Martelli, M.F.; Velardi, A. Natural killer cell alloreactivity for leukemia therapy. J. Immunother. 2005, 28, 175–182. [Google Scholar]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; Del Zotto, G.; Pietra, G.; et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef]
- Hilton, H.G.; Parham, P. Missing or altered self: Human NK cell receptors that recognize HLA-C. Immunogenetics 2017, 69, 567–579. [Google Scholar] [CrossRef]
- Enqvist, M.; Ask, E.H.; Forslund, E.; Carlsten, M.; Abrahamsen, G.; Béziat, V.; Andersson, S.; Schaffer, M.; Spurkland, A.; Bryceson, Y.; et al. Coordinated Expression of DNAM-1 and LFA-1 in Educated NK Cells. J. Immunol. 2015, 194, 4518–4527. [Google Scholar] [CrossRef]
- Anfossi, N.; André, P.; Guia, S.; Falk, C.S.; Roetynck, S.; Stewart, C.A.; Breso, V.; Frassati, C.; Reviron, D.; Middleton, D.; et al. Human NK Cell Education by Inhibitory Receptors for MHC Class I. Immunity 2006, 25, 331–342. [Google Scholar] [CrossRef]
- Béziat, V.; Descours, B.; Parizot, C.; Debré, P.; Vieillard, V. NK cell terminal differentiation: Correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS ONE 2010, 5, e11966. [Google Scholar] [CrossRef]
- Kim, S.; Poursine-Laurent, J.; Truscott, S.M.; Lybarger, L.; Song, Y.J.; Yang, L.; French, A.R.; Sunwoo, J.B.; Lemieux, S.; Hansen, T.H.; et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 2005, 436, 709–713. [Google Scholar] [CrossRef]
- Mandelboim, O.; Reyburn, H.T.; Valés-Gómez, M.; Pazmany, L.; Colonna, M.; Borsellino, G.; Strominger, J.L. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J. Exp. Med. 1996, 184, 913–922. [Google Scholar] [CrossRef]
- Foley, B.A.; De Santis, D.; Van Beelen, E.; Lathbury, L.J.; Christiansen, F.T.; Witt, C.S. The reactivity of Bw4+ HLA-B and HLA-A alleles with kir3dll: Implications for patient and donor suitability for haploidentical stem cell transplantations. Blood 2008, 112, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Hansasuta, P.; Dong, T.; Thananchai, H.; Weekes, M.; Willberg, C.; Aldemir, H.; Rowland-Jones, S.; Braud, V.M. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 2004, 34, 1673–1679. [Google Scholar] [CrossRef]
- Horowitz, A.; Djaoud, Z.; Nemat-Gorgani, N.; Blokhuis, J.; Hilton, H.G.; Béziat, V.; Malmberg, K.-J.; Norman, P.J.; Guethlein, L.A.; Parham, P. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci. Immunol. 2016, 1, eaag1672. [Google Scholar] [CrossRef] [PubMed]
- Chewning, J.H.; Gudme, C.N.; Hsu, K.C.; Selvakumar, A.; Dupont, B. KIR2DS1-Positive NK Cells Mediate Alloresponse against the C2 HLA-KIR Ligand Group In Vitro. J. Immunol. 2007, 179, 854–868. [Google Scholar] [CrossRef]
- Fauriat, C.; Ivarsson, M.A.; Ljunggren, H.; Malmberg, K.; Michae, J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood 2010, 115, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Djaoud, Z.; Willem, C.; Legrand, N.; Rettman, P.; Gagne, K.; Cesbron, A.; Retiere, C. Large Spectrum of HLA-C Recognition by Killer Ig-like Receptor (KIR)2DL2 and KIR2DL3 and Restricted C1 Specificity of KIR2DS2: Dominant Impact of KIR2DL2/KIR2DS2 on KIR2D NK Cell Repertoire Formation. J. Immunol. 2013, 191, 4778–4788. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Z.; Ko, H.L.; Shen, M.; Ren, E.C. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc. Natl. Acad. Sci. USA 2014, 111, 2662–2667. [Google Scholar] [CrossRef]
- Thiruchelvam-Kyle, L.; Hoelsbrekken, S.E.; Saether, P.C.; Bjørnsen, E.G.; Pende, D.; Fossum, S.; Daws, M.R.; Dissen, E. The Activating Human NK Cell Receptor KIR2DS2 Recognizes a β2-Microglobulin-Independent Ligand on Cancer Cells. J. Immunol. 2017, 198, 2556–2567. [Google Scholar] [CrossRef] [PubMed]
- Graef, T.; Moesta, A.K.; Norman, P.J.; Abi-Rached, L.; Vago, L.; Older Aguilar, A.M.; Gleimer, M.; Hammond, J.A.; Guethlein, L.A.; Bushnell, D.A.; et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 2009, 206, 2557–2572. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Hölzemer, A.; Martrus, G.; Chung, A.W.; Pacheco, Y.; Simoneau, C.R.; Rucevic, M.; Lamothe-Molina, P.A.; Pertel, T.; Kim, T.; et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016, 17, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, G.M.; Yamada, E.; Rampersaud, A.; Thomas, R.; Carrington, M.; McVicar, D.W. Analysis of binding of KIR3DS1*014 to HLA Suggests Distinct Evolutionary History of KIR3DS1. J. Immunol. 2011, 187, 2162–2171. [Google Scholar] [CrossRef]
- Blokhuis, J.H.; Hilton, H.G.; Guethlein, L.A.; Norman, P.J.; Nemat-Gorgani, N.; Nakimuli, A.; Chazara, O.; Moffett, A.; Parham, P. KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans. Immun. Inflamm. Dis. 2017, 5, 461–468. [Google Scholar] [CrossRef]
- Nowak, J.; Kościńska, K.; Mika-Witkowska, R.; Rogatko-Koroś, M.; Mizia, S.; Jaskuła, E.; Polak, M.; Mordak-Domagała, M.; Lange, J.; Gronkowska, A.; et al. Role of Donor Activating KIR-HLA Ligand-Mediated NK Cell Education Status in Control of Malignancy in Hematopoietic Cell Transplant Recipients. Biol. Blood Marrow Transplant. 2015, 21, 829–839. [Google Scholar] [CrossRef][Green Version]
- Sullivan, E.M.; Jeha, S.; Kang, G.; Cheng, C.; Rooney, B.; Holladay, M.; Bari, R.; Schell, S.; Tuggle, M.; Pui, C.-H.; et al. NK cell genotype and phenotype at diagnosis of acute lymphoblastic leukemia correlate with postinduction residual disease. Clin. Cancer Res. 2014, 20, 5986–5994. [Google Scholar] [CrossRef]
- Misra, M.K.; Prakash, S.; Moulik, N.R.; Kumar, A.; Agrawal, S. Genetic associations of killer immunoglobulin like receptors and class I human leukocyte antigens on childhood acute lymphoblastic leukemia among north Indians. Hum. Immunol. 2016, 77, 41–46. [Google Scholar] [CrossRef]
- Cooley, S.; Trachtenberg, E.; Bergemann, T.L.; Saeteurn, K.; Klein, J.; Le, C.T.; Marsh, S.G.E.; Guethlein, L.A.; Parham, P.; Miller, J.S.; et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood 2009, 113, 726–732. [Google Scholar] [CrossRef]
- Coudert, J.D.; Zimmer, J.; Tomasello, E.; Cebecauer, M.; Colonna, M.; Vivier, E.; Held, W. Altered NKG2D function in NK cells induced by chronic exposure to NKG2D-ligand expressing tumor cells. Blood 2005, 106, 1711–1718. [Google Scholar] [CrossRef]
- Khaznadar, Z.; Boissel, N.; Agaugué, S.; Henry, G.; Cheok, M.; Vignon, M.; Geromin, D.; Cayuela, J.-M.; Castaigne, S.; Pautas, C.; et al. Defective NK Cells in Acute Myeloid Leukemia Patients at Diagnosis Are Associated with Blast Transcriptional Signatures of Immune Evasion. J. Immunol. 2015, 195, 2580–2590. [Google Scholar] [CrossRef]
- Carlsten, M.; Björkström, N.K.; Norell, H.; Bryceson, Y.; Van Hall, T.; Baumann, B.C.; Hanson, M.; Schedvins, K.; Kiessling, R.; Ljunggren, H.G.; et al. DNAX accessory molecule-1 mediated recognition of freshly isolated ovarian carcinoma by resting natural killer cells. Cancer Res. 2007, 67, 1317–1325. [Google Scholar] [CrossRef]
- Labani-Motlagh, A.; Israelsson, P.; Ottander, U.; Lundin, E.; Nagaev, I.; Nagaeva, O.; Dehlin, E.; Baranov, V.; Mincheva-Nilsson, L. Differential expression of ligands for NKG2D and DNAM-1 receptors by epithelial ovarian cancer-derived exosomes and its influence on NK cell cytotoxicity. Tumor Biol. 2016, 37, 5455–5466. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xi, C.; Zhu, Y.; Yin, L.; Wei, J.; Zhang, J.-J.; Liu, X.-C.; Guo, S.; Fu, Y.; Miao, Y. Altered expression of CD226 and CD96 on natural killer cells in patients with pancreatic cancer. Oncotarget 2016, 7, 66586–66594. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; De Andrade, L.F.; Vuckovic, S.; Miles, K.; Ngiow, S.F.; Yong, M.C.R.; Teng, M.W.L.; Colonna, M.; Ritchie, D.S.; Chesi, M.; et al. Immunosurveillance and therapy of multiple myeloma are CD226 dependent. J. Clin. Investig. 2015, 125, 2077–2089. [Google Scholar] [CrossRef] [PubMed]
- Sivori, S.; Vacca, P.; Del Zotto, G.; Munari, E.; Mingari, M.C.; Moretta, L. Human NK cells: Surface receptors, inhibitory checkpoints, and translational applications. Cell. Mol. Immunol. 2019, 16, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Guillamón, C.F.; Martínez-Sánchez, M.V.; Gimeno, L.; Mrowiec, A.; Martínez-García, J.; Server-Pastor, G.; Martínez-Escribano, J.; Torroba, A.; Ferri, B.; Abellán, D.; et al. NK Cell Education in Tumor Immune Surveillance: DNAM-1/KIR Receptor Ratios as Predictive Biomarkers for Solid Tumor Outcome. Cancer Immunol. Res. 2018. [Google Scholar] [CrossRef]
- Guillamón, C.F.; Martínez-Sánchez, M.V.; Gimeno, L.; Campillo, J.A.; Server-Pastor, G.; Martínez-García, J.; Martínez-Escribano, J.; Torroba, A.; Ferri, B.; Abellán, D.J.; et al. Activating KIRs on Educated NK Cells Support Downregulation of CD226 and Inefficient Tumor Immunosurveillance. Cancer Immunol. Res. 2019, 7, 1307–1317. [Google Scholar] [CrossRef]
- Martínez-Sánchez, M.V.; Periago, A.; Legaz, I.; Gimeno, L.; Mrowiec, A.; Montes-Barqueros, N.R.; Campillo, J.A.; Bolarin, J.M.; Bernardo, M.V.; López-Álvarez, M.R.; et al. Overexpression of KIR inhibitory ligands (HLA-I) determines that immunosurveillance of myeloma depends on diverse and strong NK cell licensing. Oncoimmunology 2016, 5, e1093721. [Google Scholar] [CrossRef]
- Gimeno, L.; Martínez-Banaclocha, H.; Bernardo, M.V.; Bolarin, J.M.; Marín, L.; López-Hernández, R.; López-Alvarez, M.R.; Moya-Quiles, M.R.; Muro, M.; Frias-Iniesta, J.F.; et al. NKG2D Polymorphism in Melanoma Patients from Southeastern Spain. Cancers 2019, 11, 438. [Google Scholar] [CrossRef]
- Locatelli, F.; Pende, D.; Mingari, M.C.; Bertaina, A.; Falco, M.; Moretta, A.; Moretta, L. Cellular and molecular basis of haploidentical hematopoietic stem cell transplantation in the successful treatment of high-risk leukemias: Role of alloreactive NK cells. Front. Immunol. 2013, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Symons, H.J.; Leffell, M.S.; Rossiter, N.D.; Zahurak, M.; Jones, R.J.; Fuchs, E.J. Improved survival with inhibitory killer immunoglobulin receptor (KIR) gene mismatches and KIR haplotype B donors after nonmyeloablative, HLA-haploidentical bone marrow transplantation. Biol. Blood Marrow Transplant. 2010, 16, 533–542. [Google Scholar] [CrossRef]
- Nielsen, S.N.; Grell, K.; Nersting, J.; Abrahamsson, J.; Lund, B.; Kanerva, J.; Jónsson, Ó.G.; Vaitkeviciene, G.; Pruunsild, K.; Hjalgrim, L.L.; et al. DNA-thioguanine nucleotide concentration and relapse-free survival during maintenance therapy of childhood acute lymphoblastic leukaemia (NOPHO ALL2008): A prospective substudy of a phase 3 trial. Lancet Oncol. 2017, 18, 515–524. [Google Scholar] [CrossRef]
- Ullah, M.A.; Hill, G.R.; Tey, S.-K. Functional Reconstitution of Natural Killer Cells in Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 2016, 7, 144. [Google Scholar] [CrossRef]
- Ek, T.; Mellander, L.; Andersson, B.; Abrahamsson, J. Immune reconstitution after childhood acute lymphoblastic leukemia is most severely affected in the high risk group. Pediatr. Blood Cancer 2005, 44, 461–468. [Google Scholar] [CrossRef]
- Shelikhova, L.; Ilushina, M.; Shekhovtsova, Z.; Shasheleva, D.; Khismatullina, R.; Kurnikova, E.; Pershin, D.; Balashov, D.; Radygina, S.; Trakhtman, P.; et al. αβ T Cell-Depleted Haploidentical Hematopoietic Stem Cell Transplantation without Antithymocyte Globulin in Children with Chemorefractory Acute Myelogenous Leukemia. Biol. Blood Marrow Transplant. 2019, 25, e179–e182. [Google Scholar] [CrossRef]
- Sharma, A.; Rastogi, N.; Chatterjee, G.; Kapoor, R.; Nivargi, S.; Yadav, S.P. Haploidentical Stem Cell Transplantation With Posttransplant Cyclophosphamide for Pediatric Acute Leukemia is Safe and Effective. J. Pediatr. Hematol. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Katsanis, E.; Sapp, L.N.; Reid, S.C.; Reddivalla, N.; Stea, B. T-Cell Replete Myeloablative Haploidentical Bone Marrow Transplantation Is an Effective Option for Pediatric and Young Adult Patients With High-Risk Hematologic Malignancies. Front. Pediatr. 2020, 8, 282. [Google Scholar] [CrossRef] [PubMed]
- Schmiegelow, K. Maintenance therapy of childhood acute lymphoblastic leukemia: Do all roads lead to Rome? Pediatr. Blood Cancer 2020, 67, e28418. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Brady, S.W.; Ma, X.; Shen, S.; Zhang, Y.; Li, Y.; Szlachta, K.; Dong, L.; Liu, Y.; Yang, F.; et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 2020, 135, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Apps, R.; Carrington, M. Response to Comment on “Influence of HLA-C Expression Level on HIV Control. Science 2013, 341, 1175. [Google Scholar] [CrossRef][Green Version]
- Babor, F.; Manser, A.R.; Fischer, J.C.; Scherenschlich, N.; Enczmann, J.; Chazara, O.; Moffett, A.; Borkhardt, A.; Meisel, R.; Uhrberg, M. KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood 2014, 124, 2248–2251. [Google Scholar] [CrossRef]
- Bortin, M.M.; D’Amaro, J.; Bach, F.H.; Rimm, A.A.; van Rood, J.J. HLA associations with leukemia. Blood 1987, 70, 227–232. [Google Scholar] [CrossRef]
- Caruso, C.; Cammarata, G.; Sireci, G.; Modica, M.A. HLA-Cw4 association with acute lymphoblastic leukaemia in Sicilian patients. Vox Sang. 1988, 54, 57–58. [Google Scholar]
- Baldauf, C.; Kubel, M. [HLA and leukemia]. Folia Haematol. Int. Mag. Klin. Morphol. Blutforsch. 1989, 116, 123–132. [Google Scholar]
- Karabon, L.; Jedynak, A.; Giebel, S.; Wołowiec, D.; Kielbinski, M.; Woszczyk, D.; Kapelko-Slowik, K.; Kuliczkowski, K.; Frydecka, I. KIR/HLA gene combinations influence susceptibility to B-cell chronic lymphocytic leukemia and the clinical course of disease. Tissue Antigens 2011, 78, 129–138. [Google Scholar] [CrossRef]
- Arima, N.; Kanda, J.; Tanaka, J.; Yabe, T.; Morishima, Y.; Kim, S.-W.; Najima, Y.; Ozawa, Y.; Eto, T.; Kanamori, H.; et al. Homozygous HLA-C1 is Associated with Reduced Risk of Relapse after HLA-Matched Transplantation in Patients with Myeloid Leukemia. Biol. Blood Marrow Transplant. 2018, 24, 717–725. [Google Scholar] [PubMed]
- Cárceles-Álvarez, A.; Ortega-García, J.A.; López-Hernández, F.A.; Fuster-Soler, J.L.; Ramis, R.; Kloosterman, N.; Castillo, L.; Sánchez-Solís, M.; Claudio, L.; Ferris-Tortajada, J. Secondhand smoke: A new and modifiable prognostic factor in childhood acute lymphoblastic leukemias. Environ. Res. 2019, 178, 108689. [Google Scholar] [CrossRef]
- Mastaglio, S.; Wong, E.; Perera, T.; Ripley, J.; Blombery, P.; Smyth, M.J.; Koldej, R.; Ritchie, D. Natural killer receptor ligand expression on acute myeloid leukemia impacts survival and relapse after chemotherapy. Blood Adv. 2018, 2, 335–346. [Google Scholar] [PubMed]
- Epling-Burnette, P.K.; Bai, F.; Painter, J.S.; Rollison, D.E.; Salih, H.R.; Krusch, M.; Zou, J.; Ku, E.; Zhong, B.; Boulware, D.; et al. Reduced natural killer (NK) function associated with high-risk myelodysplastic syndrome (MDS) and reduced expression of activating NK receptors. Blood 2007, 109, 4816–4824. [Google Scholar] [CrossRef] [PubMed]
- Sabattini, E.; Bacci, F.; Sagramoso, C.; Pileri, S.A. WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: An overview. Pathologica 2010, 102, 83–87. [Google Scholar]




| Total (n = 102) | B-ALL (n = 70) | T-ALL (n = 16) | AML (n = 16) | |
|---|---|---|---|---|
| Age (month, mean ± SD) | 83.0 ± 49.3 | 82.5 ± 49.1 | 91.7 ± 44.9 | 76.6 ± 55.6 |
| Sex (male, n (%)) | 63 (61.8%) | 42 (60.0%) | 12 (75.0%) | 9 (56.3%) |
| Inclusion in the study | ||||
| At diagnosis, n (%) | 96 (94.1%) | 66 (94.3%) | 16 (100%) | 14 (87.5%) |
| At relapse, n (%) | 6 (5.9%) | 4 (5.7%) | 2 (12.5%) | |
| PB-WBC count at diagnosis | ||||
| <10 × 103/µL | 89 (87.3%) | 65 (92.9%) | 10 (62.5%) | 14 (87.5%) |
| 10–50 × 103/µL | 5 (4.9%) | 4 (5.7%) | 1 (6.3%) | |
| >50 × 103/µL | 8 (7.8%) | 1 (1.4%) | 6 (37.5%) | 1 (6.3%) |
| Risk stratification 1 | ||||
| Standard, n (%) | 27 (26.4%) | 23 (32.9%) | 0 (0.0%) | 3 (18.7%) |
| Intermediate, n (%) | 41 (40.2%) | 29 (41.1%) | 11 (68.7%) | 2 (12.5%) |
| High, n (%) | 34 (34.0%) | 18 (25.7%) | 5 (31.3%) | 11 (68.8%) |
| Treatments | ||||
| PETHEMA-96/2001/2005 | 13 (12.6%) | 12 (17.0%) | 1 (6.2%) | |
| LAL-SEHOP-PETHEMA-2013 | 73 (71.4%) | 58 (82.7%) | 15 (93.7%) | |
| LMA-SHOP-2007 | 10 (9.8%) | 10 (62.5%) | ||
| NOPHO-DBH-AML-2012 | 6 (5.8%) | 6 (37.5%) | ||
| HSCT, n (%) | 18 (17.6%) | 8 (11.4%) | 3 (18.8%) | 7 (43.8%) |
| Allo-HSCT, n (%) | 13 (12.7%) | 5 (7.1%) | 3 (18.8%) | 5 (31.3%) |
| Related, n (%) | 5 (38.5%) | 2 (40.0%) | 2 (66.7%) | 3 (42.9%) |
| Unrelated, n (%) | 8 (61.5%) | 3 (60.0%) | 1 (33.3%) | 4 (57.1%) |
| Haplo-HSCT, n (%) | 5 (4.9%) | 3 (4.3%) | 2 (12.5%) | |
| Clinical outcome | ||||
| Mean follow-up (months) | 41.98 ± 31.5 | 41.9 ± 32.9 | 46.6 ± 30.6 | 37.7 ± 25.9 |
| Relapse, n (%) | 9 (8.8%) | 2 (2.8%) | 2 (12.5%) | 5 (31.3%) |
| Death, n (%) | 12 (11.8%) | 7 (10.0%) | 5 (31.3%) |
| KIR Genes, n (%) | Controls (n = 83) | Patients (n = 102) | B-ALL (n = 70) | T-ALL (n = 16) | AML (n = 16) |
|---|---|---|---|---|---|
| KIR2DL1 | 80 (96.4%) | 99 (97.1%) | 68 (97.1%) | 16 (100%) | 15 (93.8%) |
| KIR2DL2/2DS2 1 | 48 (57.8%) | 49 (48.0%) | 32 (45.7%) | 7 (43.8%) | 10 (62.5%) |
| KIR2DL3 | 74 (89.2%) | 93 (91.2%) | 64 (91.4%) | 15 (93.8%) | 14 (87.5%) |
| KIR2DL5 | 44 (53.0%) | 56 (54.9%) | 35 (50.0%) | 8 (50.0%) | 13 (81.3%) |
| KIR3DL1 | 81 (97.6%) | 100 (98.0%) | 68 (97.1%) | 16 (100%) | 16 (100%) |
| KIR2DS1 | 30 (36.1%) | 38 (37.3%) | 23 (32.9%) | 7 (43.8%) | 8 (50.0%) |
| KIR2DS3 | 26 (31.3%) | 35 (34.3%) | 21 (30.0%) | 5 (31.3%) | 9 (56.3%) |
| KIR2DS4full | 35 (42.2%) | 54 (52.9%) | 34 (48.6%) | 11 (68.8%) | 9 (56.3%) |
| KIR2DS5 | 26 (31.3%) | 30 (29.4%) | 19 (27.1%) | 6 (37.5%) | 5 (31.3%) |
| KIR3DS1 | 29 (34.9%) | 38 (37.3%) | 24 (34.3%) | 7 (43.8%) | 7 (43.8%) |
| HLA-I ligands, n (%) | |||||
| HLA-A*03/A*11 | 23 (27.7%) | 30 (30.6%) | 20 (30.8%) | 4 (25.0%) | 6 (37.5%) |
| Bw4 epitope | 65 (78.3%) | 70 (68.6%) | 49 (70.0%) | 10 (62.5%) | 11 (68.8%) |
| Bw4-80T | 20 (24.1%) | 22 (21.6%) | 16 (22.9%) | 4 (25.0%) | 2 (12.5%) |
| Bw4-80I | 53 (63.9%) | 61 (59.8%) | 44 (62.9%) | 8 (50.0%) | 9 (56.3.3%) |
| HLA-C1 epitope | 61 (73.5%) | 80 (78.4%) | 56 (80.0%) | 13 (81.3%) | 11 (68.8%) |
| HLA-C2 epitope | 56 (67.5%) | 73 (71.6%) | 48 (68.6%) | 10 (62.5%) | 15 (93.8%) |
| KIR2DS4-ligands 2 | 44 (53.0%) | 71 (71.0%) 3 | 49 (72.1%) | 10 (62.5%) | 12 (75.0%) |
| KIR/HLA-ligand interactions, n (%) | |||||
| KIR2DL1/C2 | 54 (65.1%) | 70 (68.6%) | 46 (65.7%) | 10 (62.5%) | 14 (87.5%) |
| KIR2DL2/C1 | 34 (41.0%) | 38 (37.3%) | 27 (38.6%) | 5 (31.3%) | 6 (37.5%) |
| KIR2DL3/C1 | 54 (65.1%) | 73 (71.6%) | 52 (74.3%) | 12 (75.0%) | 9 (56.3%) |
| KIR3DL1/Bw4 | 63 (75.9%) | 66 (68.0%) | 45 (69.2%) | 10 (62.5%) | 11 (68.8%) |
| KIR2DS1/C2 | 22 (26.5%) | 28 (27.5%) | 16 (22.9%) | 5 (31.3%) | 7 (43.8%) |
| KIR2DS4/Ligands 1 | 18 (21.7%) | 33 (35.1%) | 23 (35.4%) | 5 (31.1%) | 6 (37.5%) |
| KIR2DS4/C*04:01 | 7 (8.4%) | 15 (15.5%) | 10 (15.4%) | 2 (15.5%) | 3 (18.8%) |
| KIR3DS1/B*51 | 3 (3.6%) | 4 (3.92%) | 2 (2.85%) | 2 (12.5%) | 0 (0.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Sánchez, M.V.; Fuster, J.L.; Campillo, J.A.; Galera, A.M.; Bermúdez-Cortés, M.; Llinares, M.E.; Ramos-Elbal, E.; Pascual-Gázquez, J.F.; Fita, A.M.; Martínez-Banaclocha, H.; et al. Expression of NK Cell Receptor Ligands on Leukemic Cells Is Associated with the Outcome of Childhood Acute Leukemia. Cancers 2021, 13, 2294. https://doi.org/10.3390/cancers13102294
Martínez-Sánchez MV, Fuster JL, Campillo JA, Galera AM, Bermúdez-Cortés M, Llinares ME, Ramos-Elbal E, Pascual-Gázquez JF, Fita AM, Martínez-Banaclocha H, et al. Expression of NK Cell Receptor Ligands on Leukemic Cells Is Associated with the Outcome of Childhood Acute Leukemia. Cancers. 2021; 13(10):2294. https://doi.org/10.3390/cancers13102294
Chicago/Turabian StyleMartínez-Sánchez, María Victoria, José Luis Fuster, José Antonio Campillo, Ana María Galera, Mar Bermúdez-Cortés, María Esther Llinares, Eduardo Ramos-Elbal, Juan Francisco Pascual-Gázquez, Ana María Fita, Helios Martínez-Banaclocha, and et al. 2021. "Expression of NK Cell Receptor Ligands on Leukemic Cells Is Associated with the Outcome of Childhood Acute Leukemia" Cancers 13, no. 10: 2294. https://doi.org/10.3390/cancers13102294
APA StyleMartínez-Sánchez, M. V., Fuster, J. L., Campillo, J. A., Galera, A. M., Bermúdez-Cortés, M., Llinares, M. E., Ramos-Elbal, E., Pascual-Gázquez, J. F., Fita, A. M., Martínez-Banaclocha, H., Galián, J. A., Gimeno, L., Muro, M., & Minguela, A. (2021). Expression of NK Cell Receptor Ligands on Leukemic Cells Is Associated with the Outcome of Childhood Acute Leukemia. Cancers, 13(10), 2294. https://doi.org/10.3390/cancers13102294