Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
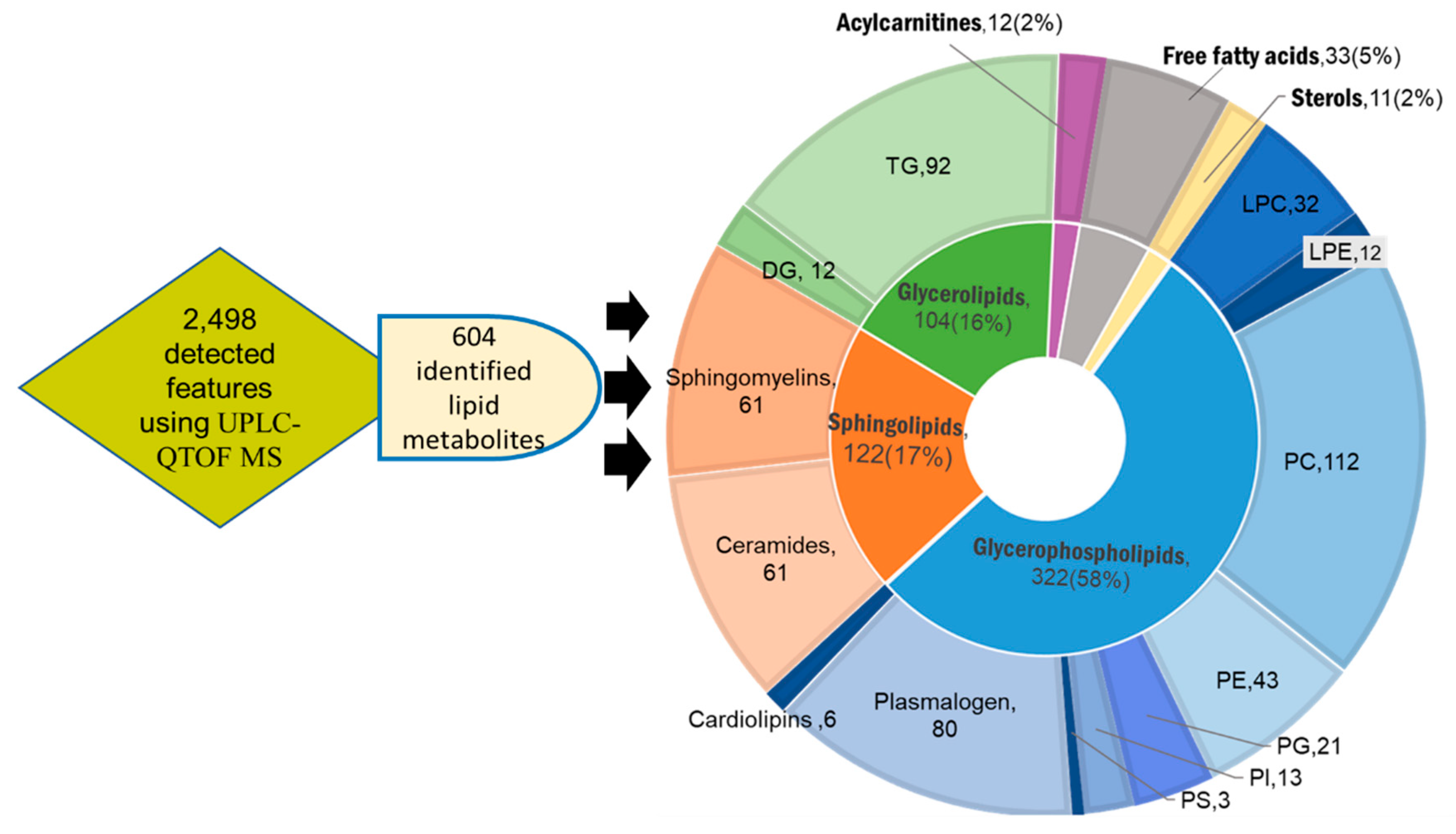
2.1. Untargeted Lipidomic Using UPLC MS-QTOF of Whole Blood and Liver Tissues of Hepatocellular Carcinoma (HCC) and Chronic Liver Disease (CLD) Patients and Healthy Control Subjects
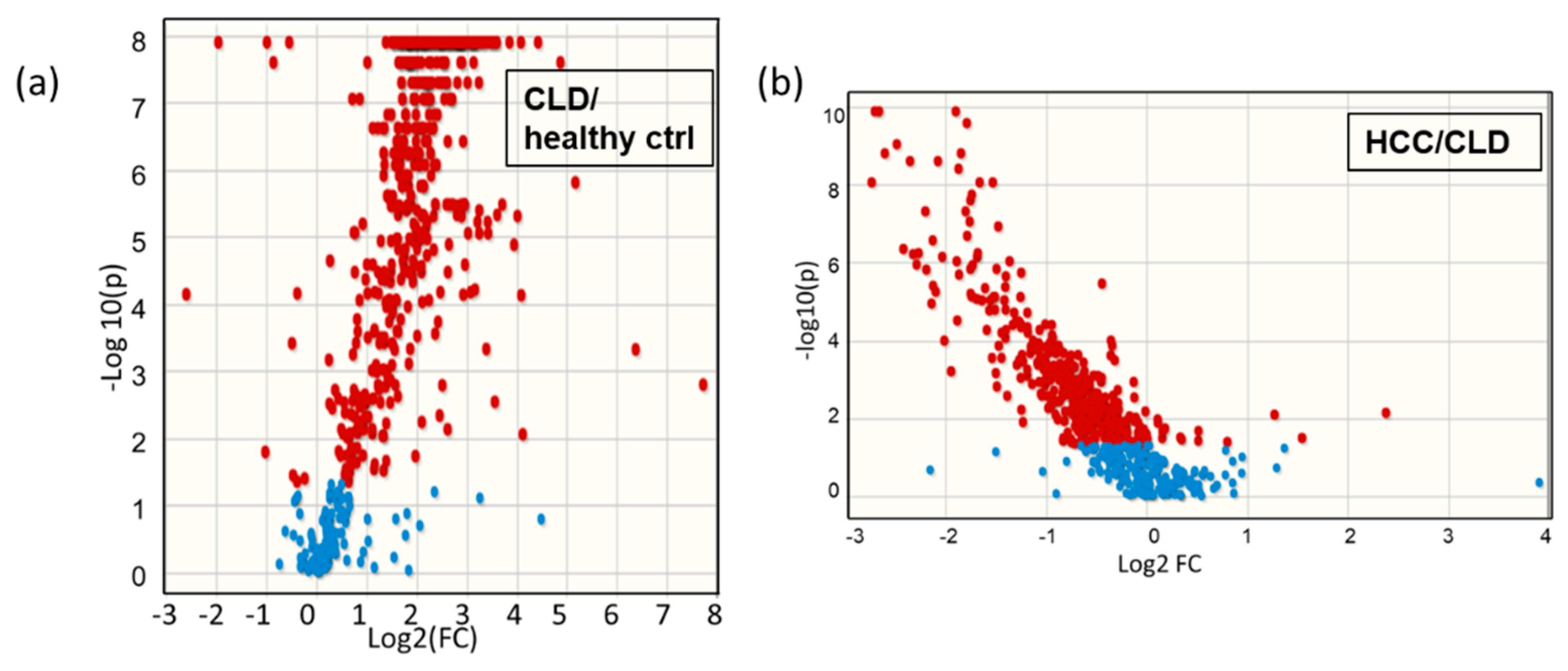
2.2. Blood Lipidomic Profiles of HCC and CLD Patients and Healthy Control Subjects
2.3. Lipidomic Profiles of Resected HCC Tumor Tissues Compared to Paired Nonmalignant Hepatic Tissues
2.4. Differential Desaturation of Lipid Species Defines Metabotypes Both in Blood and in Resected Liver Tissues
2.5. Similarities and Differences in Lipid Metabolism the Transition from CLD to HCC Disease Patients
3. Discussion
4. Materials and Methods
4.1. Participants and Collection of Clinical Samples
4.2. Sample Pretreatment
4.3. Lipidomic Data Acquisition and Data Processing
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. In The Liver; Wiley: Hoboken, NJ, USA, 2020; pp. 758–772. [Google Scholar]
- McGlynn, K.A.; Petrick, J.L.; London, W.T. Global Epidemiology of Hepatocellular Carcinoma: An Emphasis on Demographic and Regional Variability. Clin. Liver Dis. 2015, 19, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Muralidharan, P.; Raj, J.P. Update in global trends and aetiology of hepatocellular carcinoma. Wspolczesna Onkol. 2018, 22, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Ahmed, F.; Mara, K.C.; Addissie, B.D.; Allen, A.M.; Gores, G.J.; Roberts, L.R. Diabetes Is Associated with Increased Risk of Hepatocellular Carcinoma in Patients with Cirrhosis From Nonalcoholic Fatty Liver Disease. Hepatology 2020, 71, 907–916. [Google Scholar] [CrossRef]
- Tsoulfas, G. Hepatocellular carcinoma and metabolic syndrome: The times are changing and so should we. World J. Gastroenterol. 2019, 25, 3842–3848. [Google Scholar] [CrossRef]
- Henry, Z.H.; Caldwell, S.H. Obesity and Hepatocellular Carcinoma: A Complex Relationship. Gastroenterology 2015. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Green, P.; Kerr, K.F.; Berry, K. Models estimating risk of hepatocellular carcinoma in patients with alcohol or NAFLD-related cirrhosis for risk stratification. J. Hepatol. 2019, 71, 523–533. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Feng, X.; Wang, R.; Wang, Y.; Zeng, H.; Qi, J.; Zhao, H.; Li, N.; Cai, J.; et al. Contribution of hepatitis B virus and hepatitis C virus to liver cancer in China north areas: Experience of the Chinese National Cancer Center. Int. J. Infect. Dis. 2017, 65, 15–21. [Google Scholar] [CrossRef]
- Kimhofer, T.; Fye, H.; Taylor-Robinson, S.; Thursz, M.; Holmes, E. Proteomic and metabonomic biomarkers for hepatocellular carcinoma: A comprehensive review. Br. J. Cancer 2015, 112, 1141–1156. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef]
- Alamri, Z.Z. The role of liver in metabolism: An updated review with physiological emphasis. Int. J. Basic Clin. Pharmacol. 2018, 7, 2271. [Google Scholar] [CrossRef]
- Arvind, A.; Osganian, S.A.; Cohen, D.E.; Corey, K.E. Lipid and Lipoprotein Metabolism in Liver Disease; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Reddy, J.K.; Rao, M.S. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, 852–858. [Google Scholar] [CrossRef]
- Passos-Castilho, A.M.; Carvalho, V.M.; Cardozo, K.H.M.; Kikuchi, L.; Chagas, A.L.; Gomes-Gouvêa, M.S.; Malta, F.; de Seixas-Santos Nastri, A.C.; Pinho, J.R.R.; Carrilho, F.J.; et al. Serum lipidomic profiling as a useful tool for screening potential biomarkers of hepatitis B-related hepatocellular carcinoma by ultraperformance liquid chromatography-mass spectrometry. BMC Cancer 2015, 15. [Google Scholar] [CrossRef]
- Li, Z.; Guan, M.; Lin, Y.; Cui, X.; Zhang, Y.; Zhao, Z.; Zhu, J. Aberrant lipid metabolism in hepatocellular carcinoma revealed by liver lipidomics. Int. J. Mol. Sci. 2017, 18, 2550. [Google Scholar] [CrossRef]
- Chen, X.; Gao, J.; Wang, T.; Jiang, X.; Chen, J.; Liang, X.; Wu, J. Hepatocarcinoma discrimination by ratiometric lipid profiles using tip-contact sampling/ionization mass spectrometry. Anal. Chem. 2019, 91, 10376–10380. [Google Scholar] [CrossRef]
- Lin, L.; Ding, Y.; Wang, Y.; Wang, Z.; Yin, X.; Yan, G.; Zhang, L.; Yang, P.; Shen, H. Functional lipidomics: Palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology 2017, 66, 432–448. [Google Scholar] [CrossRef]
- Kessler, S.M.; Simon, Y.; Gemperlein, K.; Gianmoena, K.; Cadenas, C.; Zimmer, V.; Pokorny, J.; Barghash, A.; Helms, V.; Van Rooijen, N.; et al. Fatty acid elongation in non-alcoholic steatohepatitis and hepatocellular carcinoma. Int. J. Mol. Sci. 2014, 15, 5762–5773. [Google Scholar] [CrossRef]
- Chen, S.; Yin, P.; Zhao, X.; Xing, W.; Hu, C.; Zhou, L.; Xu, G. Serum lipid profiling of patients with chronic hepatitis B, cirrhosis, and hepatocellular carcinoma by ultra fast LC/IT-TOF MS. Electrophoresis 2013, 34, 2848–2856. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Q.; Yin, P.; Xing, W.; Wu, Z.; Chen, S.; Lu, X.; Zhang, Y.; Lin, X.; Xu, G. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal. Bioanal. Chem. 2012, 403, 203–213. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Huang, C.; Li, N.; Zou, L.; Chia, S.E.; Chen, S.; Yu, K.; Ling, Q.; Cheng, Q.; et al. Comparison of hepatic and serum lipid signatures in hepatocellular carcinoma patients leads to the discovery of diagnostic and prognostic biomarkers. Oncotarget 2018, 9, 5032–5043. [Google Scholar] [CrossRef] [PubMed]
- Krautbauer, S.; Meier, E.M.; Rein-Fischboeck, L.; Pohl, R.; Weiss, T.S.; Sigruener, A.; Aslanidis, C.; Liebisch, G.; Buechler, C. Ceramide and polyunsaturated phospholipids are strongly reduced in human hepatocellular carcinoma. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.T.; Fiehn, O.; Elfert, A.; Helal, M.; Salama, I.; El-Said, H. Sugar alcohols have a key role in pathogenesis of chronic liver disease and hepatocellular carcinoma in whole blood and liver tissues. Cancers 2020, 12, 484. [Google Scholar] [CrossRef] [PubMed]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Denkert, C.; Bucher, E.; Hilvo, M.; Salek, R.; Orešič, M.; Griffin, J.; Brockmöller, S.; Klauschen, F.; Loibl, S.; Barupal, D.K.; et al. Metabolomics of human breast cancer: New approaches for tumor typing and biomarker discovery. Genome Med. 2012, 4, 37. [Google Scholar] [CrossRef]
- Miyamoto, S.; Taylor, S.L.; Barupal, D.K.; Taguchi, A.; Wohlgemuth, G.; Wikoff, W.R.; Yoneda, K.Y.; Gandara, D.R.; Hanash, S.M.; Kim, K.; et al. Systemic metabolomic changes in blood samples of lung cancer patients identified by gas chromatography Time-Of-Flight mass spectrometry. Metabolites 2015, 5, 192–210. [Google Scholar] [CrossRef]
- El-Kenawi, A. Cancer likes it saturated. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Nguyen, L.N.; Lim, Y.-S.; Pham, L.V.; Shin, H.-Y.; Kim, Y.-S.; Hwang, S.B. Stearoyl Coenzyme A Desaturase 1 Is Associated with Hepatitis C Virus Replication Complex and Regulates Viral Replication. J. Virol. 2014, 88, 12311–12325. [Google Scholar] [CrossRef]
- Tracz-Gaszewska, Z.; Dobrzyn, P. Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers 2019, 11, 948. [Google Scholar] [CrossRef]
- Stringer, K.A.; Younger, J.G.; McHugh, C.; Yeomans, L.; Finkel, M.A.; Puskarich, M.A.; Jones, A.E.; Trexel, J.; Karnovsky, A. Whole Blood Reveals More Metabolic Detail of the Human Metabolome than Serum as Measured by 1H-NMR Spectroscopy: Implications for Sepsis Metabolomics. Shock 2015, 44, 200–208. [Google Scholar] [CrossRef]
- Amézaga, J.; Arranz, S.; Urruticoechea, A.; Ugartemendia, G.; Larraioz, A.; Louka, M.; Uriarte, M.; Ferreri, C.; Tueros, I. Altered red blood cell membrane fatty acid profile in cancer patients. Nutrients 2018, 10, 1853. [Google Scholar] [CrossRef]
- Fenton, J.I.; Gurzell, E.A.; Davidson, E.A.; Harris, W.S. Red blood cell PUFAs reflect the phospholipid PUFA composition of major organs. Prostaglandins Leukot. Essent. Fat. Acids 2016, 112, 12–23. [Google Scholar] [CrossRef]
- Nono Nankam, P.A.; Mendham, A.E.; Jaarsveld, P.J.; Adams, K.; Fortuin-de Smidt, M.C.; Clamp, L.; Blüher, M.; Goedecke, J.H. Exercise Training Alters Red Blood Cell Fatty Acid Desaturase Indices and Adipose Tissue Fatty Acid Profile in African Women with Obesity. Obesity 2020, 28, 1456–1466. [Google Scholar] [CrossRef]
- Bonet, M.L.; Ribot, J.; Palou, A. Lipid metabolism in mammalian tissues and its control by retinoic acid. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2012, 1821, 177–189. [Google Scholar] [CrossRef]
- Conraads, V. Association of lipoproteins with cytokines and cytokine receptors in heart failure patients Differences between ischaemic versus idiopathic cardiomyopathy. Eur. Heart J. 2003, 24, 2221–2226. [Google Scholar] [CrossRef]
- Tessari, P.; Coracina, A.; Cosma, A.; Tiengo, A. Hepatic lipid metabolism and non-alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 2009, 19, 291–302. [Google Scholar] [CrossRef]
- Munir, R.; Lisec, J.; Swinnen, J.V.; Zaidi, N. Lipid metabolism in cancer cells under metabolic stress. Br. J. Cancer 2019, 120, 1090–1098. [Google Scholar] [CrossRef]
- Wang, M.; Han, J.; Xing, H.; Zhang, H.; Li, Z.; Liang, L.; Li, C.; Dai, S.; Wu, M.; Shen, F.; et al. Dysregulated fatty acid metabolism in hepatocellular carcinoma. Hepatic Oncol. 2016, 3, 241–251. [Google Scholar] [CrossRef]
- Quaye, O.; Amuzu, B.G.; Adadey, S.M.; Tagoe, E.A. Effect of Hepatitis B Virus (HBV) Infection on Lipid Profile in Ghanaian Patients. Virol. Res. Treat. 2019, 10. [Google Scholar] [CrossRef]
- You, X.; Liu, F.; Zhang, T.; Li, Y.; Ye, L.; Zhang, X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis 2013, 34, 1644–1652. [Google Scholar] [CrossRef]
- Lavie, M.; Dubuisson, J. Interplay between hepatitis C virus and lipid metabolism during virus entry and assembly. Biochimie 2017, 141, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Bartenschlager, R.; Penin, F.; Lohmann, V.; André, P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2010, 19, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Hishiki, T.; Shimizu, Y.; Funami, K.; Sugiyam, K.; Miyanari, Y.; Shimotohno, K. Hepatitis C virus utilizes lipid droplet for production of infectious virus. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 1–21. [Google Scholar] [CrossRef]
- Wu, X.; Daniels, G.; Lee, P.; Monaco, M.E. Lipid metabolism in prostate cancer. Am. J. Clin. Exp. Urol. 2014, 2, 111–120. [Google Scholar]
- Blücher, C.; Stadler, S.C. Obesity and breast cancer: Current insights on the role of fatty acids and lipid metabolism in promoting breast cancer growth and progression. Front. Endocrinol. 2017, 8, 293. [Google Scholar] [CrossRef]
- Parrales, A.; Iwakuma, T. p53 as a regulator of lipid metabolism in cancer. Int. J. Mol. Sci. 2016, 17, 2074. [Google Scholar] [CrossRef]
- Ribas, V.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondria, cholesterol and cancer cell metabolism. Clin. Transl. Med. 2016, 5, 22. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Wang, X.; Dang, Z.; Jiang, Y.; Wang, X.; Yang, Z. Effect of serum triglyceride level on the prognosis of patients with hepatocellular carcinoma in the absence of cirrhosis. Lipids Health Dis. 2018, 17, 248. [Google Scholar] [CrossRef]
- Ooi, K.; Shiraki, K.; Sakurai, Y.; Morishita, Y.; Nobori, T. Clinical significance of abnormal lipoprotein patterns in liver diseases. Int. J. Mol. Med. 2005, 15, 655–660. [Google Scholar] [CrossRef]
- Cicognani, C. Serum Lipid and Lipoprotein Patterns in Patients With Liver Cirrhosis and Chronic Active Hepatitis. Arch. Intern. Med. 1997, 157, 792. [Google Scholar] [CrossRef] [PubMed]
- Lipoprotein (a) Behaviour in Patients with Hepatocellular Carcinoma—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/11675573/ (accessed on 20 October 2020).
- Lisec, J.; Jaeger, C.; Rashid, R.; Munir, R.; Zaidi, N. Cancer cell lipid class homeostasis is altered under nutrient-deprivation but stable under hypoxia. BMC Cancer 2019, 19, 501. [Google Scholar] [CrossRef] [PubMed]
- Munir, R.; Lisec, J.; Jaeger, C.; Zaidi, N. Abundance, fatty acid composition and saturation index of neutral lipids are significantly different between isogenic primary and metastatic colon cancer cell lines. bioRxiv 2020, 7, 690685. Available online: https://www.biorxiv.org/content/biorxiv/early/2019/07/04/690685.full.pdf (accessed on 20 October 2020).
- Kang, Y.P.; Yoon, J.-H.; Long, N.P.; Koo, G.-B.; Noh, H.-J.; Oh, S.-J.; Lee, S.B.; Kim, H.M.; Hong, J.Y.; Lee, W.J.; et al. Spheroid-Induced Epithelial-Mesenchymal Transition Provokes Global Alterations of Breast Cancer Lipidome: A Multi-Layered Omics Analysis. Front. Oncol. 2019, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Urso, C.J.; Jadeja, V. Saturated fatty acids in obesity-associated inflammation. J. Inflamm. Res. 2020, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lyn, R.K.; Singaravelu, R.; Kargman, S.; O’Hara, S.; Chan, H.; Oballa, R.; Huang, Z.; Jones, D.M.; Ridsdale, A.; Russell, R.S.; et al. Stearoyl-CoA desaturase inhibition blocks formation of hepatitis C virus-induced specialized membranes. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef]
- Hall, Z.; Chiarugi, D.; Charidemou, E.; Leslie, J.; Scott, E.; Pellegrinet, L.; Allison, M.; Mocciaro, G.; Anstee, Q.M.; Evan, G.I.; et al. Lipid remodelling in hepatocyte proliferation and hepatocellular carcinoma. Hepatology 2020. [Google Scholar] [CrossRef]
- Heyd, V.L.; Eynard, A.R. Effects of eicosatrienoic acid (20:3 n-9, Mead’s acid) on some promalignant-related properties of three human cancer cell lines. Prostaglandins Other Lipid Mediat. 2003, 71, 177–188. [Google Scholar] [CrossRef]
- Cabré, E.; Gassull, M.A. Polyunsaturated fatty acid deficiency in liver diseases: Pathophysiological and clinical significance. Nutrition 1996, 12, 542–548. [Google Scholar] [CrossRef]
- Hsu, P.; Shi, Y. Regulation of autophagy by mitochondrial phospholipids in health and diseases. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 114–129. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Sun, H.; Liu, X.; Zheng, Y.; Xu, D.; Wang, J.; Jia, D.; Han, X.; Liu, F.; et al. Defective Phosphatidylglycerol Remodeling Causes Hepatopathy, Linking Mitochondrial Dysfunction to Hepatosteatosis. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 763–781. [Google Scholar] [CrossRef]
- Gault, C.R.; Obeid, L.M.; Hannun, Y.A. An overview of sphingolipid metabolism: From synthesis to breakdown. Adv. Exp. Med. Biol. 2010, 688, 1–23. [Google Scholar]
- Hannun, Y.A.; Obeid, L.M. Many ceramides. J. Biol. Chem. 2011, 286, 27855–27862. [Google Scholar] [CrossRef]
- Katsume, A.; Tokunaga, Y.; Hirata, Y.; Munakata, T.; Saito, M.; Hayashi, H.; Okamoto, K.; Ohmori, Y.; Kusanagi, I.; Fujiwara, S.; et al. A serine palmitoyltransferase inhibitor blocks hepatitis C virus replication in human hepatocytes. Gastroenterology 2013, 145, 865–873. [Google Scholar] [CrossRef]
- Simon, J.; Ouro, A.; Ala-Ibanibo, L.; Presa, N.; Delgado, T.C.; Martínez-Chantar, M.L. Sphingolipids in non-alcoholic fatty liver disease and hepatocellular carcinoma: Ceramide turnover. Int. J. Mol. Sci. 2020, 21, 40. [Google Scholar] [CrossRef]
- Moro, K.; Kawaguchi, T.; Tsuchida, J.; Gabriel, E.; Qi, Q.; Yan, L.; Wakai, T.; Takabe, K.; Nagahashi, M. Ceramide species are elevated in human breast cancer and are associated with less aggressiveness. Oncotarget 2018, 9, 19874–19890. [Google Scholar] [CrossRef]
- Xu, X.; Liu, B.; Zou, P.; Zhang, Y.; You, J.; Pei, F. Silencing of LASS2/TMSG1 enhances invasion and metastasis capacity of prostate cancer cell. J. Cell. Biochem. 2014, 115, 731–743. [Google Scholar] [CrossRef]
- Hanyu, T.; Nagahashi, M.; Ichikawa, H.; Ishikawa, T.; Kobayashi, T.; Wakai, T. Expression of phosphorylated sphingosine kinase 1 is associated with diffuse type and lymphatic invasion in human gastric cancer. Surgery 2018, 163, 1301–1306. [Google Scholar] [CrossRef]
- Selzner, M.; Bielawska, A.; Morse, M.A.; Rüdiger, H.A.; Sindram, D.; Hannun, Y.A.; Clavien, P.-A. Induction of Apoptotic Cell Death and Prevention of Tumor Growth by Ceramide Analogues in Metastatic Human Colon Cancer. Cancer Res. 2001, 61, 1233–1240. [Google Scholar]
- Karahatay, S.; Thomas, K.; Koybasi, S.; Senkal, C.E.; ElOjeimy, S.; Liu, X.; Bielawski, J.; Day, T.A.; Gillespie, M.B.; Sinha, D.; et al. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): Attenuation of C18-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 2007, 256, 101–111. [Google Scholar] [CrossRef]
- Barupal, D.K.; Zhang, Y.; Shen, T.; Fan, S.; Roberts, B.S.; Fitzgerald, P.; Wancewicz, B.; Valdiviez, L.; Wohlgemuth, G.; Byram, G.; et al. A comprehensive plasma metabolomics dataset for a cohort of mouse knockouts within the international mouse phenotyping consortium. Metabolites 2019, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Liu, K.H.; Lee, D.Y.; Defelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed]
- DeFelice, B.C.; Mehta, S.S.; Samra, S.; Čajka, T.; Wancewicz, B.; Fahrmann, J.F.; Fiehn, O. Mass Spectral Feature List Optimizer (MS-FLO): A Tool To Minimize False Positive Peak Reports in Untargeted Liquid Chromatography-Mass Spectroscopy (LC-MS) Data Processing. Anal. Chem. 2017, 89, 3250–3255. [Google Scholar] [CrossRef]
- Lê Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef]
- Venny 2.1.0. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 1 August 2020).
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Mohamed, A.; Molendijk, J.; Hill, M.M. Lipidr: A Software Tool for Data Mining and Analysis of Lipidomics Datasets. J. Proteome Res. 2020, 19, 2890–2897. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, I.T.; Elfert, A.; Helal, M.; Salama, I.; El-Said, H.; Fiehn, O. Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma. Cancers 2021, 13, 88. https://doi.org/10.3390/cancers13010088
Ismail IT, Elfert A, Helal M, Salama I, El-Said H, Fiehn O. Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma. Cancers. 2021; 13(1):88. https://doi.org/10.3390/cancers13010088
Chicago/Turabian StyleIsmail, Israa T., Ashraf Elfert, Marwa Helal, Ibrahim Salama, Hala El-Said, and Oliver Fiehn. 2021. "Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma" Cancers 13, no. 1: 88. https://doi.org/10.3390/cancers13010088
APA StyleIsmail, I. T., Elfert, A., Helal, M., Salama, I., El-Said, H., & Fiehn, O. (2021). Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma. Cancers, 13(1), 88. https://doi.org/10.3390/cancers13010088





