Key MicroRNA’s and Their Targetome in Adrenocortical Cancer
Abstract
1. Introduction
2. ACC Genetic Landscape and Associated Genetic Disorders
3. Key Genetic Drivers of ACC and Their Cellular Pathways
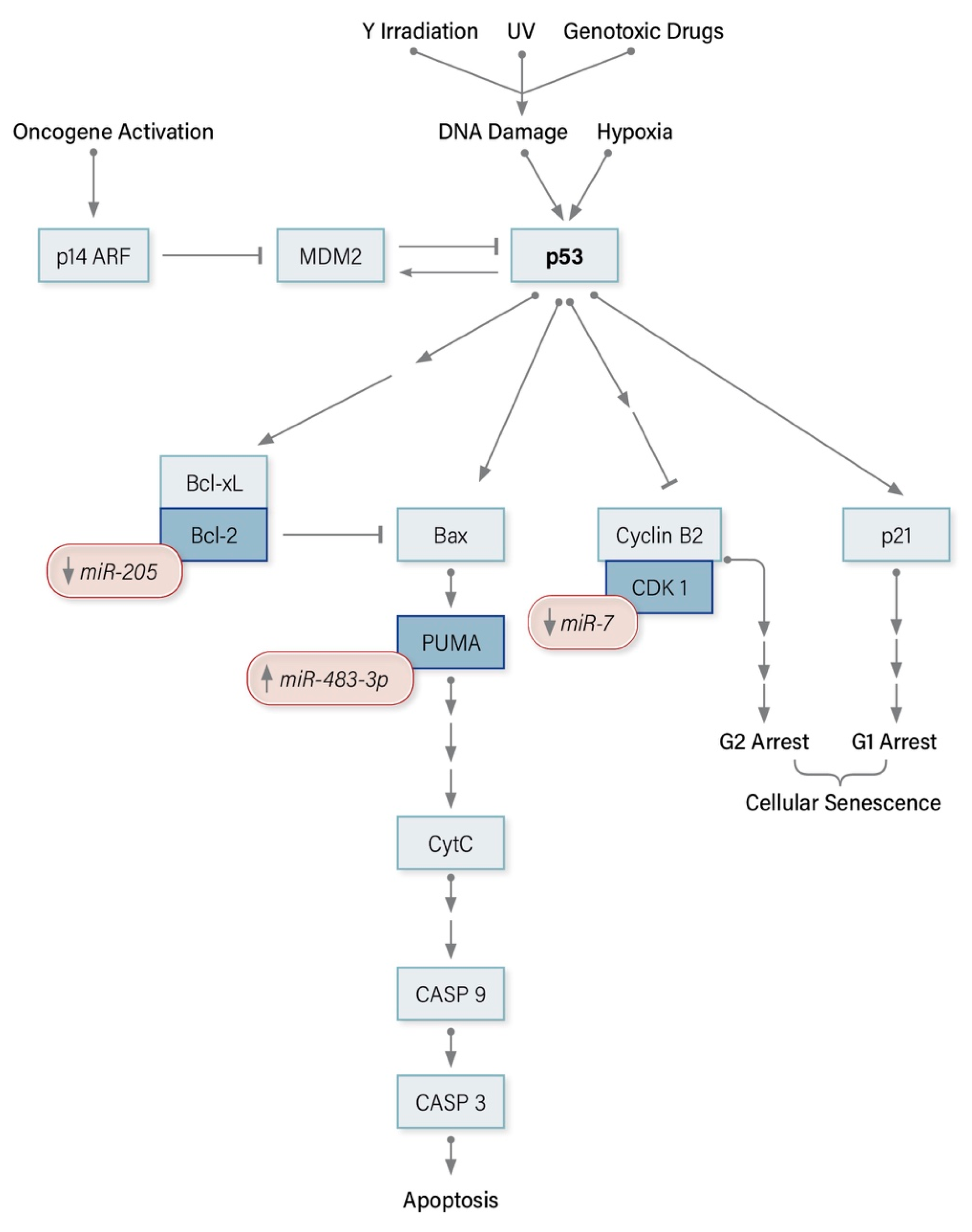
3.1. Tumor Suppressor Protein 53 (TP53)
3.2. Insulin-Like Growth Factor 2 (IGF2)
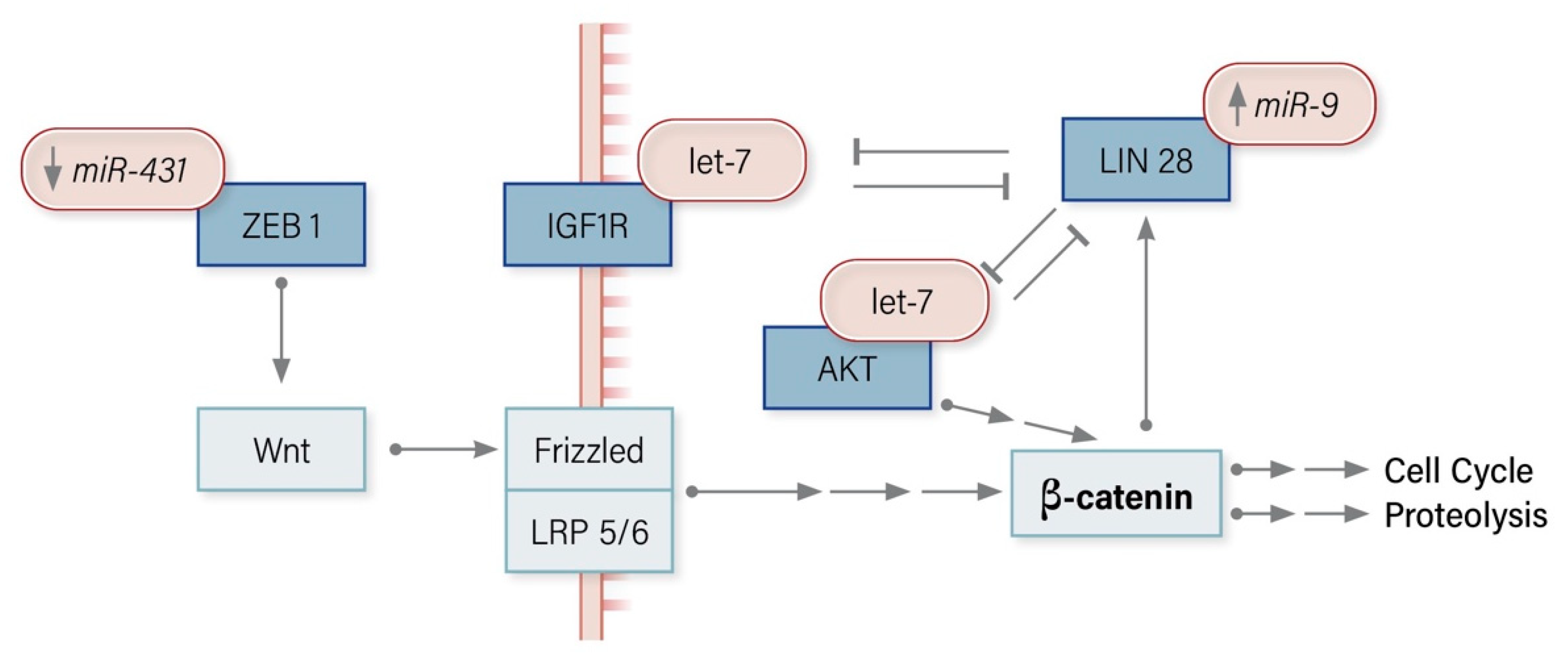
3.3. Wnt/β-Catenin Signalling Pathway
4. Overview of microRNA Structure, Biogenesis, and Function
IsomiRs and Their Emerging Significance in Cancer
5. The Unique microRNA Expression Signature of ACC and Its Clinical Significance
5.1. The microRNA Expression Signature of ACC Tissues
5.2. Circulating microRNAs as Diagnostic Biomarkers in ACC
5.3. Tissue microRNA Expression as a Prognostic Tool in ACC
6. Computational and Experimental Methods of miRNA Target Identification
7. Functional miRNA Target Relationships in ACC
7.1. Overexpressed miRNAs and Their ACCs
7.1.1. miR-9 Regulates LIN28
7.1.2. MiR-21 Regulates PDCD4
7.1.3. miR-483-3p Regulates PUMA
7.2. Underexpressed miRNAs and Their ACC Targets
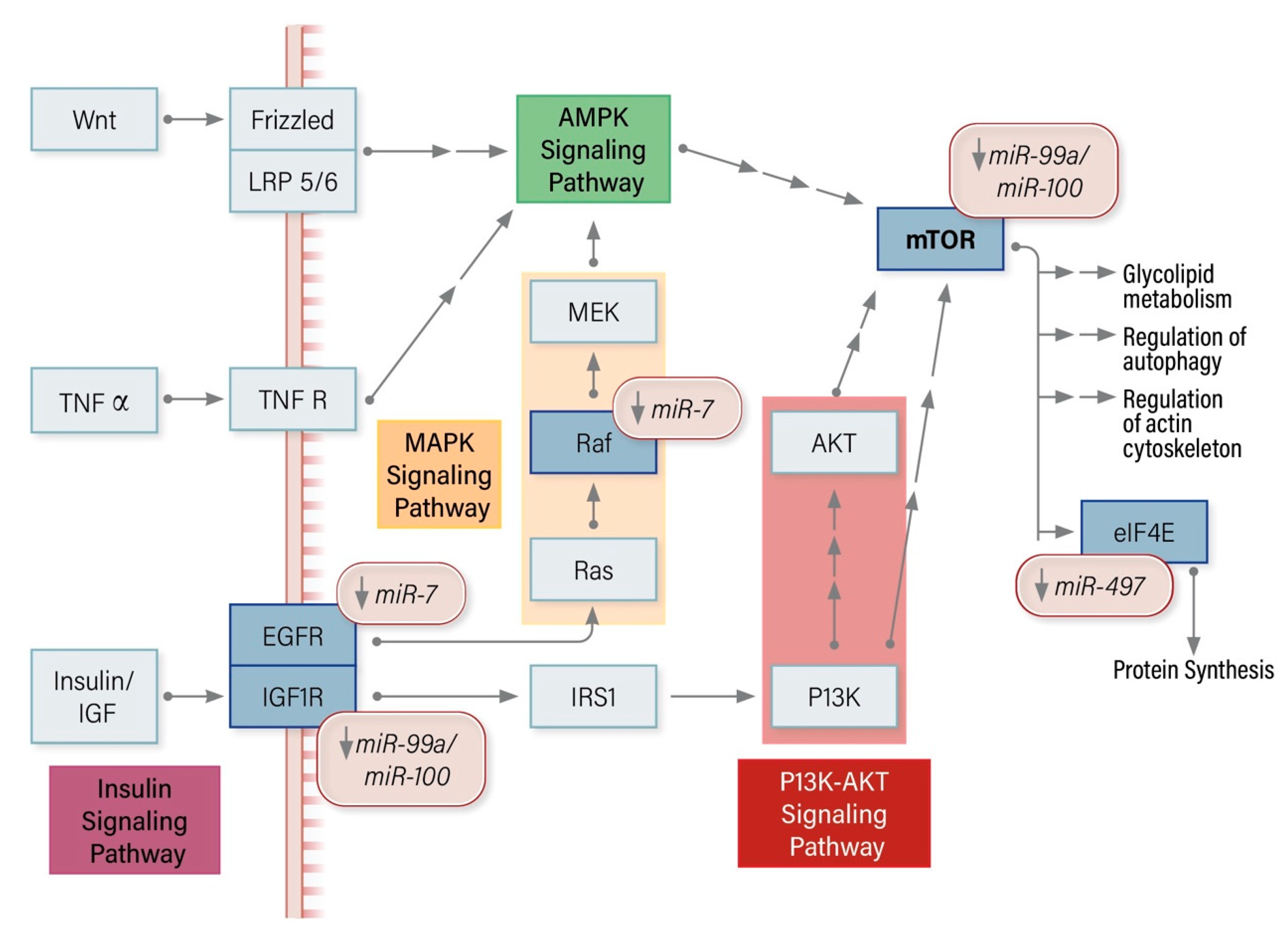
7.2.1. miR-7 Regulates Raf-1, EGFR, CDK1, PAK1, CKS2
7.2.2. miR-99a/100 Regulates IGFR1, mTOR
7.2.3. miR-205 Regulates Bcl-2
7.2.4. miR-375 Regulates MTDH
7.2.5. miR-431 Regulates ZEB1
7.2.6. miR-497 Regulates TARBP2, DICER1, MALAT1, eIF4E, SFPQ
8. miRNA Modulation of ACC Driver Pathways
8.1. miRNA Modulators of the p53 Pathway in ACC
8.2. miRNA Modulators of the mTOR Pathway in ACC
8.3. miRNA Modulators of the Wnt/Β-Catenin Pathway in ACC
9. Future Directions and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hedeland, H.; Östberg, G.; Hökfelt, B. On the Prevalence of Adrenocortical Adenomas In An Autopsy Material In Relation To Hypertension And Diabetes. Acta Medica Scand. 2009, 184, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Kebebew, E.; Reiff, E.; Duh, Q.-Y.; Clark, O.H.; McMillan, A. Extent of Disease at Presentation and Outcome for Adrenocortical Carcinoma: Have We Made Progress? World J. Surg. 2006, 30, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Koschker, A.-C.; Fassnacht, M.; Hahner, S.; Weismann, D.; Allolio, B. Adrenocortical Carcinoma—Improving Patient Care by Establishing New Structures. Exp. Clin. Endocrinol. Diabetes 2006, 114, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Luton, J.-P.; Cerdas, S.; Billaud, L.; Thomas, G.; Guilhaume, B.; Bertagna, X.; Laudat, M.-H.; Louvel, A.; Chapuis, Y.; Blondeau, P.; et al. Clinical Features of Adrenocortical Carcinoma, Prognostic Factors, and the Effect of Mitotane Therapy. N. Engl. J. Med. 1990, 322, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Sturgeon, C.; Shen, W.T.; Clark, O.H.; Duh, Q.-Y.; Kebebew, E. Risk Assessment in 457 Adrenal Cortical Carcinomas: How Much Does Tumor Size Predict the Likelihood of Malignancy? J. Am. Coll. Surg. 2006, 202, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Pommier, R.F.; Brennan, M.F. An eleven-year experience with adrenocortical carcinoma. Surgery 1992, 112, 963–971. [Google Scholar]
- Baur, J.; Büntemeyer, T.-O.; Megerle, F.; Deutschbein, T.; Spitzweg, C.; Quinkler, M.; Nawroth, P.P.; Kroiss, M.; Germer, C.-T.; Fassnacht, M.; et al. Outcome after resection of Adrenocortical Carcinoma liver metastases: A retrospective study. BMC Cancer 2017, 17, 1–10. [Google Scholar] [CrossRef]
- Bilimoria, K.Y.; Shen, W.T.; Elaraj, D.; Bentrem, D.J.; Winchester, D.J.; Kebebew, E.; Sturgeon, C. Adrenocortical carcinoma in the United States. Cancer 2008, 113, 3130–3136. [Google Scholar] [CrossRef]
- Fassnacht, P.D.M.M.; Terzolo, M.; Allolio, B.; Baudin, E.; Haak, H.; Berruti, A.; Welin, S.; Schade-Brittinger, C.; Lacroix, A.; Jarząb, M.; et al. Combination Chemotherapy in Advanced Adrenocortical Carcinoma. N. Engl. J. Med. 2012, 366, 2189–2197. [Google Scholar] [CrossRef]
- McAteer, J.P.; Huaco, J.A.; Gow, K.W. Predictors of survival in pediatric adrenocortical carcinoma: A Surveillance, Epidemiology, and End Results (SEER) program study. J. Pediatr. Surg. 2013, 48, 1025–1031. [Google Scholar] [CrossRef]
- Wasserman, J.; Novokmet, A.; Eichler-Jonsson, C.; Ribeiro, R.C.; Rodriguez-Galindo, C.; Zambetti, G.P.; Malkin, D. Prevalence and Functional Consequence of TP53 Mutations in Pediatric Adrenocortical Carcinoma: A Children’s Oncology Group Study. J. Clin. Oncol. 2015, 33, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Varley, J.M.; McGown, G.; Thorncroft, M.; James, L.A.; Margison, G.P.; Forster, G.; Evans, D.G.R.; Harris, M.; Kelsey, A.M.; Birch, J.M. Are There Low-Penetrance TP53 Alleles? Evidence from Childhood Adrenocortical Tumors. Am. J. Hum. Genet. 1999, 65, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, L.J.M.; Heinze, B.; Fassnacht, M.; Willenberg, H.S.; Quinkler, M.; Reisch, N.; Zink, M.; Allolio, B.; Hahner, S. TP53Germline Mutations in Adult Patients with Adrenocortical Carcinoma. J. Clin. Endocrinol. Metab. 2012, 97, E476–E485. [Google Scholar] [CrossRef]
- Figueiredo, B.C.; Sandrini, R.; Zambetti, G.P.; Pereira, R.M.; Cheng, C.; Liu, W.; Lacerda, L.; A Pianovski, M.; Michalkiewicz, E.; Jenkins, J.; et al. Penetrance of adrenocortical tumours associated with the germline TP53 R337H mutation. J. Med Genet. 2005, 43, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Soon, P.S.H.; Libé, R.; Benn, D.E.; Gill, A.; Shaw, J.; Sywak, M.S.; Groussin, L.; Bertagna, X.; Gicquel, C.; Bertherat, J.; et al. Loss of Heterozygosity of 17p13, With Possible Involvement of ACADVL and ALOX15B, in the Pathogenesis of Adrenocortical Tumors. Ann. Surg. 2008, 247, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Lapunzina, P. Risk of Tumorigenesis in Overgrowth Sydndromes: A Comprehensive Review. Am. J. Med. Genet. Part C Semin. Med. Genet. 2005, 137C, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Gicquel, C.; Raffin-Sanson, M.-L.; Gaston, V.; Bertagna, X.; Plouin, P.F.; Schlumberger, M.; Louvel, A.; Luton, J.P.; Le Bouc, Y. Structural and Functional Abnormalities at 11p15 are associated with the Malignant Phenotype in Sporadic Adrenocortical Tumors: Study on a Series of 82 Tumors. J. Clin. Endocrinol. Metab. 1997, 82, 2559–2565. [Google Scholar] [CrossRef]
- Shiroky, J.S.; Lerner-Ellis, J.P.; Govindarajan, A.; Urbach, D.R.; Devon, K.M. Characteristics of Adrenal Masses in Familial Adenomatous Polyposis. Dis. Colon Rectum 2018, 61, 679–685. [Google Scholar] [CrossRef]
- Gaujoux, S.; Grabar, S.; Fassnacht, M.; Ragazzon, B.; Launay, P.; Libé, R.; Chokri, I.; Audebourg, A.; Royer, B.; Sbiera, S.; et al. β-Catenin Activation is Associated with Specific Clinical and Pathologic Characteristics and a Poor Outcome in Adrenocortical Carcinoma. Clin. Cancer Res. 2011, 17, 206–211. [Google Scholar] [CrossRef]
- Else, T. Association of adrenocortical carcinoma with familial cancer susceptibility syndromes. Mol. Cell. Endocrinol. 2012, 351, 66–70. [Google Scholar] [CrossRef]
- Gatta-Cherifi, B.; Chabre, O.; Murat, A.; Niccoli, P.; Cardot-Bauters, C.; Rohmer, V.; Young, J.; Delemer, B.; Du Boullay, H.; Verger, M.F.; et al. Adrenal involvement in MEN1. Analysis of 715 cases from the Groupe d’etude des Tumeurs Endocrines database. Eur. J. Endocrinol. 2011, 166, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Langer, P.D.P.; Cupisti, K.; Bartsch, D.K.; Nies, C.; Goretzki, P.E.; Rothmund, M.; Röher, H.D. Adrenal Involvement in Multiple Endocrine Neoplasia Type 1. World J. Surg. 2002, 26, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.J.; Pichurin, P.N.; Hines, J.M.; Singh, R.J.; Grebe, S.K.; Bancos, I. Adrenal Cortical Carcinoma Associated With Lynch Syndrome: A Case Report and Review of Literature. J. Endocr. Soc. 2019, 3, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Menon, R.K.; Ferraù, F.; Kurzawinski, T.R.; Rumsby, G.; Freeman, A.; Amin, Z.; Korbonits, M.; Chung, T.-T.L.L. Adrenal cancer in neurofibromatosis type 1: Case report and DNA analysis. Endocrinol. Diabetes Metab. Case Rep. 2014, 2014, 140074. [Google Scholar] [CrossRef] [PubMed]
- Bertherat, J. Adrenocortical cancer in Carney complex: A paradigm of endocrine tumor progression or an association of genetic predisposing factors? J. Clin. Endocrinol. Metab. 2012, 97, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Boil. 2019, 20, 199–210. [Google Scholar] [CrossRef]
- Baugh, E.H.; Ke, H.; Levine, A.J.; A Bonneau, R.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2017, 25, 154–160. [Google Scholar] [CrossRef]
- Petitjean, A.; Mathe, E.; Kato, S.; Ishioka, C.; Tavtigian, S.V.; Hainaut, P.; Olivier, M. Impact of mutant p53 functional properties onTP53mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007, 28, 622–629. [Google Scholar] [CrossRef]
- Libé, R.; Groussin, L.; Tissier, F.; Elie, C.; René-Corail, F.; Fratticci, A.; Jullian, E.; Beck-Peccoz, P.; Bertagna, X.; Gicquel, C.; et al. Somatic TP53 Mutations Are Relatively Rare among Adrenocortical Cancers with the Frequent 17p13 Loss of Heterozygosity. Clin. Cancer Res. 2007, 13, 844–850. [Google Scholar] [CrossRef]
- Wasserman, J.D.; Zambetti, G.P.; Malkin, D. Towards an understanding of the role of p53 in adrenocortical carcinogenesis. Mol. Cell. Endocrinol. 2012, 351, 101–110. [Google Scholar] [CrossRef]
- Jacks, T.; Remington, L.; Williams, B.O.; Schmitt, E.M.; Halachmi, S.; Bronson, R.T.; Weinberg, R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Boil. 1994, 4, 1–7. [Google Scholar] [CrossRef]
- Lang, G.A.; Iwakuma, T.; Suh, Y.-A.; Liu, G.; Rao, V.; Parant, J.M.; Valentin-Vega, Y.A.; Terzian, T.; Caldwell, L.C.; Strong, L.C.; et al. Gain of Function of a p53 Hot Spot Mutation in a Mouse Model of Li-Fraumeni Syndrome. Cell 2004, 119, 861–872. [Google Scholar] [CrossRef]
- Else, T.; Trovato, A.; Kim, A.C.; Wu, Y.; Ferguson, D.O.; Kuick, R.D.; Lucas, P.C.; Hammer, G.D. Genetic p53 Deficiency Partially Rescues the Adrenocortical Dysplasia Phenotype at the Expense of Increased Tumorigenesis. Cancer Cell 2009, 15, 465–476. [Google Scholar] [CrossRef]
- Brice, A.L.; E Cheetham, J.; Bolton, V.N.; Hill, N.C.; Schofield, P.N. Temporal changes in the expression of the insulin-like growth factor II gene associated with tissue maturation in the human fetus. Development 1989, 106, 543–554. [Google Scholar]
- Giordano, T.J.; Thomas, D.G.; Kuick, R.; Lizyness, M.; Misek, D.E.; Smith, A.L.; Sanders, D.; Aljundi, R.T.; Gauger, P.G.; Thompson, N.W.; et al. Distinct Transcriptional Profiles of Adrenocortical Tumors Uncovered by DNA Microarray Analysis. Am. J. Pathol. 2003, 162, 521–531. [Google Scholar] [CrossRef]
- Gicquel, C.; Bertagna, X.; Gaston, V.; Coste, J.; Louvel, A.; Baudin, E.; Bertherat, J.; Chapuis, Y.; Duclos, J.M.; Schlumberger, M.; et al. Molecular markers and long-term recurrences in a large cohort of patients with sporadic adrenocortical tumors. Cancer Res. 2001, 61, 6762–6767. [Google Scholar] [PubMed]
- Giordano, T.J.; Kuick, R.; Else, T.; Gauger, P.G.; Vinco, M.; Bauersfeld, J.; Sanders, D.; Thomas, D.G.; Doherty, G.M.; Hammer, G. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin. Cancer Res. 2009, 15, 668–676. [Google Scholar] [CrossRef] [PubMed]
- De Fraipont, F.; El Atifi, M.; Cherradi, N.; Le Moigne, G.; Defaye, G.; Houlgatte, R.; Bertherat, J.; Bertagna, X.; Plouin, P.-F.; Baudin, E.; et al. Gene Expression Profiling of Human Adrenocortical Tumors Using Complementary Deoxyribonucleic Acid Microarrays Identifies Several Candidate Genes as Markers of Malignancy. J. Clin. Endocrinol. Metab. 2005, 90, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.-H.; Suppola, S.; Liu, J.; Heikkilä, P.; Jänne, J.; Voutilainen, R. Association of H19 Promoter Methylation with the Expression of H19 and IGF-II Genes in Adrenocortical Tumors. J. Clin. Endocrinol. Metab. 2002, 87, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kahri, A.I.; Heikkila, P.; Ilvesmäki, V.; Voutilainen, R. H19 and Insulin-Like Growth Factor-II Gene Expression in Adrenal Tumors and Cultured Adrenal Cells. J. Clin. Endocrinol. Metab. 1995, 80, 492–496. [Google Scholar] [PubMed]
- Glover, A.; Zhao, J.T.; Ip, J.C.; Lee, J.C.; Robinson, B.G.; Gill, A.J.; Soon, P.S.H.; Sidhu, S.B. Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocrine-Related Cancer 2015, 22, 99–109. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, X.; Zhu, X.; Liu, F.-C.; Ye, F.; Wu, D.-H.; Zhong, P. Bioinformatic analysis of long non-coding RNA-associated competing endogenous RNA network in adrenocortical carcinoma. Transl. Cancer Res. 2019, 8, 2175–2186. [Google Scholar] [CrossRef]
- Soon, P.S.; McDonald, K.L.; Robinson, B.G.; Sidhu, S.B. Molecular Markers and the Pathogenesis of Adrenocortical Cancer. Oncology 2008, 13, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.S.; Monteiro, M.P.; Costa, M.M.; Moreira, Â.; Alves, M.G.; Oliveira, P.F.; Jarak, I.; Pignatelli, D. IGF2 role in adrenocortical carcinoma biology. Endocrine 2019, 66, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Drelon, C.; Berthon, A.S.; Ragazzon, B.; Tissier, F.; Bandiera, R.; Sahut-Barnola, I.; De Joussineau, C.; Batisse-Lignier, M.; Lefrançois-Martinez, A.-M.; Bertherat, J.; et al. Analysis of the Role of Igf2 in Adrenal Tumour Development in Transgenic Mouse Models. PLoS ONE 2012, 7, e44171. [Google Scholar] [CrossRef] [PubMed]
- Heaton, J.H.; Wood, M.A.; Kim, A.C.; Lima, L.O.; Barlaskar, F.M.; Almeida, M.Q.; Fragoso, M.; Kuick, R.; Lerario, A.; Simon, D.P.; et al. Progression to adrenocortical tumorigenesis in mice and humans through insulin-like growth factor 2 and β-catenin. Am. J. Pathol. 2012, 181, 1017–1033. [Google Scholar] [CrossRef]
- Fassnacht, M.; Berruti, A.; Baudin, E.; Demeure, M.J.; Gilbert, J.; Haak, H.; Kroiss, M.; Quinn, D.I.; Hesseltine, E.; Ronchi, C.L.; et al. Linsitinib (OSI-906) versus placebo for patients with locally advanced or metastatic adrenocortical carcinoma: A double-blind, randomised, phase 3 study. Lancet Oncol. 2015, 16, 426–435. [Google Scholar] [CrossRef]
- Haluska, P.; Worden, F.; Olmos, D.; Yin, D.; Schteingart, D.; Batzel, G.N.; Paccagnella, M.L.; De Bono, J.S.; Gualberto, A.; Hammer, G.D. Safety, tolerability, and pharmacokinetics of the anti-IGF-1R monoclonal antibody figitumumab in patients with refractory adrenocortical carcinoma. Cancer Chemother. Pharmacol. 2009, 65, 765–773. [Google Scholar] [CrossRef][Green Version]
- Naing, A.; Lorusso, P.; Fu, S.; Hong, D.; Chen, H.X.; A Doyle, L.; Phan, A.T.; Habra, M.A.; Kurzrock, R. Insulin growth factor receptor (IGF-1R) antibody cixutumumab combined with the mTOR inhibitor temsirolimus in patients with metastatic adrenocortical carcinoma. Br. J. Cancer 2013, 108, 826–830. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Assié, G.; Letouzé, E.; Fassnacht, M.; Jouinot, A.; Luscap, W.; Barreau, O.; Omeiri, H.; Rodriguez, S.; Perlemoine, K.; Rene-Corail, F.; et al. Integrated genomic characterization of adrenocortical carcinoma. Nat. Genet. 2014, 46, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, R.; Backman, S.; Åkerström, T.; Hellman, P.; Björklund, P. Comprehensive analysis of CTNNB1 in adrenocortical carcinomas: Identification of novel mutations and correlation to survival. Sci. Rep. 2018, 8, 8610. [Google Scholar] [CrossRef] [PubMed]
- Berthon, A.S.; Sahut-Barnola, I.; Lambert-Langlais, S.; De Joussineau, C.; Damon-Soubeyrand, C.; Louiset, E.; Taketo, M.M.; Tissier, F.; Bertherat, J.; Lefrançois-Martinez, A.-M.; et al. Constitutive β-catenin activation induces adrenal hyperplasia and promotes adrenal cancer development. Hum. Mol. Genet. 2010, 19, 1561–1576. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; A Grässer, F.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, B. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Caramuta, S.; Lee, L.; Özata, D.M.; Akçakaya, P.; Xie, H.; Höög, A.; Zedenius, J.; Bäckdahl, M.; Larsson, C.; Lui, W.-O. Clinical and functional impact of TARBP2 over-expression in adrenocortical carcinoma. Endocrine-Related Cancer 2013, 20, 551–564. [Google Scholar] [CrossRef]
- Hibio, N.; Hino, K.; Shimizu, E.; Nagata, Y.; Ui-Tei, K. Stability of miRNA 5′terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy. Sci. Rep. 2012, 2, srep00996. [Google Scholar] [CrossRef]
- Hammond, S.M.; Kuner, R.; Köhr, G.; Grünewald, S.; Eisenhardt, G.; Bach, A.; Kornau, H.-C. Argonaute2, a Link Between Genetic and Biochemical Analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Pantano, L.; Banez-Coronel, M.; Llorens, F.; Miñones-Moyano, E.; Porta, S.; Sumoy, L.; Ferrer, I.; Estivill, X. A myriad of miRNA variants in control and Huntington’s disease brin regions detected by massively parallel sequencing. Nucleic Acids Res. 2010, 38, 7219–7235. [Google Scholar] [CrossRef]
- Neilsen, C.T.; Goodall, G.J.; Bracken, C.P. IsomiRs – the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012, 28, 544–549. [Google Scholar] [CrossRef]
- Manzano, M.; Forte, E.; Raja, A.N.; Schipma, M.J.; Gottwein, E. Divergent target recognition by coexpressed 5′-isomiRs of miR-142-3p and selective viral mimicry. RNA 2015, 21, 1606–1620. [Google Scholar] [CrossRef]
- Moore, M.J.; Scheel, T.K.H.; Luna, J.M.; Park, C.Y.; Fak, J.J.; Nishiuchi, E.; Rice, C.M.; Darnell, R.B. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015, 6, 8864. [Google Scholar] [CrossRef]
- Chan, Y.-T.; Lin, Y.-C.; Lin, R.-J.; Kuo, H.-H.; Thang, W.C.; Chiu, K.-P.; Yu, A.L. Concordant and Discordant Regulation of Target Genes by miR-31 and Its Isoforms. PLoS ONE 2013, 8, e58169. [Google Scholar] [CrossRef]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.R.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Z.; Chen, P.; Wu, M. Tumor classification and biomarker discovery based on the 5′isomiR expression level. BMC Cancer 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Lan, C.; Peng, H.; McGowan, E.M.; Hutvagner, G.; Li, J. An isomiR expression panel based novel breast cancer classification approach using improved mutual information. BMC Med. Genom. 2018, 11, 118. [Google Scholar] [CrossRef]
- Koperski, L.; Kotlarek, M.; Swierniak, M.; Kolanowska, M.; Kubiak, A.; Górnicka, B.; Jazdzewski, K.; Wójcicka, A. Next-generation sequencing reveals microRNA markers of adrenocortical tumors malignancy. Oncotarget 2017, 8, 49191–49200. [Google Scholar] [CrossRef]
- Soon, P.; Tacon, L.J.; Gill, A.J.; Bambach, C.P.; Sywak, M.S.; Campbell, P.R.; Yeh, M.W.; Wong, S.G.; Clifton-Bligh, R.J.; Robinson, B.G.; et al. miR-195 and miR-483-5p Identified as Predictors of Poor Prognosis in Adrenocortical Cancer. Clin. Cancer Res. 2009, 15, 7684–7692. [Google Scholar] [CrossRef]
- Özata, D.M.; Caramuta, S.; Velàzquez-Fernàndez, D.; Akçakaya, P.; Xie, H.; Höög, A.; Zedenius, J.; Bäckdahl, M.; Larsson, C.; Lui, W.-O. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocrine-Related Cancer 2011, 18, 643–655. [Google Scholar] [CrossRef]
- Patterson, E.E.; Holloway, A.K.; Weng, J.; Fojo, T.; Kebebew, E. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer 2010, 117, 1630–1639. [Google Scholar] [CrossRef]
- Chabre, O.; Libé, R.; Assié, G.; Barreau, O.; Bertherat, J.; Bertagna, X.; Feige, J.-J.; Cherradi, N. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocrine-Related Cancer 2013, 20, 579–594. [Google Scholar] [CrossRef]
- Feinmesser, M.; Benbassat, C.; Meiri, E.; Benjamin, H.; Lebanony, D.; Lebenthal, Y.; De Vries, L.; Drozd, T.; Spector, Y. Specific MicroRNAs Differentiate Adrenocortical Adenomas from Carcinomas and Correlate With Weiss Histopathologic System. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 522–531. [Google Scholar] [CrossRef]
- Tömböl, Z.; Szabó, P.M.; Molnár, V.; Wiener, Z.; Tölgyesi, G.; Horányi, J.; Riesz, P.; Reismann, P.; Patocs, A.; Liko, I.; et al. Integrative molecular bioinformatics study of human adrenocortical tumors: microRNA, tissue-specific target prediction, and pathway analysis. Endocrine-Related Cancer 2009, 16, 895–906. [Google Scholar] [CrossRef]
- Schmitz, K.J.; Helwig, J.; Bertram, S.; Sheu, S.Y.; Suttorp, A.C.; Seggewiß, J.; Willscher, E.; Walz, M.K.; Worm, K.; Schmid, K.W. Differential expression of microRNA-675, microRNA-139-3p and microRNA-335 in benign and malignant adrenocortical tumours. J. Clin. Pathol. 2011, 64, 529–535. [Google Scholar] [CrossRef]
- Duregon, E.; Rapa, I.; Votta, A.; Giorcelli, J.; Daffara, F.; Terzolo, M.; Scagliotti, G.V.; Volante, M.; Papotti, M. MicroRNA expression patterns in adrenocortical carcinoma variants and clinical pathologic correlations. Hum. Pathol. 2014, 45, 1555–1562. [Google Scholar] [CrossRef]
- Zheng, S.; Cherniack, A.D.; Dewal, N.; Moffitt, R.A.; Danilova, L.; Murray, B.A.; Lerario, A.M.; Else, T.; Knijnenbury, T.A.; Girielli, G.; et al. Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer Cell 2016, 29, 723–736. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Decmann, A.; Perge, P.; Turai, P.I.; Patocs, A.; Igaz, P. Non-Coding RNAs in Adrenocortical Cancer: From Pathogenesis to Diagnosis. Cancers 2020, 12, 461. [Google Scholar] [CrossRef] [PubMed]
- Szabó, D.R.; Luconi, M.; Szabó, P.M.; Tóth, M.; Szücs, N.; Horányi, J.; Nagy, Z.; Mannelli, M.; Patocs, A.; Rácz, K.; et al. Analysis of circulating microRNAs in adrenocortical tumors. Lab. Investig. 2013, 94, 331–339. [Google Scholar] [CrossRef]
- Patel, D.; Boufraqech, M.; Jain, M.; Zhang, L.; He, M.; Gesuwan, K.; Gulati, N.; Nilubol, N.; Fojo, T.; Kebebew, E. MiR-34a and miR-483-5p are candidate serum biomarkers for adrenocortical tumors. Surgery 2013, 154, 1224–1229. [Google Scholar] [CrossRef]
- Shen, J.; Wang, A.; Wang, Q.; Gurvich, I.; Siegel, A.B.; Remotti, H.; Santella, R.M. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: MiR-483-5p as a potential biomarker. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2364–2373. [Google Scholar] [CrossRef]
- Lamichhane, S.R.; Thachil, T.; Gee, H.; Milic, N. Circulating MicroRNAs as Prognostic Molecular Biomarkers in Human Head and Neck Cancer: A Systematic Review and Meta-Analysis. Dis. Markers 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorshid, H.R.K.; Ghaffari, S.H. Why have microRNA biomarkers not been translated from bench to clinic? Future Oncol. 2019, 15, 801–803. [Google Scholar] [CrossRef]
- Farina, N.H.; Wood, M.E.; Perrapato, S.D.; Francklyn, C.S.; Stein, G.S.; Stein, J.L.; Lian, J. Standardizing analysis of circulating microRNA: Clinical and biological relevance. J. Cell. Biochem. 2014, 115, 805–811. [Google Scholar] [CrossRef]
- Benz, F.; Roderburg, C.; Cardenas, D.V.; Vucur, M.; Gautheron, J.; Koch, A.; Zimmermann, H.; Janssen, J.; Nieuwenhuijsen, L.; Luedde, M.; et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp. Mol. Med. 2013, 45, e42. [Google Scholar] [CrossRef]
- Marabita, F.; De Candia, P.; Torri, A.; Tegnér, J.; Abrignani, S.; Rossi, R.L. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Briefings Bioinform. 2015, 17, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Jouinot, A.; Bertherat, J. MANAGEMENT OF ENDOCRINE DISEASE: Adrenocortical carcinoma: Differentiating the good from the poor prognosis tumors. Eur. J. Endocrinol. 2018, 178, R215–R230. [Google Scholar] [CrossRef] [PubMed]
- Faria, A.M.; Sbiera, S.; Ribeiro, T.C.; Soares, I.C.; Mariani, B.M.; Freire, D.S.; De Sousa, G.R.; Lerario, A.M.; Ronchi, C.L.; Deutschbein, T.; et al. Expression of LIN28 and its regulatory microRNAs in adult adrenocortical cancer. Clin. Endocrinol. 2014, 82, 481–488. [Google Scholar] [CrossRef]
- Streicher, K.L.; Zhu, W.; Lehmann, K.P.; Georgantas, R.W.; A Morehouse, C.; Brohawn, P.; A Carrasco, R.; Xiao, Z.; A Tice, D.; Higgs, B.W.; et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 2011, 31, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef] [PubMed]
- Ab Mutalib, N.-S.; Sulaiman, S.A.; Jamal, R. Computational Tools for microRNA Target Prediction. In Computational Epigenetics and Diseases; Elsevier: Amsterdam, The Netherlands, 2019; pp. 79–105. [Google Scholar]
- Zhang, Y.; Zang, Q.; Zhang, H.; Ban, R.; Yang, Y.; Iqbal, F.; Li, A.; Shi, Q. DeAnnIso: A tool for online detection and annotation of isomiRs from small RNA sequencing data. Nucleic Acids Res. 2016, 44, W166–W175. [Google Scholar] [CrossRef]
- Hammell, M. Computational methods to identify miRNA targets. Semin. Cell Dev. Boil. 2010, 21, 738–744. [Google Scholar] [CrossRef]
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Briefings Bioinform. 2012, 15, 1–19. [Google Scholar] [CrossRef]
- Thomson, D.W.; Bracken, C.P.; Goodall, G. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef]
- Martinez-Sanchez, A.; Murphy, C.L. MicroRNA Target Identification—Experimental Approaches. Boil. 2013, 2, 189–205. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y. Current experimental strategies for intracellular target identification of microRNA. ExRNA 2019, 1, 6. [Google Scholar] [CrossRef]
- Mockly, S.; Seitz, H. Inconsistencies and Limitations of Current MicroRNA Target Identification Methods. In Breast Cancer; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2019; Volume 1970, pp. 291–314. [Google Scholar]
- Khafaei, M.; Rezaie, E.; Mohammadi, A.; Gerdehsang, P.S.; Ghavidel, S.; Kadkhoda, S.; Zahra, A.Z.; Forouzanfar, N.; Arabameri, H.; Tavallaei, M. miR-9: From function to therapeutic potential in cancer. J. Cell. Physiol. 2019, 234, 14651–14665. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-H.; Tsao, C.-J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yang, W.; Ma, J.; Zhang, H.; Li, Z.; Zhang, L.; Liu, J.; Han, Z.; Wang, H.; Hong, L. Role of miR-483 in digestive tract cancers: From basic research to clinical value. J. Cancer 2018, 9, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-L.; Huang, L.; Wang, L.; Tong, B.-D.; Wei, Q.; Ding, X.-S. Potential role of miR-139-5p in cancer diagnosis, prognosis and therapy. Oncol. Lett. 2017, 14, 1215–1222. [Google Scholar] [CrossRef]
- Agosta, C.; Laugier, J.; Guyon, L.; Denis, J.; Bertherat, J.; Libe, R.; Boisson, B.; Sturm, N.; Feige, J.-J.; Chabre, O.; et al. MiR-483-5p and miR-139-5p promote aggressiveness by targeting N-myc downstream-regulated gene family members in adrenocortical cancer. Int. J. Cancer 2018, 143, 944–957. [Google Scholar] [CrossRef]
- De Sousa, G.R.V.; Ribeiro, T.C.; Faria, A.M.; Mariani, B.M.P.; Lerario, A.M.; Zerbini, M.C.N.; Soares, I.C.; Wakamatsu, A.; Alves, V.A.F.; Mendonca, B.B.; et al. Low DICER1 expression is associated with poor clinical outcome in adrenocortical carcinoma. Oncotarget 2015, 6, 22724–22733. [Google Scholar] [CrossRef]
- Zhou, J.; Ng, S.-B.; Chng, W.J. LIN28/LIN28B: An emerging oncogenic driver in cancer stem cells. Int. J. Biochem. Cell Boil. 2013, 45, 973–978. [Google Scholar] [CrossRef]
- Zhong, X.; Li, N.; Liang, S.; Huang, Q.; Coukos, G.; Zhang, L. Identification of MicroRNAs Regulating Reprogramming FactorLIN28in Embryonic Stem Cells and Cancer Cells. J. Boil. Chem. 2010, 285, 41961–41971. [Google Scholar] [CrossRef]
- Lima, C.R.; Gomes, C.C.; Santos, M. Role of microRNAs in endocrine cancer metastasis. Mol. Cell. Endocrinol. 2017, 456, 62–75. [Google Scholar] [CrossRef]
- Romero, D.G.; Plonczynski, M.W.; Carvajal, C.A.; Gomez-Sanchez, E.P.; Gomez-Sanchez, C.E. Microribonucleic Acid-21 Increases Aldosterone Secretion and Proliferation in H295R Human Adrenocortical Cells. Endocrinology 2008, 149, 2477–2483. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.A.; Rasheed, S.A.K.; A Nikolova, D.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2007, 27, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- Talotta, F.; Cimmino, A.; Matarazzo, M.R.; Casalino, L.; De Vita, G.; D’Esposito, M.; Di Lauro, R.; Verde, P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 2008, 28, 73–84. [Google Scholar] [CrossRef]
- Veronese, A.; Lupini, L.; Consiglio, J.; Visone, R.; Ferracin, M.; Fornari, F.; Zanesi, N.; Alder, H.; D’Elia, G.; Gramantieri, L.; et al. Oncogenic role of miR-483-3p at the IGF2/483 Locus. Cancer Res. 2010, 70, 3140–3149. [Google Scholar] [CrossRef] [PubMed]
- Nowek, K.; Wiemer, E.A.C.; Jongen-Lavrencic, M. The versatile nature of miR-9/9* in human cancer. Oncotarget 2018, 9, 20838–20854. [Google Scholar] [CrossRef]
- Jiang, Y.; Jia, Y.; Zhang, L. Role of programmed cell death 4 in diseases: A double-edged sword. Cell. Mol. Immunol. 2017, 14, 884–886. [Google Scholar] [CrossRef][Green Version]
- Hikisz, P.; Kiliańska, Z.M. Puma, a critical mediator of cell death—One decade on from its discovery. Cell. Mol. Boil. Lett. 2012, 17, 646–669. [Google Scholar] [CrossRef]
- Glover, A.; Zhao, J.T.; Gill, A.J.; Weiss, J.; Mugridge, N.; Kim, E.; Feeney, A.L.; Ip, J.C.; Reid, G.; Clarke, S.; et al. microRNA-7 as a tumor suppressor and novel therapeutic for adrenocortical carcinoma. Oncotarget 2015, 6, 36675–36688. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, W.; Hu, W.; Xu, W.; Xiao, G.; Nie, Q.; Ouyang, K.; Chen, S. MicroRNA-205 suppresses the growth of adrenocortical carcinoma SW-13 cells via targeting Bcl-2. Oncol. Rep. 2015, 34, 3104–3110. [Google Scholar] [CrossRef]
- Finnerty, J.R.; Wang, W.-X.; Hebert, S.S.; Wilfred, B.R.; Mao, G.; Nelson, P.T. The miR-15/107 Group of MicroRNA Genes: Evolutionary Biology, Cellular Functions, and Roles in Human Diseases. J. Mol. Boil. 2010, 402, 491–509. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, B.; Wang, H.-Y.; Chang, A.; Zheng, X.S.; Bo, H. Emerging Role of MicroRNAs in mTOR Signaling. Cell. Mol. Life Sci. 2017, 74, 2613–2625. [Google Scholar] [CrossRef] [PubMed]
- Doghman, M.; El Wakil, A.; Cardinaud, B.; Thomas, E.; Wang, J.; Zhao, W.; Valle, M.P.-D.; Figueiredo, B.; Zambetti, G.; Lalli, E. Regulation of IGF—mTOR Signalling by miRNA in Childhood Adrenocortical Tumors. In Proceedings of the Endocrine Society’s 92nd Annual Meeting, San Diego, CA, USA, 19–22 June 2010; Volume 70, p. OR20-4. [Google Scholar] [CrossRef]
- Jain, M.; Zhang, L.; Boufraqech, M.; Liu, J.O.; Bussey, K.J.; Demeure, M.J.; Wu, X.; Su, L.; Pacak, K.; Stratakis, C.A.; et al. ZNF367 Inhibits Cancer Progression and Is Targeted by miR-195. PLoS ONE 2014, 9, e101423. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cao, Y.; Su, T.; Jiang, Y.; Jiang, L.; Zhou, W.; Zhang, C.; Wang, W.; Ning, G. Downregulation of miR-375 in aldosterone-producing adenomas promotes tumour cell growth via MTDH. Clin. Endocrinol. 2015, 83, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Kwok, G.T.; Zhao, J.T.; Glover, A.; Gill, A.J.; Clifton-Bligh, R.; Robinson, B.G.; Ip, J.C.; Sidhu, S.B. microRNA-431 as a Chemosensitizer and Potentiator of Drug Activity in Adrenocortical Carcinoma. Oncology 2019, 24, e241–e250. [Google Scholar] [CrossRef]
- Hassan, N.; Zhao, J.; Glover, A.R.; Robinson, B.G.; Sidhu, S.B. Reciprocal interplay of miR-497 and MALAT1 promotes tumourigenesis of adrenocortical cancer. Endocrine-Related Cancer 2019, 27, 677–688. [Google Scholar] [CrossRef]
- Kalinowski, F.; Brown, R.A.; Ganda, C.; Giles, K.M.; Epis, M.R.; Horsham, J.L.; Leedman, P.J. microRNA-7: A tumor suppressor miRNA with therapeutic potential. Int. J. Biochem. Cell Boil. 2014, 54, 312–317. [Google Scholar] [CrossRef]
- Zhao, J.; Tao, Y.; Zhou, Y.; Qin, N.; Chen, C.; Tian, D.; Xu, L. MicroRNA-7: A promising new target in cancer therapy. Cancer Cell Int. 2015, 15, 103. [Google Scholar] [CrossRef]
- Das, A.; Reis, F.; Mishra, P.K. mTOR Signaling in Cardiometabolic Disease, Cancer, and Aging 2018. Oxidative Med. Cell. Longev. 2019, 2019, 1–3. [Google Scholar] [CrossRef]
- Jin, Y.; Tymen, S.D.; Chen, D.; Fang, Z.J.; Zhao, Y.; Dragas, D.; Dai, Y.; Marucha, P.T.; Zhou, X. MicroRNA-99 Family Targets AKT/mTOR Signaling Pathway in Dermal Wound Healing. PLoS ONE 2013, 8, e64434. [Google Scholar] [CrossRef]
- Tsai, T.-F.; Lin, J.-F.; Chou, K.-Y.; Lin, Y.-C.; Chen, H.-E.; Hwang, T.I.-S. miR-99a-5p acts as tumor suppressor via targeting to mTOR and enhances RAD001-induced apoptosis in human urinary bladder urothelial carcinoma cells. OncoTargets Ther. 2018, 11, 239–252. [Google Scholar] [CrossRef]
- Yin, H.; Ma, J.; Chen, L.; Piao, S.; Zhang, Y.; Zhang, S.; Ma, H.; Li, Y.; Qu, Y.; Wang, X.; et al. MiR-99a Enhances the Radiation Sensitivity of Non-Small Cell Lung Cancer by Targeting mTOR. Cell. Physiol. Biochem. 2018, 46, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.J.; Tait, S.W. Targeting BCL-2 regulated apoptosis in cancer. Open Boil. 2018, 8, 180002. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wei, Y.; Kang, Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin. Cancer Res. 2009, 15, 5615–5620. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Zeng, T.; Huang, D.; Liu, Z.; Huang, S.; Liu, J.; Qu, Z. MicroRNA-431 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the ZEB1-mediated epithelial-mensenchymal transition. FEBS Open Bio 2015, 5, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar] [CrossRef]
| Genetic Syndrome | Inheritance | Mutated Gene/s | Cellular Pathway/s Affected | Gene Locus | ACC Penetrance | Reference/s |
|---|---|---|---|---|---|---|
| Li Fraumeni Syndrome | Autosomal Dominant | TP53 | Cell cycle | 17p13.1 | 10% | [14] |
| Beckwith-Weidemann Syndrome | Sporadic | IGF2/H19 * CDKN1C/KCNQ1OT1 * | PI3K/TGF-β | 11p15 | 7% | [16] |
| Familial Adenomatous Polyposis | Autosomal Dominant | APC | Wnt/β-Catenin | 5q22.2 | 3% | [18] |
| Multiple Endocrine Neoplasia Type 1 | Autosomal Dominant | MEN1 | Cell Cycle | 11q13 | 1–5% | [21,22] |
| Lynch Syndrome | Autosomal Dominant | MLH1 MSH2 MSH6 PMS2 | DNA mismatch repair | 3p22.2 2p21 2p16.3 7p22 | 14 case reports | [23] |
| Neurofibromatosis Type 1 | Autosomal Dominant | NF1 | MAPK/ERK | 17q11.2 | 9 case reports | [24] |
| Carney Complex | Autosomal Dominant | CNC1 (PRKAR1A) | cAMP | 17q22-24 | 2 case reports | [25] |
| Year of Publication | Methodology | Tissue Samples | Upregulated miRNA | Downregulated miRNA | Reference | ||
|---|---|---|---|---|---|---|---|
| 2009 | TLDA | 7 ACC, 19 ACA, 10 NAC | miR-184 miR-210 miR-503 | miR-214 miR-511 miR-375 | [78] | ||
| 2009 | Microarray VC: RT-q-PCR | 22 ACC, 27 ACA, 6 NAC VC (10 ACC, 9 ACA) | miR-483-5p miR-503 | miR-7 miR-195 | miR-335 | [73] | |
| 2011 | Microarray VC: RT-q-PCR | 25 ACC, 43 ACA, 10 NAC | miR-483-3p miR-483-5p | miR-210 miR-21 | miR-195 miR-497 | [74] | |
| 2011 | Microarray VC: RT-q-PCR | 10 ACC, 26 ACA VC (31 ACC, 35 ACA, 21 NAC) | miR-483-5p | miR-195 miR-125b | miR-100 | [75] | |
| 2011 | TLDA VC: RT-q-PCR | 7 ACC, 9 ACA, 4 NAC VC (16 ACC) | miR-139-5p | miR-139-3p miR-675 | miR-335 | [79] | |
| 2013 | Microarray | 12 ACC, 6 NAC VC (18 ACC, 10 ACA, 3 NAC) | miR-483-5p miR-503 miR-210 miR-542-5p | miR-320a miR-93 miR-148b | miR-195 miR-335 miR-497 | miR-199a-5p miR-199a-3p | [76] |
| 2014 | RT-q-PCR | 51 ACC, 47 ACA | miR-483-3p miR-483-5p miR-210 | miR-195 | [80] | ||
| 2014 | RNA sequencing | 45 ACC, 3 NAC | miR-34b-5p miR-410 miR-483-3p miR-483-5p miR-503 | miR-506-3p miR-506-5p miR-508-3p miR-508-5p miR-510 | miR-511 miR-214-3p miR-485-3p miR-497 miR-195 | [51] | |
| 2015 | Microarray VC: RT-q-PCR | 8 ACC, 25 ACA VC (11 ACC, 4 ACA) | miR-503 | miR-34a | miR-497 | [77] | |
| 2016 | RNA sequencing | 79 ACC, 120 NAC | miR-10-5p miR-483-5p miR-22-3p miR-508-3p miR-509-3p | miR-509-5p miR-340 miR-146a miR-21-3p miR-21-5p | [81] | ||
| 2017 | RNA sequencing | 7 ACC, 8 ACA, 8 NAC VC (8 ACC, 10 ACA, 10 NAC) | miR-503-5p miR-450a-5p miR-542-5p miR-483-3p miR-542-3p miR-450b-5p miR-210 miR-483-5p | miR-421 miR-424-3p miR-424-5p miR-598 miR-148b-3p miR-184 miR-128 | [72] | ||
| microRNA | Functional Role in ACC | Molecular Target | Evidence for Regulatory Interaction | Reference/s |
|---|---|---|---|---|
| miR-9 | Associated with reduced DFS and increased recurrence in clinical data | LIN28 | Weak protein expression pattern in aggressive ACC Established reporter assays in non-ACC models | [109,110,111] |
| miR-21 | ↑ cellular proliferation | PCDC4 | Inverse correlation in expression Reporter assays in non-ACC cell models | [112,113,114,115] |
| miR-139-5p | Associated with anchorage-independent colony formation | NDRG4 | Inverse correlation in expression Reporter assays in non-ACC cell models | [116] |
| miR-483-3p | ↑ cellular proliferation ↓ apoptosis | PUMA | Inverse correlation in expression Reporter assays in non-ACC models | [74,117] |
| miR-483-5p | Associated with anchorage-independent colony formation | NDRG2 | Inverse correlation in expression Reporter assays in non-ACC models | [116] |
| MicroRNA | Functional Role in ACC | Molecular Target | Evidence for Regulatory Interaction | Reference/s |
|---|---|---|---|---|
| miR-7 | ↓ cellular proliferation ↑ G1 cell cycle arrest ↓ H295R xenograft growth in vivo | Raf-1 * EGFR * CDK1 PAK1 CKS2 | Reporter assays * Inverse correlation in expression | [121] |
| miR-99a | IGFR1 mTOR | Inverse correlation in expression Reporter assays in non-ACC models | [125] | |
| miR-100 | IGFR1 mTOR | Inverse correlation in expression Reporter assays in non-ACC models | [125] | |
| miR-195 | ↓ cellular proliferation ↑ cellular invasion ↑ apoptosis | TARBP2 DICER1 | Inverse correlation in expression Ago-2 IP | [58,74] |
| ZNF367 | Reporter assays in SW13 cells | [126] | ||
| miR-205 | ↓ cellular proliferation ↑ apoptosis ↓ SW13 xenograft tumor growth in vivo | Bcl-2 | Reporter assays | [122] |
| miR-375 | ↓ cellular proliferation | MTDH | Reporter assays Inverse correlation in expression | [127] |
| miR-431 | ↑ cellular sensitivity to doxorubicin and mitotane | ZEB1 | Inverse correlation in expression in miRNA overexpressing doxorubicin treated H295R cells. Reporter assays in non-ACC models | [128] |
| miR-497 | ↓ cellular proliferation ↑ apoptosis ↑ G1 cell cycle arrest | TARBP2 DICER1 | Inverse correlation in expressionAgo-2 IP | [58,74] |
| MALAT1 eIF4E SFPQ | Inverse correlation in expression Reporter assays | [129] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chehade, M.; Bullock, M.; Glover, A.; Hutvagner, G.; Sidhu, S. Key MicroRNA’s and Their Targetome in Adrenocortical Cancer. Cancers 2020, 12, 2198. https://doi.org/10.3390/cancers12082198
Chehade M, Bullock M, Glover A, Hutvagner G, Sidhu S. Key MicroRNA’s and Their Targetome in Adrenocortical Cancer. Cancers. 2020; 12(8):2198. https://doi.org/10.3390/cancers12082198
Chicago/Turabian StyleChehade, Marthe, Martyn Bullock, Anthony Glover, Gyorgy Hutvagner, and Stan Sidhu. 2020. "Key MicroRNA’s and Their Targetome in Adrenocortical Cancer" Cancers 12, no. 8: 2198. https://doi.org/10.3390/cancers12082198
APA StyleChehade, M., Bullock, M., Glover, A., Hutvagner, G., & Sidhu, S. (2020). Key MicroRNA’s and Their Targetome in Adrenocortical Cancer. Cancers, 12(8), 2198. https://doi.org/10.3390/cancers12082198







