Prognostic Value of CD200R1 mRNA Expression in Head and Neck Squamous Cell Carcinoma
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
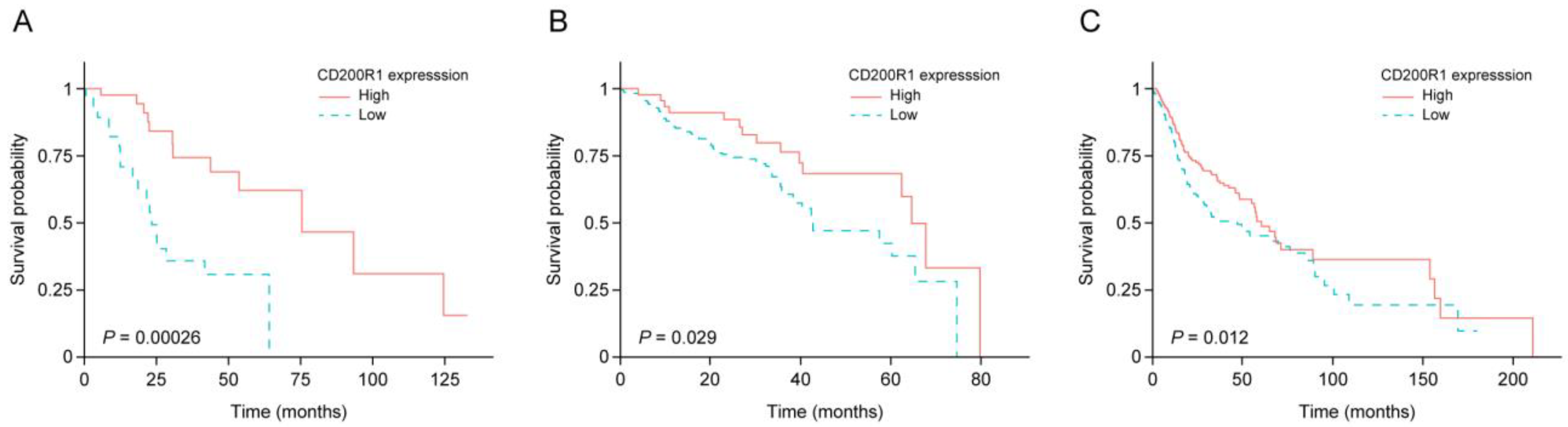
2.2. IRG Expression and Survival of Patients with HNSCC
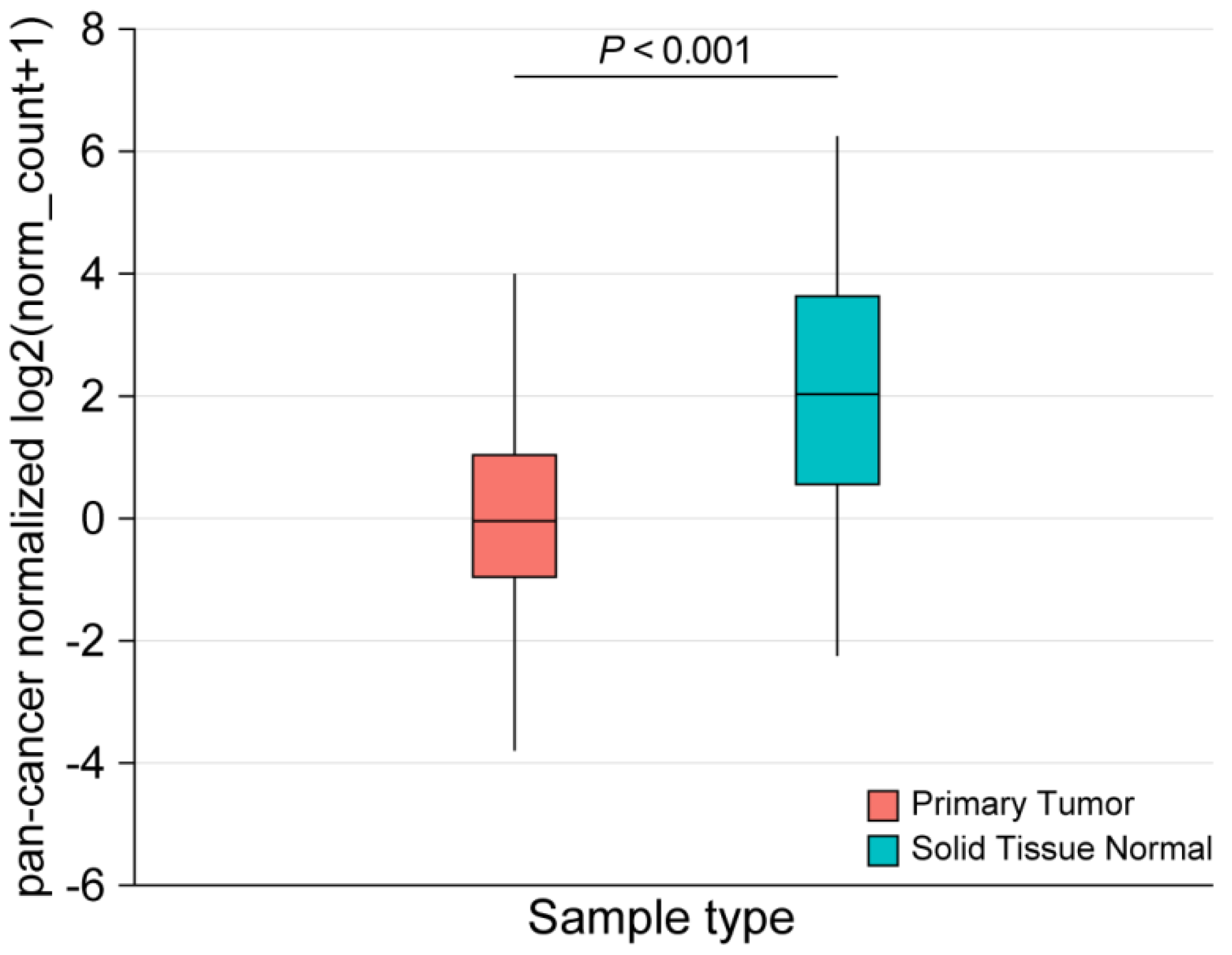
2.3. Performance of CD200R1 mRNA Expression as a Biomarker
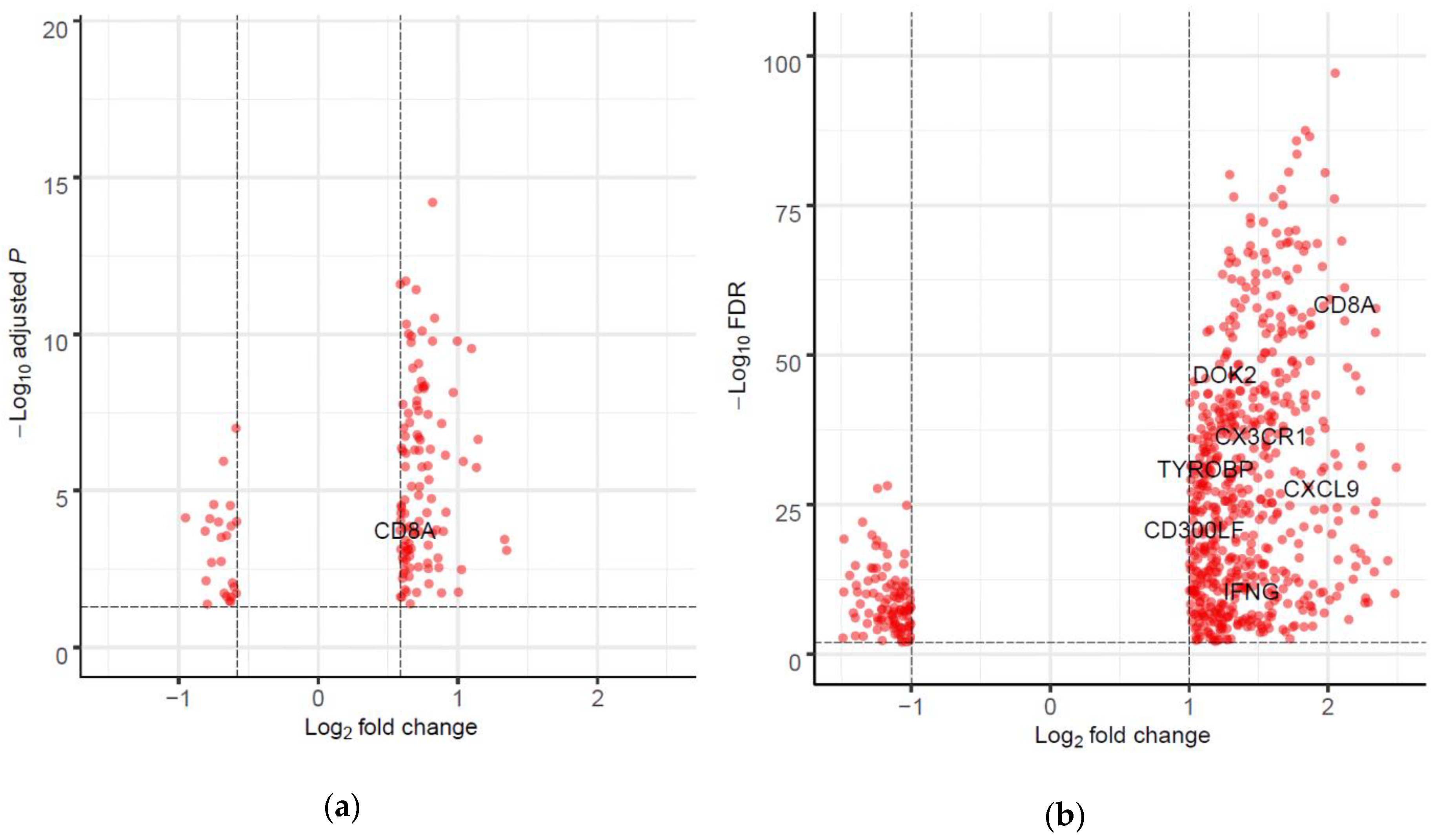
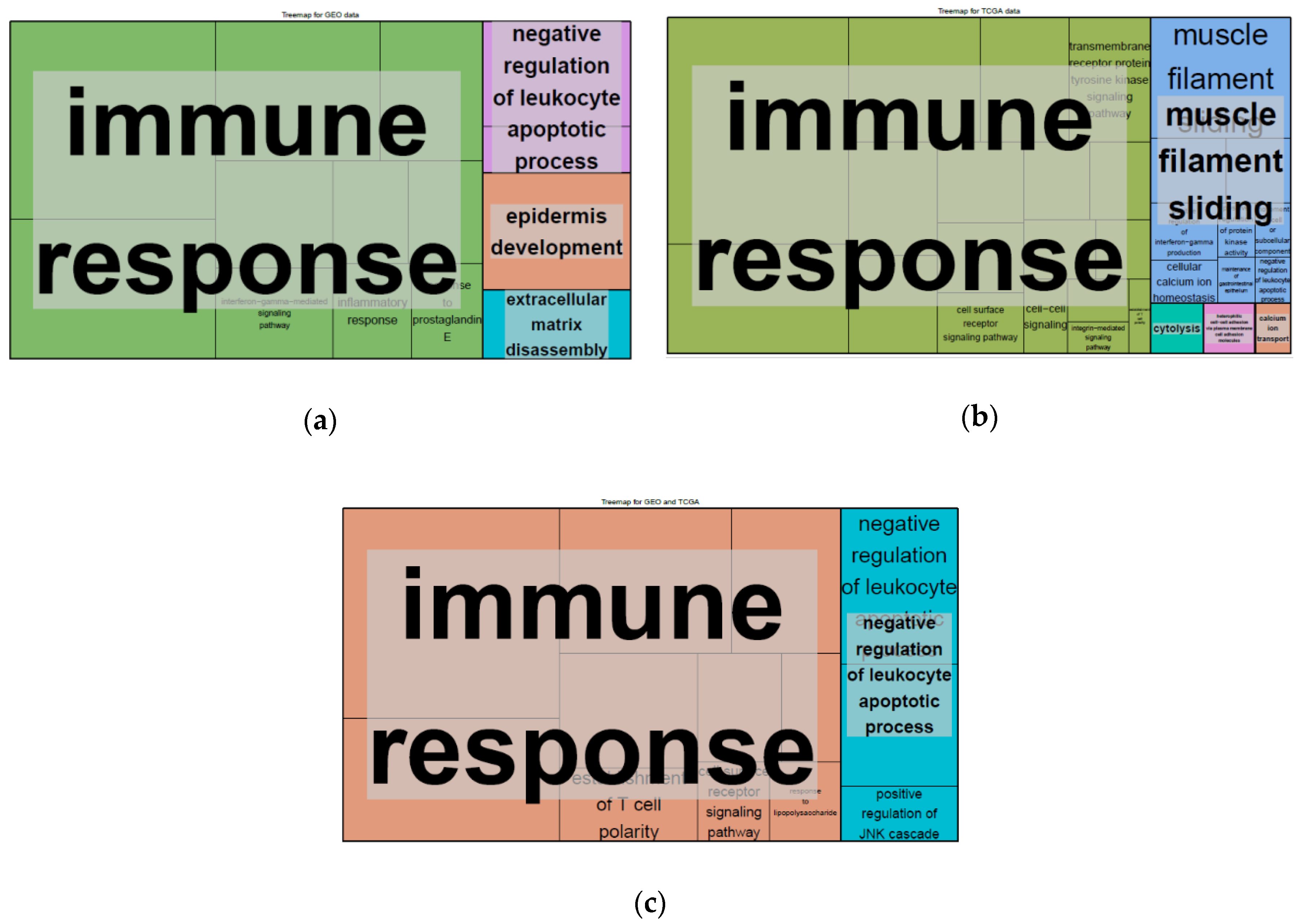
2.4. Differentially Expressed Genes (DEGs) and Gene set Enrichment Analysis
2.5. Interaction between CD200R1 and IRGs
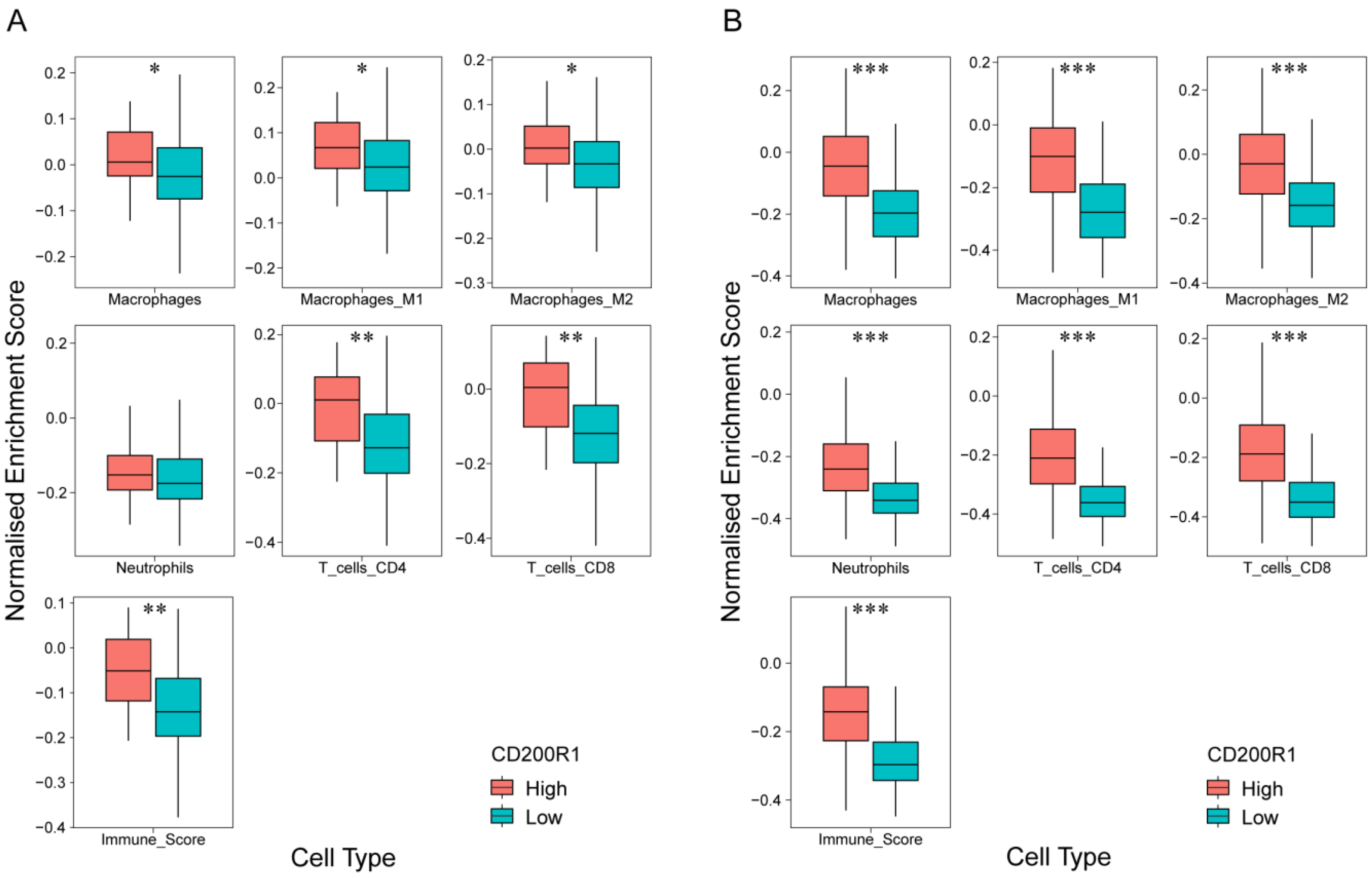
2.6. Correlation of CD200R1 with Immune Cell Signatures
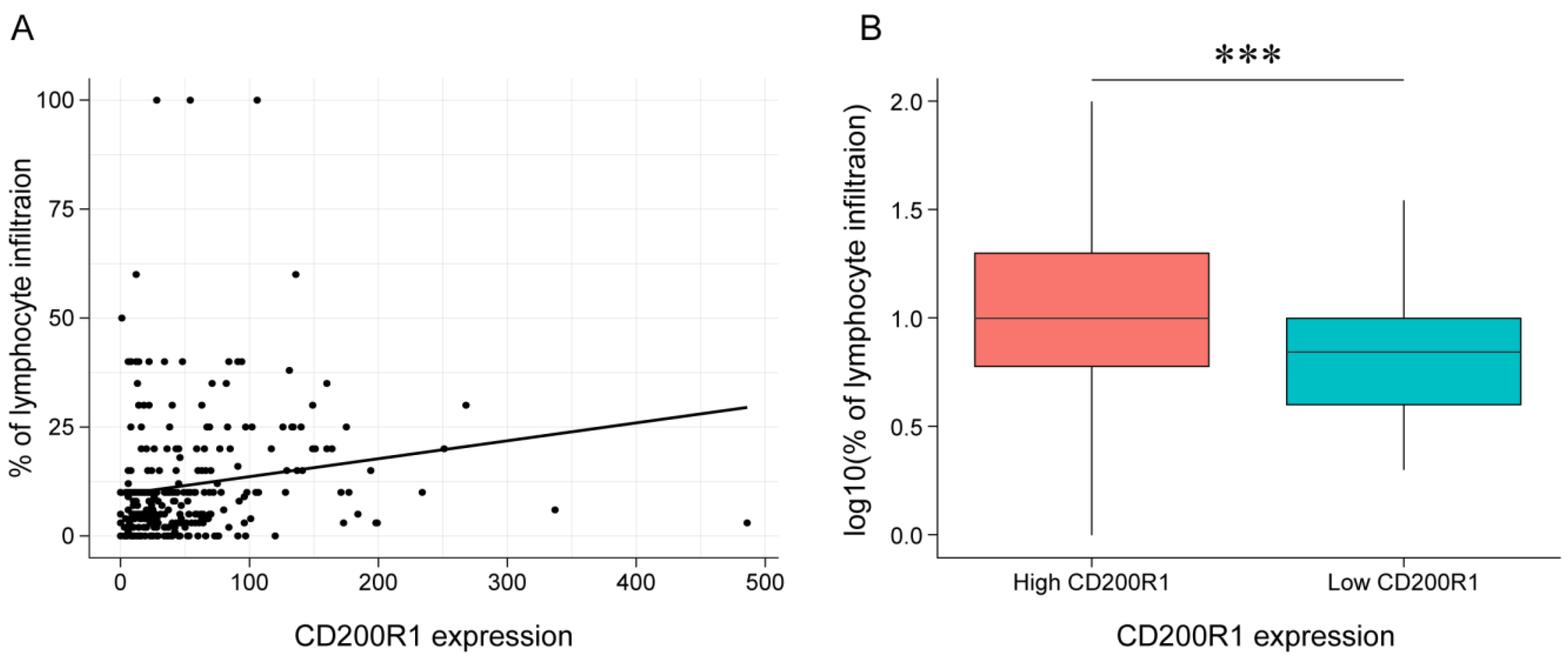
2.7. Association between CD200R1 and Lymphocyte Infiltration
3. Discussion
4. Materials and Methods
4.1. Patients and Data Collection
4.2. Nanostring Assay
4.3. TCGA and GEO Data Analysis
4.4. Statistical Methods
4.5. DEG Screening and Analysis
4.6. Immune Cell Analysis with In Silico Deconvolution and Microscopic Examination
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef]
- Kang, H.; Kiess, A.; Chung, C.H. Emerging biomarkers in head and neck cancer in the era of genomics. Nat. Rev. Clin. Oncol. 2015, 12, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.H.; Thomas, G.; Ottensmeier, C.H.; King, E.V. Importance of the immune system in head and neck cancer. Head Neck 2019, 41, 2789–2800. [Google Scholar] [CrossRef] [PubMed]
- Almangush, A.; Heikkinen, I.; Makitie, A.A.; Coletta, R.D.; Laara, E.; Leivo, I.; Salo, T. Prognostic biomarkers for oral tongue squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2017, 117, 856–866. [Google Scholar] [CrossRef]
- Kerrebijn, J.D.; Balm, A.J.; Knegt, P.P.; Meeuwis, C.A.; Drexhage, H.A. Macrophage and dendritic cell infiltration in head and neck squamous-cell carcinoma; an immunohistochemical study. Cancer Immunol. Immunother. 1994, 38, 31–37. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Burtness, B.; Mehra, R.; Weiss, J.; Berger, R.; Eder, J.P.; Heath, K.; McClanahan, T.; Lunceford, J.; Gause, C.; et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (keynote-012): An open-label, multicentre, phase 1b trial. Lancet Oncol. 2016, 17, 956–965. [Google Scholar] [CrossRef]
- Wansom, D.; Light, E.; Worden, F.; Prince, M.; Urba, S.; Chepeha, D.B.; Cordell, K.; Eisbruch, A.; Taylor, J.; D’Silva, N.; et al. Correlation of cellular immunity with human papillomavirus 16 status and outcome in patients with advanced oropharyngeal cancer. Arch. Otolaryngol. Head Neck Surg. 2010, 136, 1267–1273. [Google Scholar] [CrossRef]
- Spector, M.E.; Bellile, E.; Amlani, L.; Zarins, K.; Smith, J.; Brenner, J.C.; Rozek, L.; Nguyen, A.; Thomas, D.; McHugh, J.B.; et al. Prognostic value of tumor-infiltrating lymphocytes in head and neck squamous cell carcinoma. JAMA Otolaryngol Head Neck Surg. 2019. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; He, M.Y.; Zhu, L.F.; Yang, C.C.; Zhou, M.L.; Wang, Q.; Zhang, W.; Zheng, Y.Y.; Wang, D.M.; Xu, Z.Q.; et al. Tumor-associated macrophages correlate with the clinicopathological features and poor outcomes via inducing epithelial to mesenchymal transition in oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 12. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Park, S.; Park, S.Y.; Kim, G.; Park, S.M.; Cho, J.-W.; Kim, D.H.; Park, Y.M.; Koh, Y.W.; Kim, H.R.; et al. Single-cell transcriptome analysis reveals tox as a promoting factor for t cell exhaustion and a predictor for anti-pd-1 responses in human cancer. Genome Med. 2020, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. The ucsc xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv 2019. [Google Scholar] [CrossRef]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. String: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Liu, J.Q.; Talebian, F.; Wu, L.; Liu, Z.; Li, M.S.; Wu, L.; Zhu, J.; Markowitz, J.; Carson, W.E., 3rd; Basu, S.; et al. A critical role for cd200r signaling in limiting the growth and metastasis of cd200+ melanoma. J. Immunol. 2016, 197, 1489–1497. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.Q.; Talebian, F.; El-Omrani, H.Y.; Khattabi, M.; Yu, L.; Bai, X.F. Tumor expression of cd200 inhibits il-10 production by tumor-associated myeloid cells and prevents tumor immune evasion of ctl therapy. Eur. J. Immunol. 2010, 40, 2569–2579. [Google Scholar] [CrossRef]
- Jenmalm, M.C.; Cherwinski, H.; Bowman, E.P.; Phillips, J.H.; Sedgwick, J.D. Regulation of myeloid cell function through the cd200 receptor. J. Immunol. 2006, 176, 191–199. [Google Scholar] [CrossRef]
- Minas, K.; Liversidge, J. Is the cd200/cd200 receptor interaction more than just a myeloid cell inhibitory signal? Crit. Rev. Immunol. 2006, 26, 213–230. [Google Scholar] [CrossRef]
- Talebian, F.; Liu, J.Q.; Liu, Z.; Khattabi, M.; He, Y.; Ganju, R.; Bai, X.F. Melanoma cell expression of cd200 inhibits tumor formation and lung metastasis via inhibition of myeloid cell functions. PLoS ONE 2012, 7, e31442. [Google Scholar] [CrossRef]
- Zgodzinski, W.; Grywalska, E.; Surdacka, A.; Zinkiewicz, K.; Majewski, M.; Szczepanek, D.; Wallner, G.; Rolinski, J. Surface cd200 and cd200r antigens on lymphocytes in advanced gastric cancer: A new potential target for immunotherapy. Arch. Med. Sci. 2018, 14, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Erin, N.; Tanriover, G.; Curry, A.; Akman, M.; Duymus, O.; Gorczynski, R. Cd200fc enhances anti-tumoral immune response and inhibits visceral metastasis of breast carcinoma. Oncotarget 2018, 9, 19147–19158. [Google Scholar] [CrossRef] [PubMed]
- Erin, N.; Podnos, A.; Tanriover, G.; Duymus, O.; Cote, E.; Khatri, I.; Gorczynski, R.M. Bidirectional effect of cd200 on breast cancer development and metastasis, with ultimate outcome determined by tumor aggressiveness and a cancer-induced inflammatory response. Oncogene 2015, 34, 3860–3870. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Vallon-Eberhard, A.; Zigmond, E.; Farache, J.; Shezen, E.; Shakhar, G.; Ludwig, A.; Lira, S.A.; Jung, S. In vivo structure/function and expression analysis of the cx3c chemokine fractalkine. Blood 2011, 118, e156–e167. [Google Scholar] [CrossRef]
- Lee, M.; Lee, Y.; Song, J.; Lee, J.; Chang, S.Y. Tissue-specific role of cx3cr1 expressing immune cells and their relationships with human disease. Immune Netw. 2018, 18, e5. [Google Scholar] [CrossRef]
- Yan, Y.; Cao, S.; Liu, X.; Harrington, S.M.; Bindeman, W.E.; Adjei, A.A.; Jang, J.S.; Jen, J.; Li, Y.; Chanana, P.; et al. Cx3cr1 identifies pd-1 therapy-responsive cd8+ t cells that withstand chemotherapy during cancer chemoimmunotherapy. JCI Insight 2018, 3, e97828. [Google Scholar] [CrossRef]
- Dangaj, D.; Bruand, M.; Grimm, A.J.; Ronet, C.; Barras, D.; Duttagupta, P.A.; Lanitis, E.; Duraiswamy, J.; Tanyi, J.L.; Benencia, F.; et al. Cooperation between constitutive and inducible chemokines enables t cell engraftment and immune attack in solid tumors. Cancer Cell 2019, 35, 885–900 e810. [Google Scholar] [CrossRef]
- Zaidi, M.R.; Merlino, G. The two faces of interferon-gamma in cancer. Clin. Cancer Res. 2011, 17, 6118–6124. [Google Scholar] [CrossRef]
- Lahmidi, S.; Yousefi, M.; Dridi, S.; Duplay, P.; Pearson, A. Dok-1 and dok-2 are required to maintain herpes simplex virus 1-specific cd8(+) t cells in a murine model of ocular infection. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Mihrshahi, R.; Barclay, A.N.; Brown, M.H. Essential roles for dok2 and rasgap in cd200 receptor-mediated regulation of human myeloid cells. J. Immunol. 2009, 183, 4879–4886. [Google Scholar] [CrossRef]
- Lanier, L.L.; Bakker, A.B. The itam-bearing transmembrane adaptor dap12 in lymphoid and myeloid cell function. Immunol. Today 2000, 21, 611–614. [Google Scholar] [CrossRef]
- Kobayashi, M.; Konishi, H.; Sayo, A.; Takai, T.; Kiyama, H. Trem2/dap12 signal elicits proinflammatory response in microglia and exacerbates neuropathic pain. J. Neurosci. 2016, 36, 11138–11150. [Google Scholar] [CrossRef] [PubMed]
- Voss, O.H.; Tian, L.; Murakami, Y.; Coligan, J.E.; Krzewski, K. Emerging role of cd300 receptors in regulating myeloid cell efferocytosis. Mol. Cell Oncol. 2015, 2, e964625. [Google Scholar] [CrossRef] [PubMed]
- Korver, W.; Zhao, X.; Singh, S.; Pardoux, C.; Zhao, J.; Guzman, M.L.; Sen, S.; Yonkovich, S.; Liu, S.; Zhan, X.; et al. Monoclonal antibodies against irem-1: Potential for targeted therapy of aml. Leukemia 2009, 23, 1587–1597. [Google Scholar] [CrossRef]
- Erin, N.; Dilmac, S.; Curry, A.; Duymus, O.; Tanriover, G.; Prodeus, A.; Gariepy, J.; Gorczynski, R.M. Cd200 mimetic aptamer peg-m49 markedly increases the therapeutic effects of pegylated liposomal doxorubicin in a mouse model of metastatic breast carcinoma: An effect independent of cd200 receptor 1. Cancer Immunol. Immunother. 2020, 69, 103–114. [Google Scholar] [CrossRef]
- Mahadevan, D.; Lanasa, M.C.; Farber, C.; Pandey, M.; Whelden, M.; Faas, S.J.; Ulery, T.; Kukreja, A.; Li, L.; Bedrosian, C.L.; et al. Phase i study of samalizumab in chronic lymphocytic leukemia and multiple myeloma: Blockade of the immune checkpoint cd200. J. Immunother. Cancer 2019, 7, 227. [Google Scholar] [CrossRef]
- Lim, S.M.; Cho, S.H.; Hwang, I.G.; Choi, J.W.; Chang, H.; Ahn, M.J.; Park, K.U.; Kim, J.W.; Ko, Y.H.; Ahn, H.K.; et al. Investigating the feasibility of targeted next-generation sequencing to guide the treatment of head and neck squamous cell carcinoma. Cancer Res. Treat. 2019, 51, 300–312. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Wichmann, G.; Rosolowski, M.; Krohn, K.; Kreuz, M.; Boehm, A.; Reiche, A.; Scharrer, U.; Halama, D.; Bertolini, J.; Bauer, U.; et al. The role of hpv rna transcription, immune response-related gene expression and disruptive tp53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int. J. Cancer 2015, 137, 2846–2857. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. Geoquery: A bridge between the gene expression omnibus (geo) and bioconductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Lausen, B.; Hothorn, T.; Bretz, F.; Schumacher, M. Assessment of optimal selected prognostic factors. Biom. J. 2004, 46, 364–374. [Google Scholar] [CrossRef]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for cox’s proportional hazards model via coordinate descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Uno, H.; Cai, T.; Pencina, M.J.; D’Agostino, R.B.; Wei, L.J. On the c-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 2011, 30, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. Tcgabiolinks: An r/bioconductor package for integrative analysis of tcga data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Choi, K.; Ratner, N. Igeak: An interactive gene expression analysis kit for seamless workflow using the r/shiny platform. BMC Genomics 2019, 20, 177. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, A.; Cast, O.; Miller, M.L. Comprehensive benchmarking and integration of tumor microenvironment cell estimation methods. Cancer Res. 2019, 79, 6238–6246. [Google Scholar] [CrossRef]
| Patient Characteristics | No. | % | High CD200R1 Expression No./Subgroup (%) | |
|---|---|---|---|---|
| Total | 71 | 100 | 43/71 (60.5) | |
| Sex | Male | 58 | 81.6 | 35/58 (60.3) |
| Female | 13 | 18.4 | 8/13 (61.5) | |
| Age | <58 years | 31 | 43.6 | 19/31 (61.2) |
| ≥58 years | 40 | 56.3 | 24/40 (60.0) | |
| Smoking | Never | 20 | 28.1 | 10/20 (50.0) |
| Yes | 51 | 71.8 | 33/51 (64.7) | |
| Tumor location | Oral cavity | 26 | 36.6 | 15/26 (57.6) |
| Oropharynx | 17 | 23.9 | 11/17 (64.7) | |
| Hypopharynx | 10 | 14 | 7/10 (70.0) | |
| Larynx | 12 | 16.9 | 8/12 (66.7) | |
| Other | 6 | 8.5 | 2/6 (33.3) | |
| Stage | I | 11 | 15.5 | 8/11 (72.7) |
| II | 13 | 18.3 | 8/13 (61.5) | |
| III | 17 | 23.9 | 8/17 (47.0) | |
| IV | 30 | 42.3 | 19/30 (63.3) | |
| Human papilloma virus | Negative | 47 | 66.2 | 26/47 (55.3) |
| Positive | 12 | 16.9 | 10/12 (83.3) | |
| Unknown | 12 | 16.9 | 7/12 (58.3) | |
| Factors | Korean Cohort | GEO (GSE65858) | TCGA HNSCC | |||
|---|---|---|---|---|---|---|
| p Value | Hazard Ratio (95% CI) | p Value | Hazard Ratio (95% CI) | p Value | Hazard Ratio (95% CI) | |
| CD200R1 (high vs. low) | <0.01 * | 0.19 (0.06–0.58) | <0.05 * | 0.54 (0.29–0.99) | <0.05 * | 0.72 (0.53–0.98) |
| Smoking (Yes vs. never) | >0.1 | 1.67 (0.37–7.48) | >0.1 | 0.97 (0.55–1.70) | - | - |
| T stage (T3 + T4 vs. T1 + T2) | >0.1 | 1.12 (0.95–1.31) | 0.001 * | 2.81 (1.49–5.30) | <0.001 * | 1.74 (1.25–2.43) |
| N stage (N+ vs. N0) | >0.1 | 3.06 (0.53–17.7) | >0.1 | 1.14 (0.63–2.03) | <0.0001 * | 2.00 (1.43–2.78) |
| M stage (M1 vs. M0) | >0.1 | 6.27 (0.36–115.9) | >0.1 | 1.91 (0.76–4.75) | >0.1 | 1.01 (0.98–1.05) |
| Stage (III, IV vs. I, II) | >0.1 | 0.52 (0.09–3.01) | >0.1 | 0.93 (0.33–2.59) | - | - |
| Tumor grade (2, 3 vs. 0, 1) | >0.1 | 0.94 (0.81–1.09) | - | - | - | - |
| Sex (F vs. M) | >0.1 | 2.89 (0.84–9.90) | >0.1 | 1.00 (0.58–1.73) | >0.1 | 0.81 (0.58–1.12) |
| Age | >0.1 | 1.00 (0.95–1.06) | >0.1 | 1.18 (0.76–1.84) | >0.1 | 1.33 (0.97–1.80) |
| Factors | Korean Cohort | GEO Data | TCGA HNSCC Data | ||||||
|---|---|---|---|---|---|---|---|---|---|
| C-index | 95% CI | AUC | C-index | 95% CI | AUC | C-index | 95% CI | AUC | |
| CD200R1 | 0.59 | 0.41–0.77 | 0.63 | 0.58 | 0.53–0.63 | 0.58 | 0.53 | 0.49–0.57 | 0.54 |
| T stage | 0.55 | 0.42–0.69 | 0.50 | 0.58 | 0.51–0.65 | 0.58 | 0.55 | 0.52–0.59 | 0.58 |
| N stage | 0.55 | 0.41–0.70 | 0.59 | 0.53 | 0.45–0.60 | 0.55 | 0.56 | 0.52–0.59 | 0.58 |
| Age | 0.52 | 0.39–0.66 | 0.50 | 0.64 | 0.43–0.81 | 0.62 | 0.52 | 0.47–0.56 | 0.53 |
| Sex | 0.54 | 0.44–0.64 | 0.59 | 0.58 | 0.42–0.74 | 0.58 | 0.59 | 0.49–0.57 | 0.54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.; Lee, Y.-G.; Ko, Y.H.; Cho, J.H.; Choi, J.-K.; Park, K.U.; Kang, E.J.; Lee, K.-W.; Lim, S.M.; Kim, J.-S.; et al. Prognostic Value of CD200R1 mRNA Expression in Head and Neck Squamous Cell Carcinoma. Cancers 2020, 12, 1777. https://doi.org/10.3390/cancers12071777
Chang H, Lee Y-G, Ko YH, Cho JH, Choi J-K, Park KU, Kang EJ, Lee K-W, Lim SM, Kim J-S, et al. Prognostic Value of CD200R1 mRNA Expression in Head and Neck Squamous Cell Carcinoma. Cancers. 2020; 12(7):1777. https://doi.org/10.3390/cancers12071777
Chicago/Turabian StyleChang, Hyun, Yun-Gyoo Lee, Yoon Ho Ko, Jang Ho Cho, Jong-Kwon Choi, Keon Uk Park, Eun Joo Kang, Keun-Wook Lee, Sun Min Lim, Jin-Soo Kim, and et al. 2020. "Prognostic Value of CD200R1 mRNA Expression in Head and Neck Squamous Cell Carcinoma" Cancers 12, no. 7: 1777. https://doi.org/10.3390/cancers12071777
APA StyleChang, H., Lee, Y.-G., Ko, Y. H., Cho, J. H., Choi, J.-K., Park, K. U., Kang, E. J., Lee, K.-W., Lim, S. M., Kim, J.-S., Lee, H. W., Kim, M. K., Hwang, I. G., Kim, S., Nam, B.-H., & Kim, H. R. (2020). Prognostic Value of CD200R1 mRNA Expression in Head and Neck Squamous Cell Carcinoma. Cancers, 12(7), 1777. https://doi.org/10.3390/cancers12071777





