Modification of the Histone Landscape with JAK Inhibition in Myeloproliferative Neoplasms
Simple Summary
Abstract
1. Introduction
2. Results
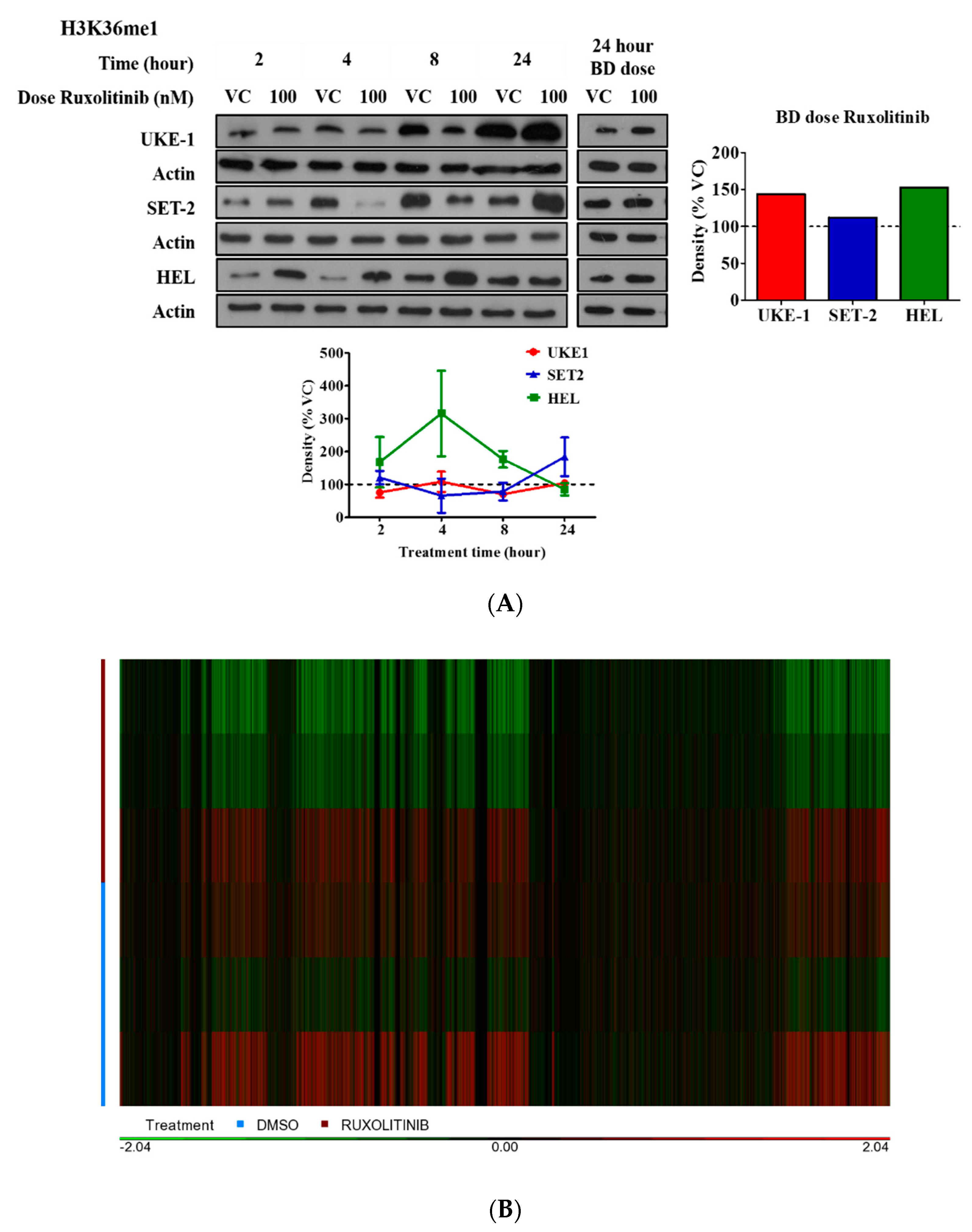
2.1. Effective Ruxolitinib Concentration in Cell Line Culture
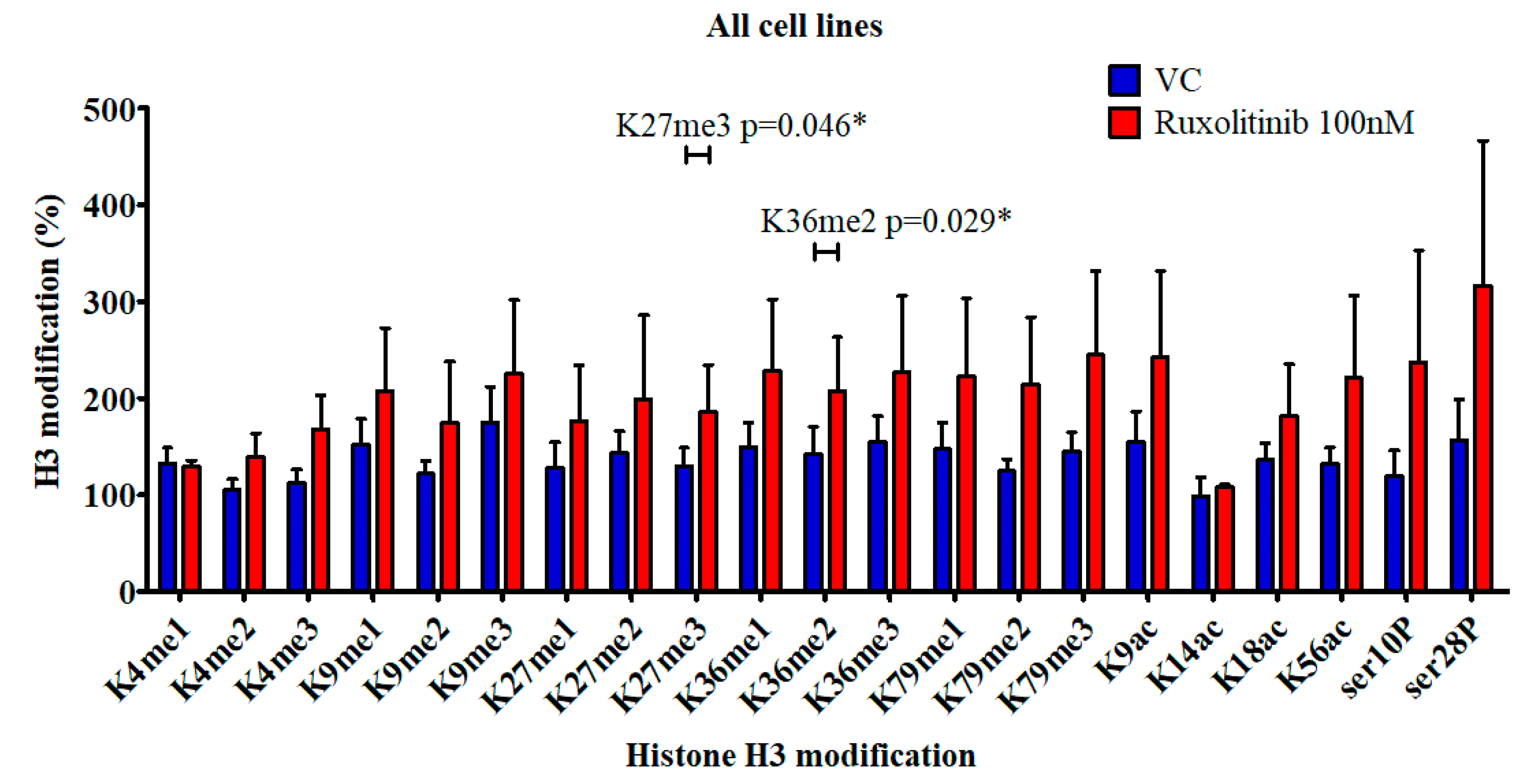
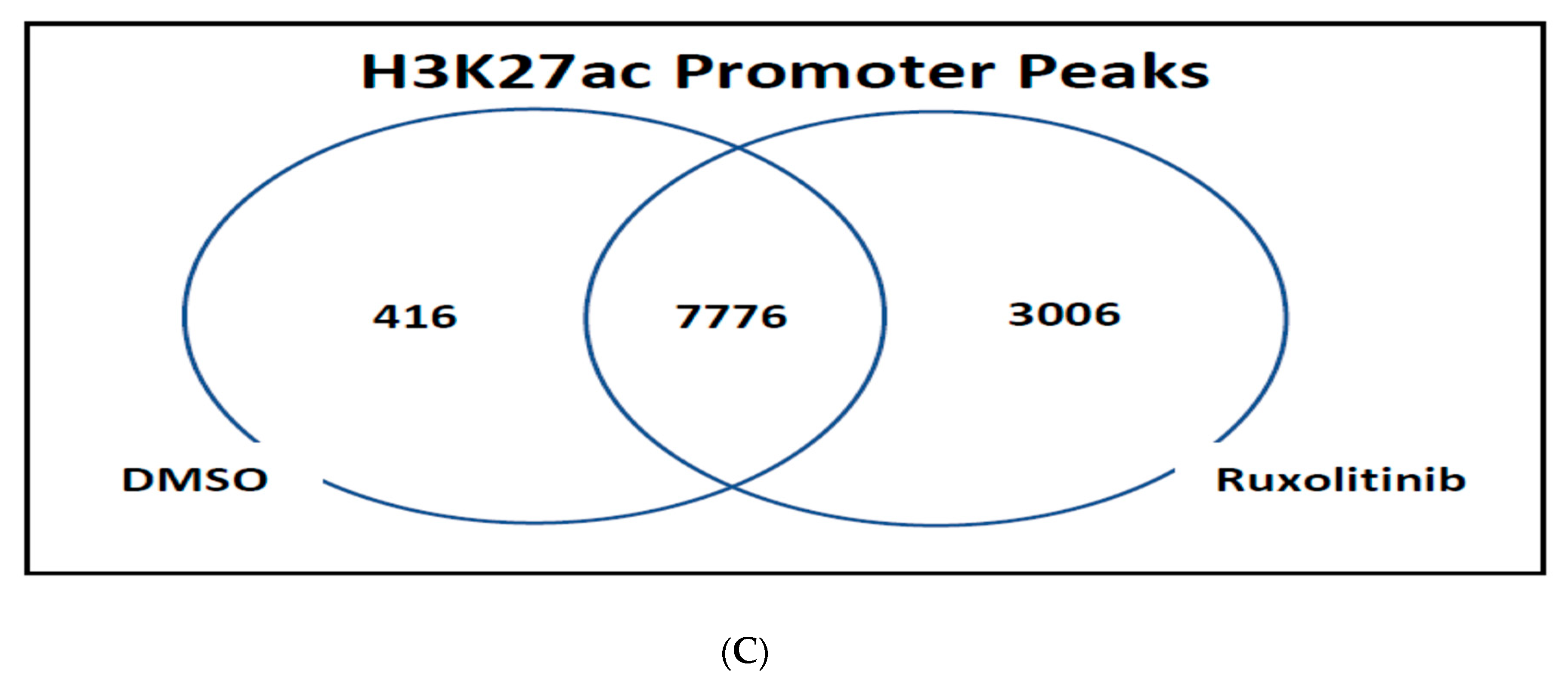
2.2. Histone Modification in Cell Culture
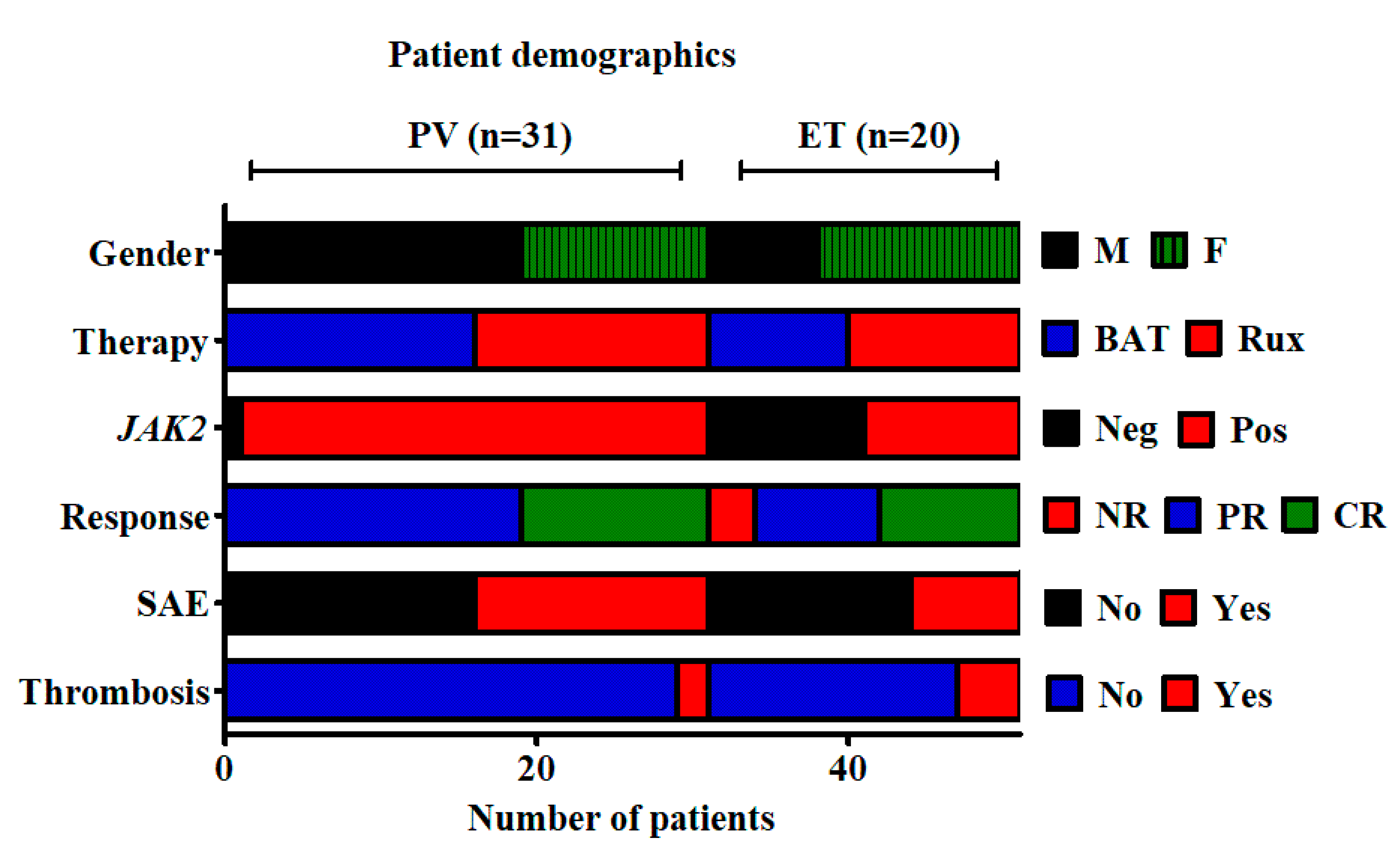
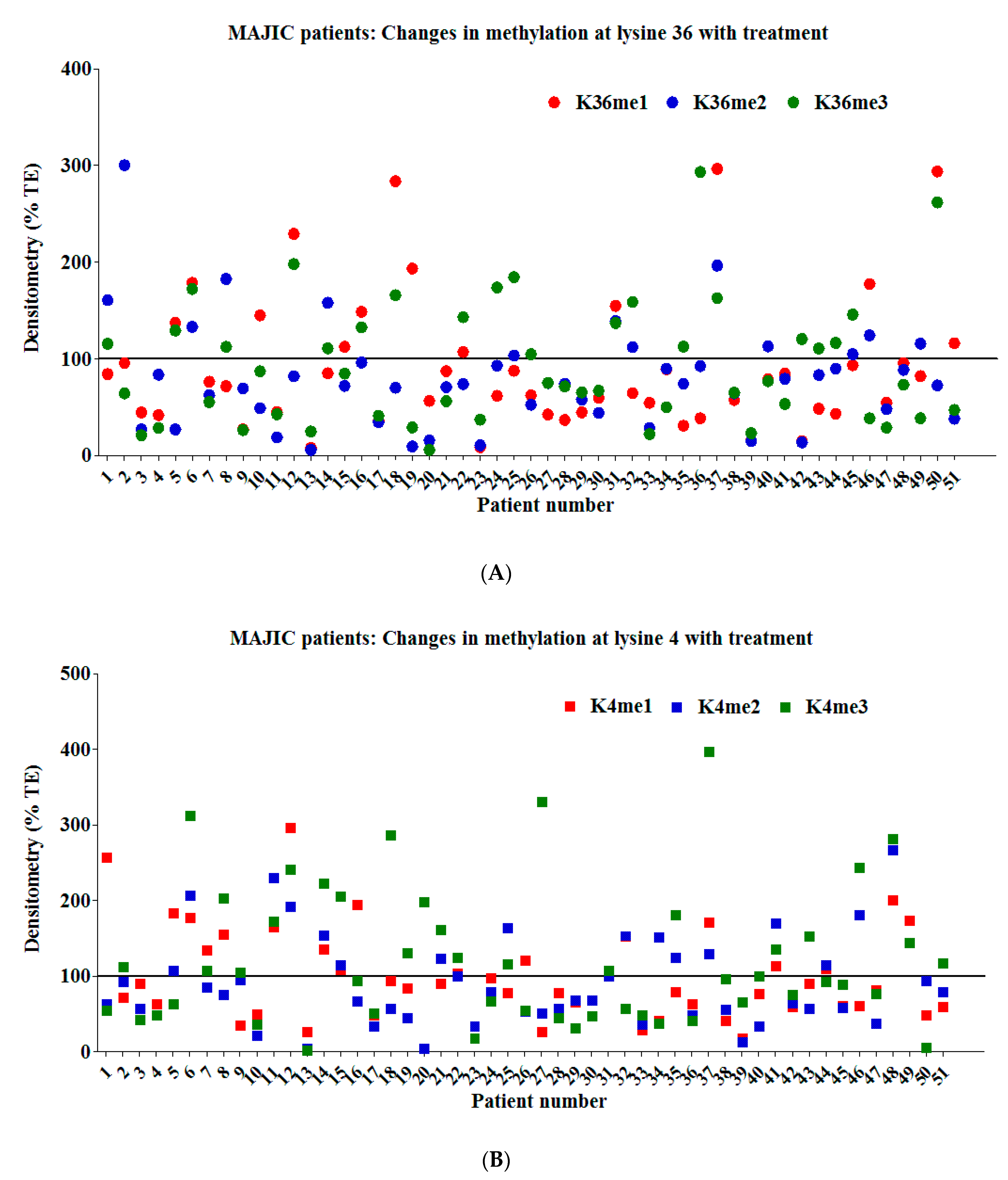
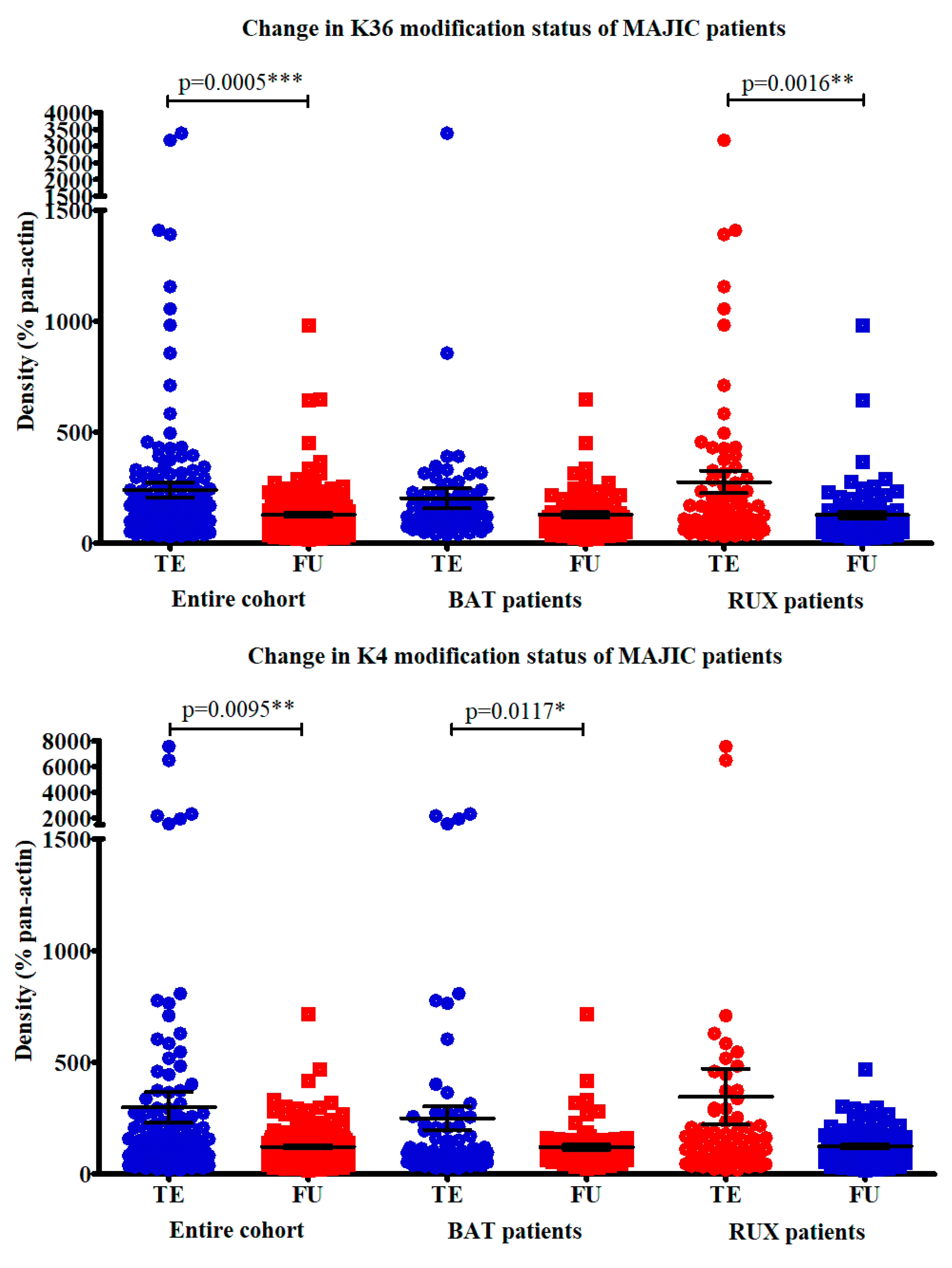
2.3. Examination of Candidate Histone Modifications in Patient Samples from the MAJIC Clinical Trial
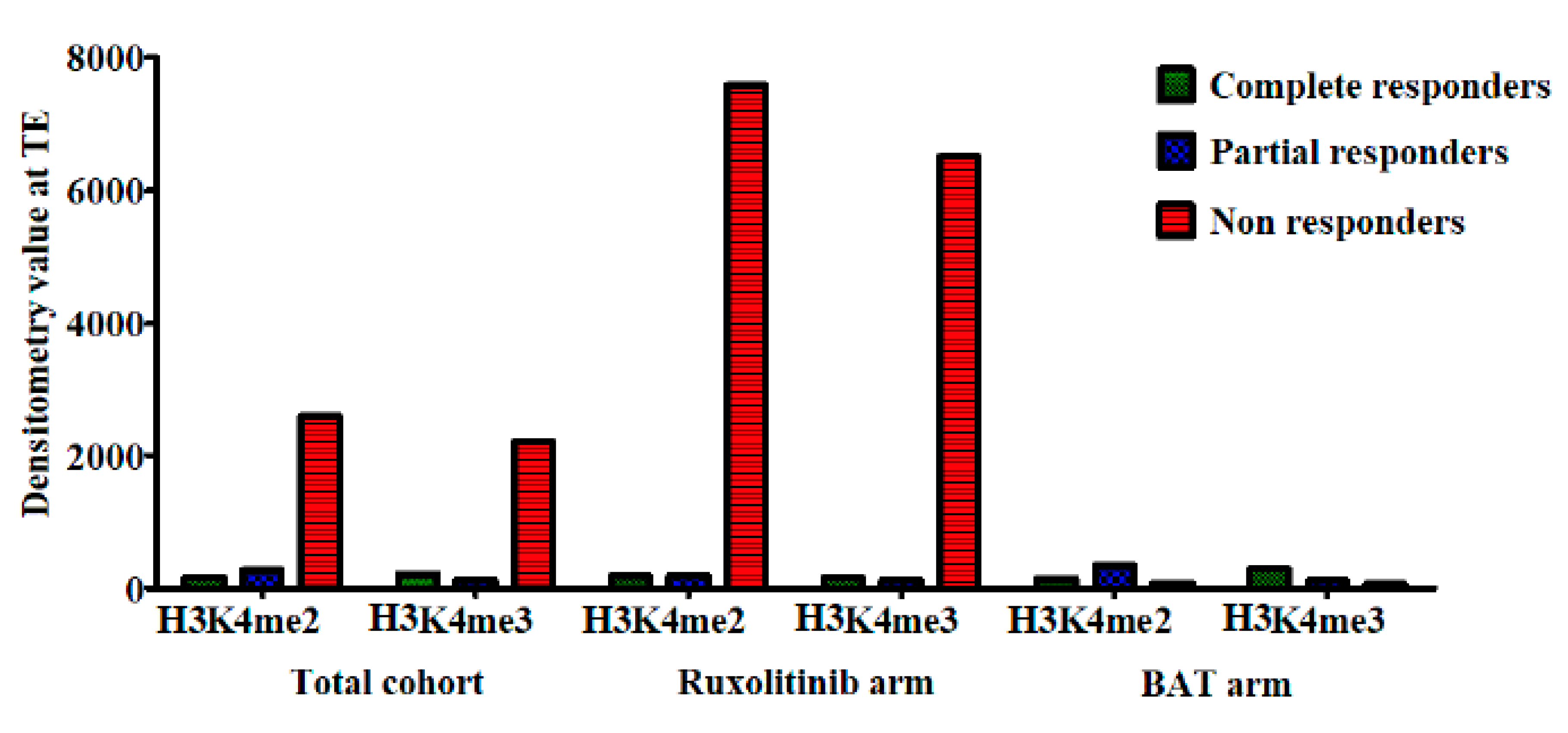
2.4. Correlation of Histone Modifications with Treatment Response
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Drug Treatment
4.2. Western Blotting
4.3. Histone Modification Assay
4.4. RNA and Chromatin Immunoprecipitation(CHIP) Sequencing
4.5. Ethical Approval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethics Statement
References
- Baxter, E.J.; Scott, L.M.; Campbell, P.J.; East, C.; Fourouclas, N.; Swanton, S.; Vassiliou, G.S.; Bench, J.A.; Boyd, E.M.; Curtin, N.; et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005, 365, 1054–1061. [Google Scholar] [CrossRef]
- Pikman, Y.; Lee, B.H.; Mercher, T.; McDowell, E.; Ebert, B.L.; Gozo, M.; Cuker, A.; Wernig, G.; Moore, S.; Galinsky, I.; et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006, 3, e270. [Google Scholar] [CrossRef] [PubMed]
- Nangalia, J.; Massie, C.E.; Baxter, E.J.; Nice, F.; Gundem, G.; Wedge, D.C.; Avezov, E.; Li, J.; Kollmann, K.; Kent, D.G.; et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 2013, 369, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- McPherson, S.; McMullin, M.F.; Mills, K. Epigenetics in Myeloproliferative Neoplasms. J. Cell Mol. Med. 2017, 21, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Nangalia, J.; Green, A.R. Myeloproliferative neoplasms: From origins to outcomes. Blood 2017, 130, 2475–2483. [Google Scholar] [CrossRef]
- Grinfeld, J.; Nangalia, J.; Green, A.R. Molecular determinants of pathogenesis and clinical phenotype in myeloproliferative neoplasms. Haematologica 2017, 102, 7–17. [Google Scholar] [CrossRef]
- Grinfeld, J.; Nangalia, J.; Baxter, E.J.; Wedge, D.C.; Angelopoulos, N.; Cantrill, R.; Godfrey, A.L.; Papaemmanuil, E.; Gundem, G.; MacLean, C.; et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N. Engl. J. Med. 2018, 379, 1416–1430. [Google Scholar] [CrossRef]
- O’Sullivan, J.; Mead, A.J. Heterogeneity in myeloproliferative neoplasms: Causes and consequences. Adv. Biol Regul. 2019, 71, 55–68. [Google Scholar] [CrossRef]
- Verstovsek, S.; Mesa, R.A.; Gotlib, J.; Levy, R.S.; Gupta, V.; DiPersio, J.F.; Catalano, J.V.; Deininger, M.; Miller, C.; Silver, R.T.; et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N. Engl. J. Med. 2012, 366, 799–807. [Google Scholar] [CrossRef]
- Vannucchi, A.M.; Kiladjian, J.J.; Griesshammer, M.; Masszi, T.; Durrant, S.; Passamonti, F.; Harrison, C.N.; Pane, F.; Zachee, P.; Mesa, M.; et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N. Engl. J. Med. 2015, 372, 426–435. [Google Scholar] [CrossRef]
- Passamonti, F.; Maffioli, M. The role of JAK2 inhibitors in MPNs 7 years after approval. Blood 2018, 131, 2426–2435. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Verstovsek, S.; Guglielmelli, P.; Griesshammer, M.; Burn, T.C.; Naim, A.; Paranagama, D.; Marker, M.; Gadbaw, B.; Kiladjian, J.-J. Ruxolitinib reduces JAK2 p. V617F allele burden in patients with polycythemia vera enrolled in the RESPONSE study. Ann. Hematol. 2017, 96, 1113–1120. [Google Scholar] [PubMed]
- Harrison, C.; Stalbovskaya, V.; Vannucchi, A.M.; Kiladjian, J.-J.; Al-Ali, H.; Gisslinger, H.; Knoops, L.; Cervantes, F.; Jones, M.M.; Sun, K.; et al. Long-term findings from COMFORT-II, a phase 3 study of ruxolitinib vs best available therapy for myelofibrosis. Leukemia 2016, 30, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Kvasnicka, H.M.; Thiele, J.; Bueso-Ramos, C.E.; Sun, W.; Cortes, J.; Kantarjian, H.M.; Verstovsek, S. Long-term effects of ruxolitinib versus best available therapy on bone marrow fibrosis in patients with myelofibrosis. J. Hematol. Oncol. 2018, 11, 42. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Pardanani, A. Serious adverse events during ruxolitinib treatment discontinuation in patients with myelofibrosis. Mayo Clin. Proc. 2011, 86, 1188–1191. [Google Scholar] [CrossRef]
- O’Sullivan, J.M.; Hamblin, A.; Yap, C.; Fox, S.; Boucher, R.; Panchal, A.; Alimam, S.; Dreau, H.M.; Howard, K.; Ware, P.; et al. The poor outcome in high molecular risk, hydroxycarbamide-resistant/intolerant ET is not ameliorated by ruxolitinib. Blood 2019, 134, 2107–2111. [Google Scholar] [CrossRef]
- Cervantes, F.; Pereira, A. Does ruxolitinib prolong the survival of patients with myelofibrosis? Blood 2017, 129, 832–837. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Kribelbauer, J.F.; Lu, X.J.; Rohs, R.; Mann, R.S.; Bussemaker, H.J. Toward a Mechanistic Understanding of DNA Methylation Readout by Transcription Factors. J. Mol. Biol. 2019, 432, 1801–1815. [Google Scholar]
- Pérez, C.; Pascual, M.; Martin-Subero, J.I.; Bellosillo, B.; Segura, V.; Delabesse, E.; Álvarez, S.; Larrayoz, M.J.; Rifón, J.; Cigudosa, J.C.; et al. Aberrant DNA methylation profile of chronic and transformed classic Philadelphia-negative myeloproliferative neoplasms. Haematologica 2013, 98, 1414–1420. [Google Scholar] [CrossRef]
- McPherson, S.; Greenfield, G.; Andersen, C.; Grinfeld, J.; Hasselbalch, H.C.; Nangalia, J.; Mills, K.; McMullin, M.F. Methylation age as a correlate for allele burden, disease status, and clinical response in myeloproliferative neoplasm patients treated with vorinostat. Exp. Hematol. 2019, 79, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, D.; Sokol, K.; Bhalla, S.; Rampal, R.; Mascarenhas, J.O. Implications of Mutation Profiling in Myeloid Malignancies-PART 1: Myelodysplastic Syndromes and Acute Myeloid Leukemia. Oncology (Williston Park) 2018, 32, e38–e44. [Google Scholar] [PubMed]
- Schischlik, F.; Jäger, R.; Rosebrock, F.; Hug, E.; Schuster, M.K.; Holly, R.; Fuchs, E.; Feenstra, J.D.M.; Bogner, E.; Gisslinger, B.; et al. Mutational landscape of the transcriptome offers putative targets for immunotherapy of myeloproliferative neoplasms. Blood 2019, 134, 199–210. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, X.; Perna, F.; Wang, L.; Koppikar, P.; Abdel-Wahab, O.; Harr, M.W.; Levine, R.L.; Xu, H.; Tefferi, A.; et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 2011, 19, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; McMullin, M.F.; Ejerblad, E.; Zweegman, S.; Harrison, C.; Fernandes, S.; Bareford, D.; Knapper, S.; Samuelsson, J.; Löfvenberg, E.; et al. A phase II study of vorinostat (MK-0683) in patients with polycythaemia vera and essential thrombocythaemia. Br. J. Haematol. 2013, 162, 498–508. [Google Scholar] [CrossRef]
- Rampal, R.K.; Mascarenhas, J.O.; Kosiorek, H.E.; Price, L.; Berenzon, D.; Hexner, E.; Abboud, C.N.; Kremyanskaya, M.; Weinberg, R.S.; Salama, M.E.; et al. Safety and efficacy of combined ruxolitinib and decitabine in accelerated and blast-phase myeloproliferative neoplasms. Blood Adv. 2018, 2, 3572–3580. [Google Scholar] [CrossRef]
- Harrison, C.; Mead, A.J.; Panchal, A.; Fox, S.; Yap, C.; Gbandi, E.; Houlton, A.; Alimam, S.; Ewing, J.; Wood, M.; et al. Ruxolitinib vs best available therapy for ET intolerant or resistant to hydroxycarbamide. Blood 2017, 130, 1889–1897. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Rinke, J.; Chase, A.; Cross, N.C.P.; Hochhaus, A.; Ernst, T. EZH2 in Myeloid Malignancies. Cells 2020, 9, 1639. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; Beek, M.V.D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greenfield, G.; McPherson, S.; Smith, J.; Mead, A.; Harrison, C.; Mills, K.; McMullin, M.F. Modification of the Histone Landscape with JAK Inhibition in Myeloproliferative Neoplasms. Cancers 2020, 12, 2669. https://doi.org/10.3390/cancers12092669
Greenfield G, McPherson S, Smith J, Mead A, Harrison C, Mills K, McMullin MF. Modification of the Histone Landscape with JAK Inhibition in Myeloproliferative Neoplasms. Cancers. 2020; 12(9):2669. https://doi.org/10.3390/cancers12092669
Chicago/Turabian StyleGreenfield, Graeme, Suzanne McPherson, James Smith, Adam Mead, Claire Harrison, Ken Mills, and Mary Frances McMullin. 2020. "Modification of the Histone Landscape with JAK Inhibition in Myeloproliferative Neoplasms" Cancers 12, no. 9: 2669. https://doi.org/10.3390/cancers12092669
APA StyleGreenfield, G., McPherson, S., Smith, J., Mead, A., Harrison, C., Mills, K., & McMullin, M. F. (2020). Modification of the Histone Landscape with JAK Inhibition in Myeloproliferative Neoplasms. Cancers, 12(9), 2669. https://doi.org/10.3390/cancers12092669






