Preoperative Fasting and General Anaesthesia Alter the Plasma Proteome
Abstract
1. Introduction
2. Results
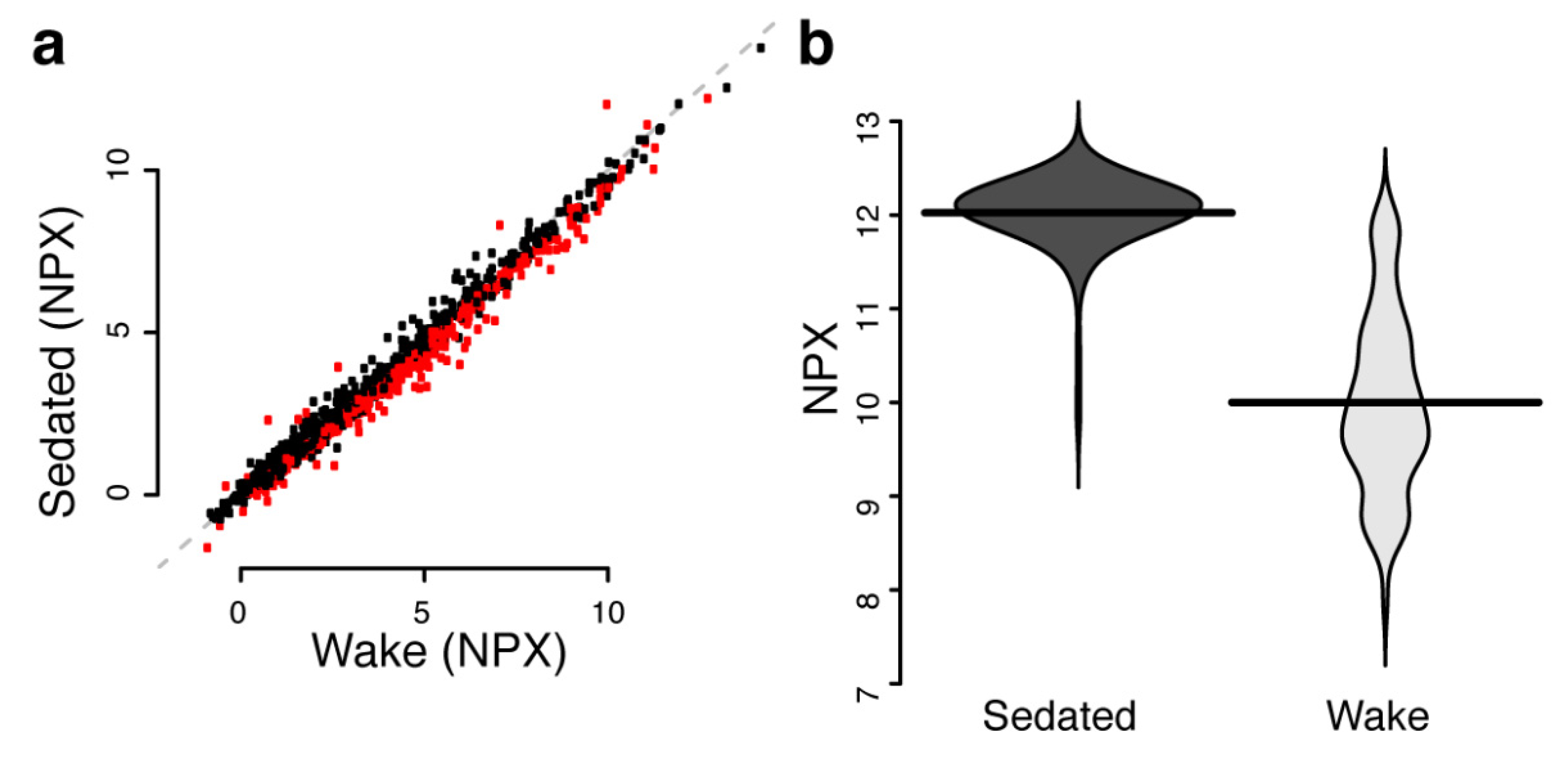
2.1. Effects of Sampling at Surgery Compared at Time of Diagnosis
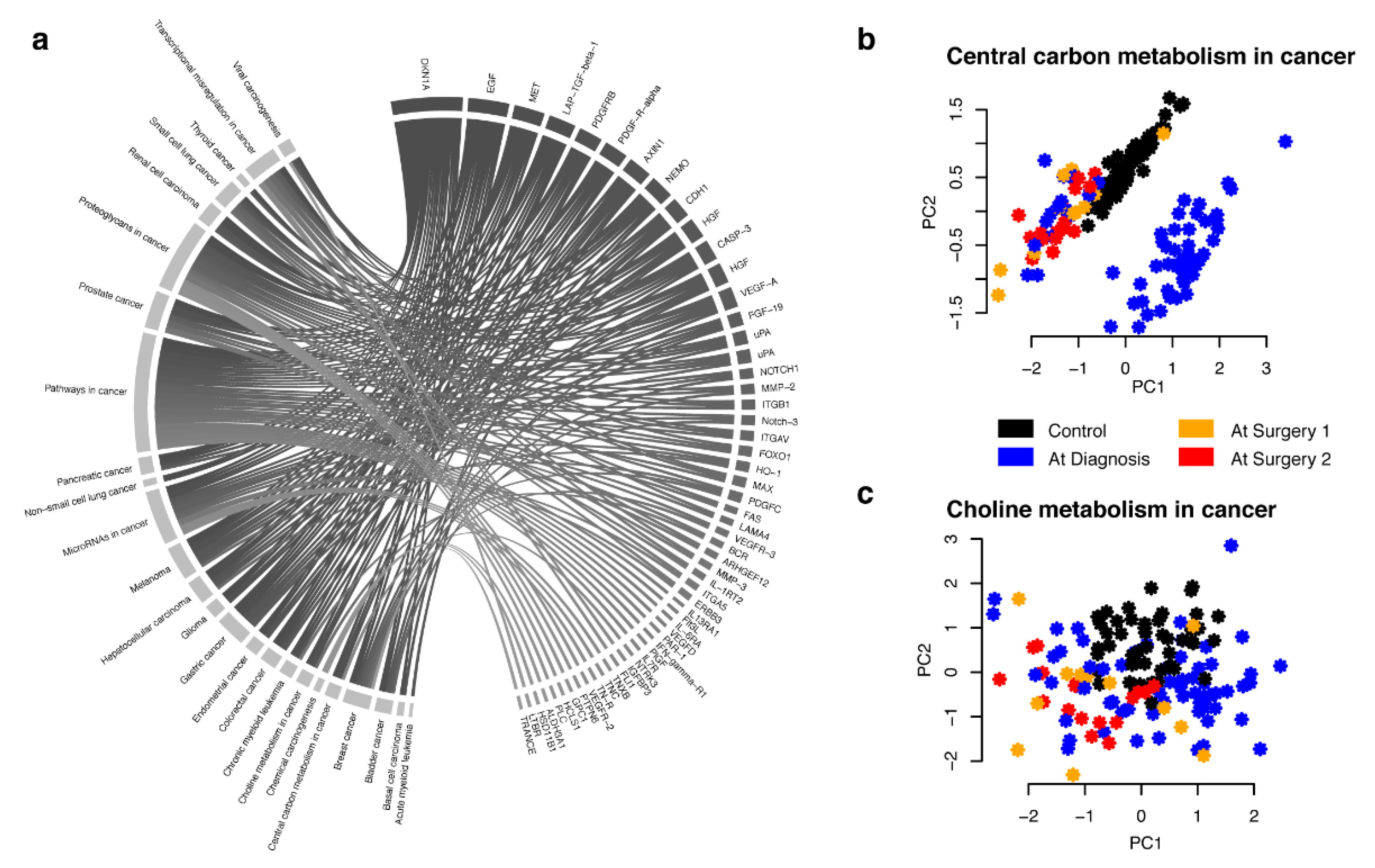
2.2. Combined Analysis and Replication of Previously Published Results
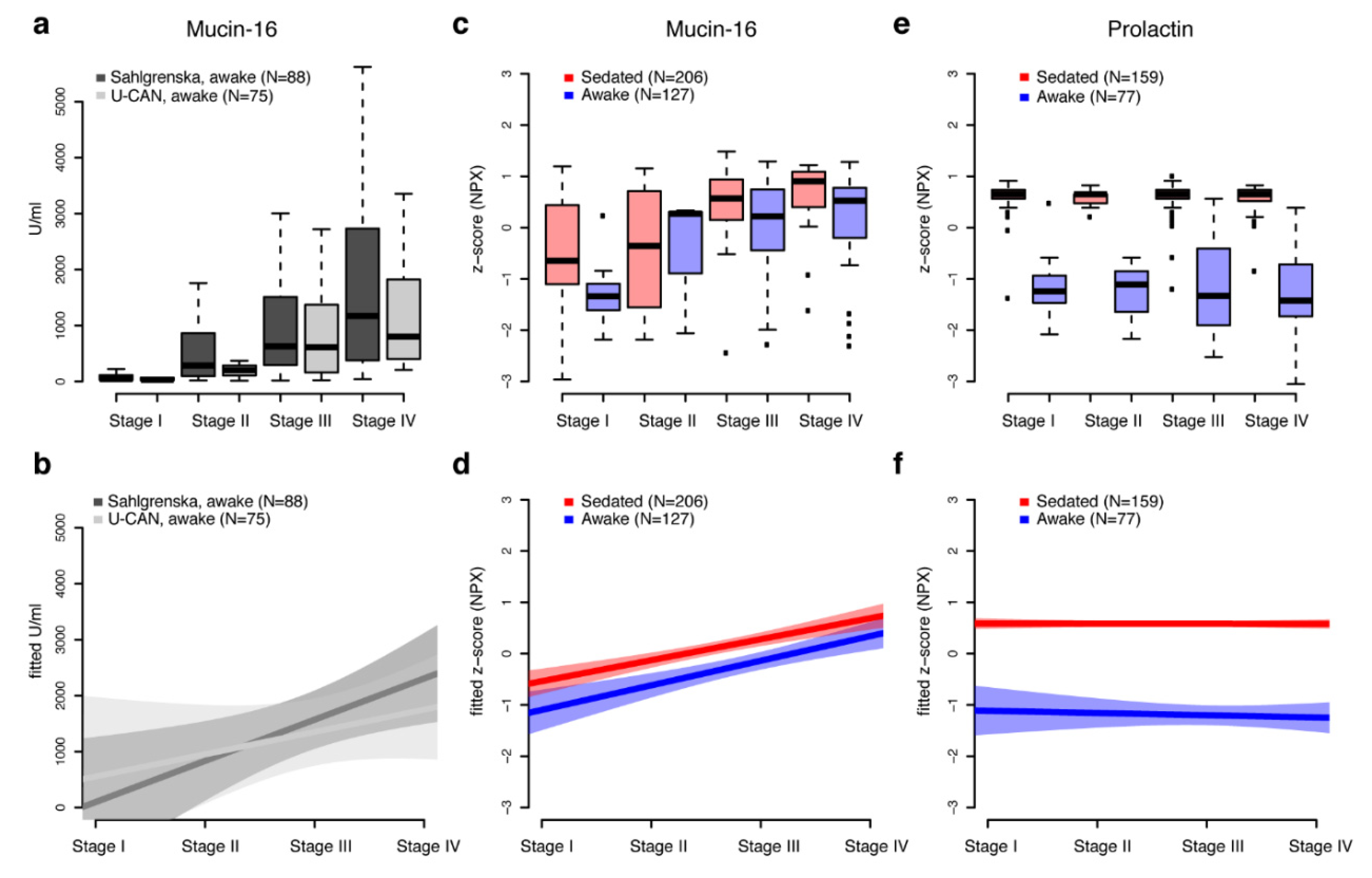
2.3. Stage-Independent Effects by Sedation on Mucin-16 for Ovarian Cancer
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. Plasma Proteome Characterization
4.3. Statistical Analysis
4.4. Mouse Human Ortholog Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Enroth, S.; Bosdotter Enroth, S.; Johansson, A.; Gyllensten, U. Effect of genetic and environmental factors on protein biomarkers for common non-communicable disease and use of personally normalized plasma protein profiles (PNPPP). Biomarkers 2015, 20, 355–364. [Google Scholar] [CrossRef]
- Enroth, S.; Johansson, A.; Enroth, S.B.; Gyllensten, U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat. Commun. 2014, 5, 4684. [Google Scholar] [CrossRef] [PubMed]
- Enroth, S.; Maturi, V.; Berggrund, M.; Enroth, S.B.; Moustakas, A.; Johansson, A.; Gyllensten, U. Systemic and specific effects of antihypertensive and lipid-lowering medication on plasma protein biomarkers for cardiovascular diseases. Sci. Rep. 2018, 8, 5531. [Google Scholar] [CrossRef] [PubMed]
- Stenemo, M.; Teleman, J.; Sjostrom, M.; Grubb, G.; Malmstrom, E.; Malmstrom, J.; Nimeus, E. Cancer associated proteins in blood plasma: Determining normal variation. Proteomics 2016, 16, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.M.; Cooper, J.D.; Penninx, B.W.; Bahn, S. Variation in serum biomarkers with sex and female hormonal status: Implications for clinical tests. Sci. Rep. 2016, 6, 26947. [Google Scholar] [CrossRef]
- Sundkvist, A.; Myte, R.; Boden, S.; Enroth, S.; Gyllensten, U.; Harlid, S.; van Guelpen, B. Targeted plasma proteomics identifies a novel, robust association between cornulin and Swedish moist snuff. Sci. Rep. 2018, 8, 2320. [Google Scholar] [CrossRef]
- Enroth, S.; Hallmans, G.; Grankvist, K.; Gyllensten, U. Effects of Long-Term Storage Time and Original Sampling Month on Biobank Plasma Protein Concentrations. EBioMedicine 2016, 12, 309–314. [Google Scholar] [CrossRef]
- Shabihkhani, M.; Lucey, G.M.; Wei, B.; Mareninov, S.; Lou, J.J.; Vinters, H.V.; Singer, E.J.; Cloughesy, T.F.; Yong, W.H. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin. Biochem. 2014, 47, 258–266. [Google Scholar] [CrossRef]
- Nygren, J. The metabolic effects of fasting and surgery. Best Pract. Res. Clin. Anaesthesiol. 2006, 20, 429–438. [Google Scholar] [CrossRef]
- Nygren, J.; Carlsson-Skwirut, C.; Brismar, K.; Thorell, A.; Ljungqvist, O.; Bang, P. Insulin infusion increases levels of free IGF-I and IGFBP-3 proteolytic activity in patients after surgery. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E736–E741. [Google Scholar] [CrossRef]
- Goldstein, I.; Baek, S.; Presman, D.M.; Paakinaho, V.; Swinstead, E.E.; Hager, G.L. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 2017, 27, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Kahn, N.; Riedlinger, J.; Roessler, M.; Rabe, C.; Lindner, M.; Koch, I.; Schott-Hildebrand, S.; Herth, F.J.; Schneider, M.A.; Meister, M.; et al. Blood-sampling collection prior to surgery may have a significant influence upon biomarker concentrations measured. Clin. Proteom. 2015, 12, 19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thorpe, J.D.; Duan, X.; Forrest, R.; Lowe, K.; Brown, L.; Segal, E.; Nelson, B.; Anderson, G.L.; McIntosh, M.; Urban, N. Effects of blood collection conditions on ovarian cancer serum markers. PLoS ONE 2007, 2, e1281. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C., Jr.; Klug, T.L.; St John, E.; Jenison, E.; Niloff, J.M.; Lazarus, H.; Berkowitz, R.S.; Leavitt, T.; Griffiths, C.T.; Parker, L.; et al. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. N. Engl. J. Med. 1983, 309, 883–887. [Google Scholar] [CrossRef]
- Soletormos, G.; Duffy, M.J.; Hassan, S.O.A.; Verheijen, R.H.; Tholander, B.; Bast, R.C., Jr.; Gaarenstroom, K.N.; Sturgeon, C.M.; Bonfrer, J.M.; Petersen, P.H.; et al. Clinical Use of Cancer Biomarkers in Epithelial Ovarian Cancer: Updated Guidelines From the European Group on Tumor Markers. Int. J. Gynecol. Cancer 2016, 26, 43–51. [Google Scholar] [CrossRef]
- Lycke, M.; Kristjansdottir, B.; Sundfeldt, K. A multicenter clinical trial validating the performance of HE4, CA125, risk of ovarian malignancy algorithm and risk of malignancy index. Gynecol. Oncol. 2018, 151, 159–165. [Google Scholar] [CrossRef]
- Hagen, C.; Brandt, M.R.; Kehlet, H. Prolactin, LH, FSH, GH and cortisol response to surgery and the effect of epidural analgesia. Acta Endocrinol. 1980, 94, 151–154. [Google Scholar] [CrossRef]
- Enroth, S.; Berggrund, M.; Lycke, M.; Broberg, J.; Lundberg, M.; Assarsson, E.; Olovsson, M.; Stålberg, K.; Sundfeldt, K.; Gyllensten, U. High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer. Commun. Biol. 2019, 2, 221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Shen, Q.; Bjorkesten, J.; Galli, J.; Ekman, D.; Broberg, J.; Nordberg, N.; Tillander, A.; Kamali-Moghaddam, M.; Tybring, G.; Landegren, U. Strong impact on plasma protein profiles by precentrifugation delay but not by repeated freeze-thaw cycles, as analyzed using multiplex proximity extension assays. Clin. Chem. Lab. Med. 2018, 56, 582–594. [Google Scholar] [CrossRef] [PubMed]
- Betsou, F.; Lehmann, S.; Ashton, G.; Barnes, M.; Benson, E.E.; Coppola, D.; DeSouza, Y.; Eliason, J.; Glazer, B.; Guadagni, F.; et al. Standard preanalytical coding for biospecimens: Defining the sample PREanalytical code. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Arbous, M.S.; Grobbee, D.E.; van Kleef, J.W.; de Lange, J.J.; Spoormans, H.H.; Touw, P.; Werner, F.M.; Meursing, A.E. Mortality associated with anaesthesia: A qualitative analysis to identify risk factors. Anaesthesia 2001, 56, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Zdolsek, H.J.; Vegfors, M.; Lindahl, T.L.; Tornquist, T.; Bortnik, P.; Hahn, R.G. Hydroxyethyl starches and dextran during hip replacement surgery: Effects on blood volume and coagulation. Acta Anaesthesiol. Scand. 2011, 55, 677–685. [Google Scholar] [CrossRef]
- Pan, S.; Brentnall, T.A.; Chen, R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics 2015, 15, 2705–2715. [Google Scholar] [CrossRef]
- Ilic, M.; Ilic, I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016, 22, 9694–9705. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Che, S.P.; Kurywchak, P.; Tavormina, J.L.; Gansmo, L.B.; Correa de Sampaio, P.; Tachezy, M.; Bockhorn, M.; Gebauer, F.; Haltom, A.R.; et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol. 2017, 18, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, A.; Chang, D.K.; Nones, K.; Corbo, V.; Patch, A.M.; Bailey, P.; Lawlor, R.T.; Johns, A.L.; Miller, D.K.; Mafficini, A.; et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017, 543, 65–71. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, S.; Zhang, B.; Ni, Q.; Yu, X.; Xu, J. Circulating biomarkers for early diagnosis of pancreatic cancer: Facts and hopes. Am. J. Cancer Res. 2018, 8, 332–353. [Google Scholar] [PubMed]
- Wingren, C.; Sandstrom, A.; Segersvard, R.; Carlsson, A.; Andersson, R.; Lohr, M.; Borrebaeck, C.A. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012, 72, 2481–2490. [Google Scholar] [CrossRef] [PubMed]
- Gerdtsson, A.S.; Wingren, C.; Persson, H.; Delfani, P.; Nordstrom, M.; Ren, H.; Wen, X.; Ringdahl, U.; Borrebaeck, C.A.; Hao, J. Plasma protein profiling in a stage defined pancreatic cancer cohort–Implications for early diagnosis. Mol. Oncol. 2016, 10, 1305–1316. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, V.; Arumugam, T.; Wang, H.; Logsdon, C.D. Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer Res. 2008, 68, 7811–7818. [Google Scholar] [CrossRef]
- Herreros-Villanueva, M.; Bujanda, L. Non-invasive biomarkers in pancreatic cancer diagnosis: What we need versus what we have. Ann. Transl. Med. 2016, 4, 134. [Google Scholar] [CrossRef]
- Glimelius, B.; Melin, B.; Enblad, G.; Alafuzoff, I.; Beskow, A.; Ahlstrom, H.; Bill-Axelson, A.; Birgisson, H.; Bjor, O.; Edqvist, P.H.; et al. U-CAN: A prospective longitudinal collection of biomaterials and clinical information from adult cancer patients in Sweden. Acta Oncol. 2018, 57, 187–194. [Google Scholar] [CrossRef]
- Enroth, S.; Berggrund, M.; Lycke, M.; Lundberg, M.; Assarsson, E.; Olovsson, M.; Stalberg, K.; Sundfeldt, K.; Gyllensten, U. A two-step strategy for identification of plasma protein biomarkers for endometrial and ovarian cancer. Clin. Proteom. 2018, 15, 38. [Google Scholar] [CrossRef]
- Region Västra Götaland. Gothia Forum för Klinisk Forskning: Biobank Väst. Available online: https://www.gothiaforum.com (accessed on 12 March 2019).
- Partheen, K.; Kristjansdottir, B.; Sundfeldt, K. Evaluation of ovarian cancer biomarkers HE4 and CA-125 in women presenting with a suspicious cystic ovarian mass. J Gynecol. Oncol. 2011, 22, 244–252. [Google Scholar] [CrossRef]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Bucht Thorsen, S.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-Plex PEA Immunoassay Exhibiting High Sensitivity, Specificity, and Excellent Scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef]
- Assarsson, E.; Lundberg, M. Development and Validation of Customized PEA Biomarker Panels with Clinical Utility. Advancing Precision Medicine: Current and Future Proteogenomic Strategies for Biomarker Discovery and Development; Science/AAAS: Washington, DC, USA, 2017; pp. 32–36. [Google Scholar]
- Champely, S. pwr: Basic Functions for Power Analysis, 1.2-2. 2018. Available online: https://CRAN.R-project.org/package=pwr (accessed on 16 May 2019).
- Kampstra, P. Beanplot: A Boxplot Alternative for Visual Comparison of Distributions. J. Stat. Softw. 2008, 28, 1–9. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Giron, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
| State | Nr. | Sex a | Age (Mean +/sd) | Age Pval b | Panel 1–5 c | 42-Plex d | Panel 6–11 e | Ovarian Cancer Stages | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | ||||||||
| At Diagnosis | 77 | 1.0 | 58.8 (12.5) | 0.17 | 50 e | 77 | 77 | 10 | 3 | 40 | 24 |
| At Surgery 1st | 159 | 1.0 | 61.8 (11.9) | 0.17 | 159 | 100 | 35 | 9 | 96 | 19 | |
| At Surgery 2nd | 91 | 1.0 | 61.8 (12.6) | 0.38 | 79 | 35 | 7 | 33 | 4 | ||
| 47 | 11 | 36 | |||||||||
| Control | 50 | 1.0 | 58.4 (17.6) | 0.39 | 50 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gyllensten, U.; Bosdotter Enroth, S.; Stålberg, K.; Sundfeldt, K.; Enroth, S. Preoperative Fasting and General Anaesthesia Alter the Plasma Proteome. Cancers 2020, 12, 2439. https://doi.org/10.3390/cancers12092439
Gyllensten U, Bosdotter Enroth S, Stålberg K, Sundfeldt K, Enroth S. Preoperative Fasting and General Anaesthesia Alter the Plasma Proteome. Cancers. 2020; 12(9):2439. https://doi.org/10.3390/cancers12092439
Chicago/Turabian StyleGyllensten, Ulf, Sofia Bosdotter Enroth, Karin Stålberg, Karin Sundfeldt, and Stefan Enroth. 2020. "Preoperative Fasting and General Anaesthesia Alter the Plasma Proteome" Cancers 12, no. 9: 2439. https://doi.org/10.3390/cancers12092439
APA StyleGyllensten, U., Bosdotter Enroth, S., Stålberg, K., Sundfeldt, K., & Enroth, S. (2020). Preoperative Fasting and General Anaesthesia Alter the Plasma Proteome. Cancers, 12(9), 2439. https://doi.org/10.3390/cancers12092439





