Functional Effects of let-7g Expression in Colon Cancer Metastasis
Abstract
:1. Introduction
2. Results
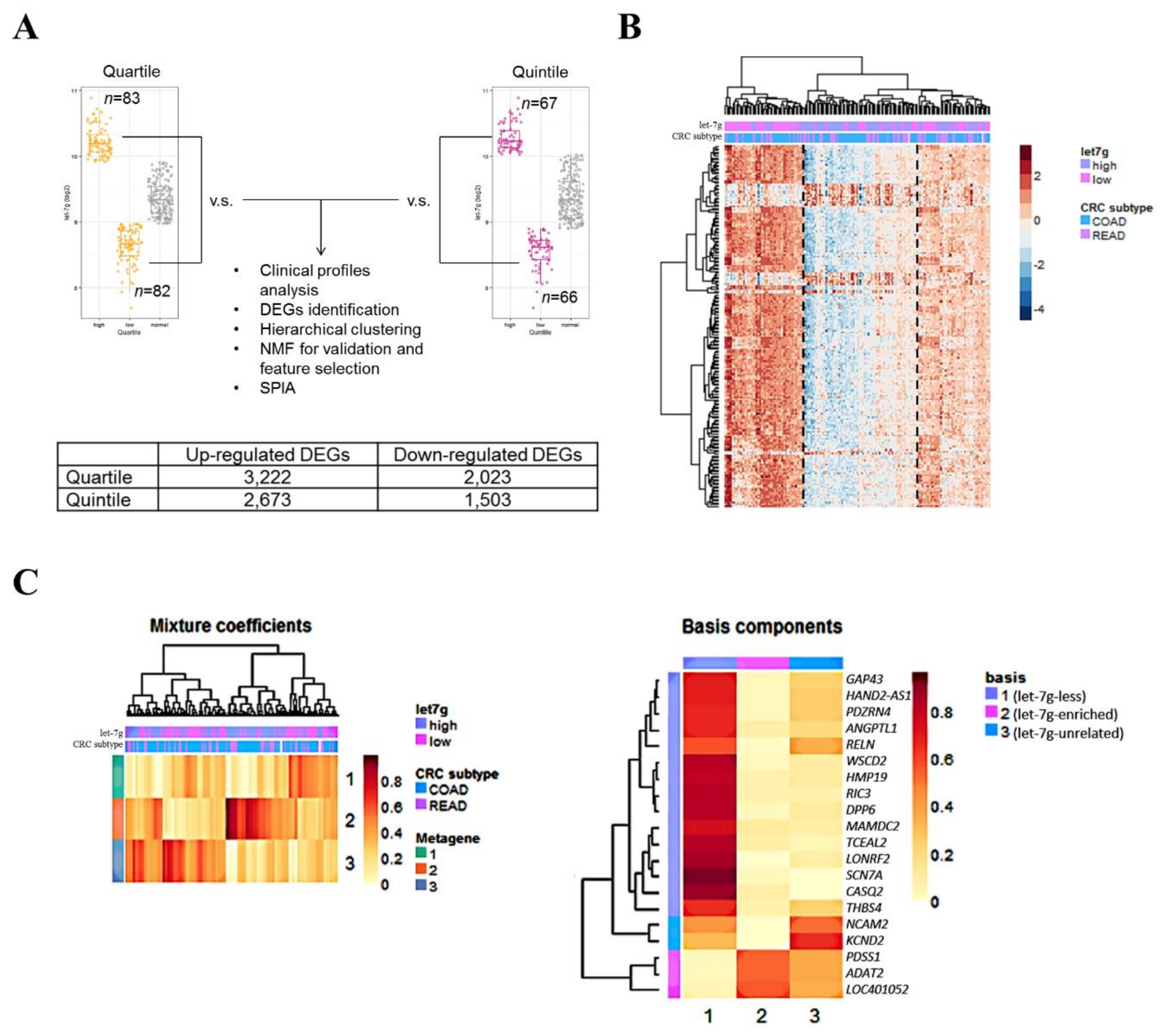
2.1. Correlation between let-7g Expression and Clinical Profiles in CRC Patients
2.2. Let-7g-Associated Transcriptomic Profiles Reveal Three CRC Subpopulations
2.3. Targets of let-7g were Significantly Enriched in let-7g-Associated Profiles
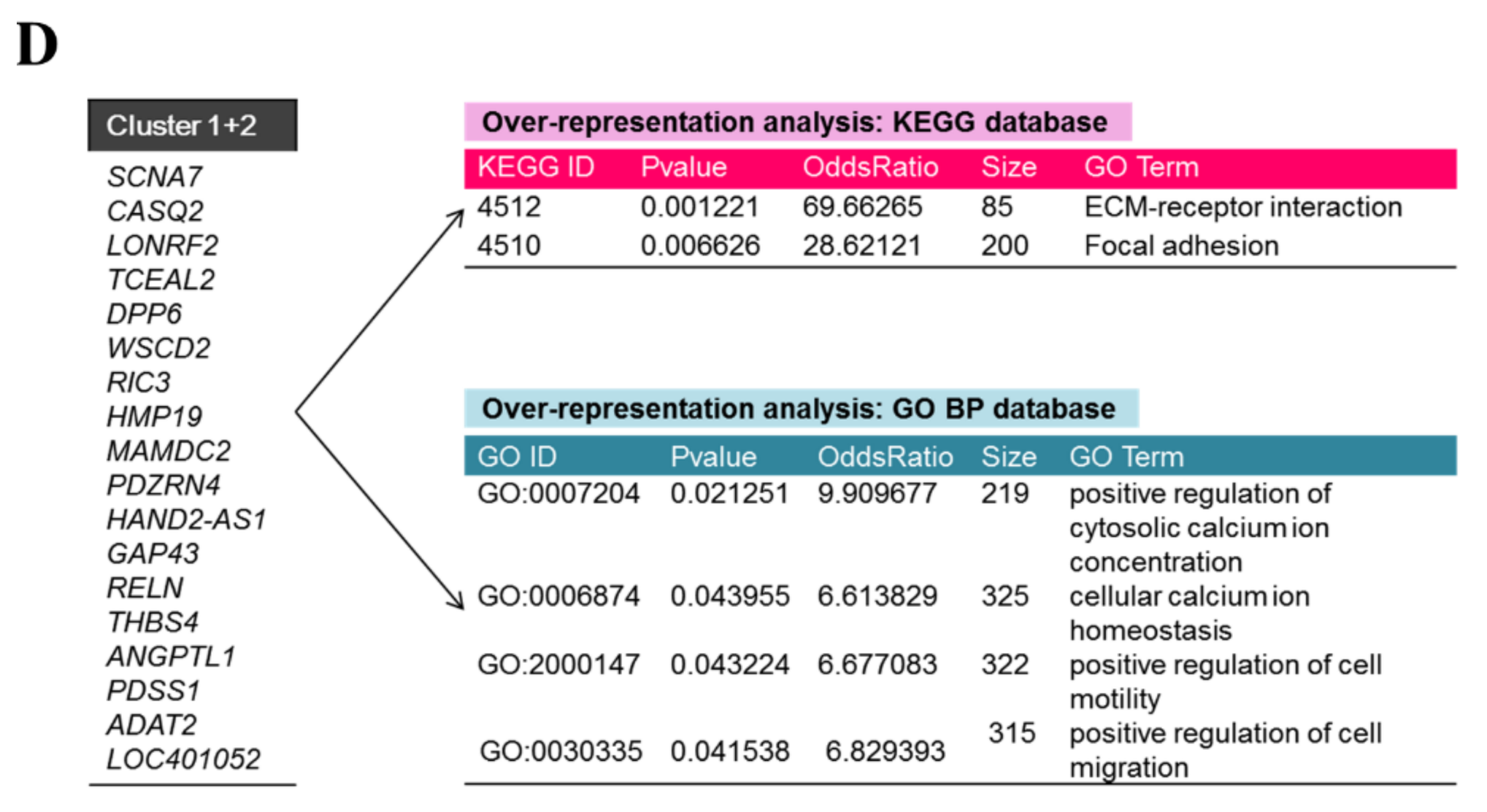
2.4. Prospecting let-7g-Associated Biological Features in CRC Patients
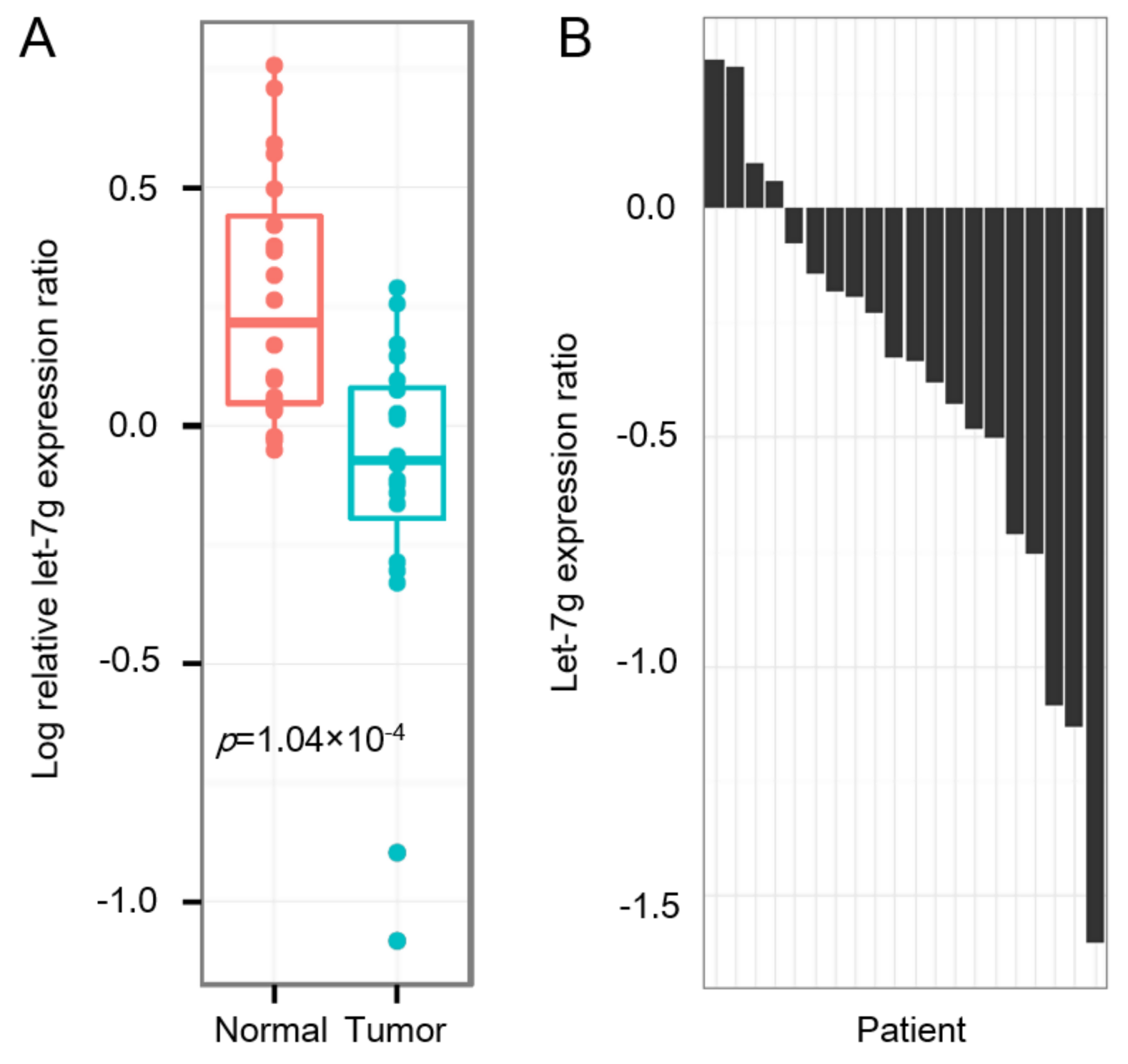
2.5. Reduced let-7g in CRC Specimens
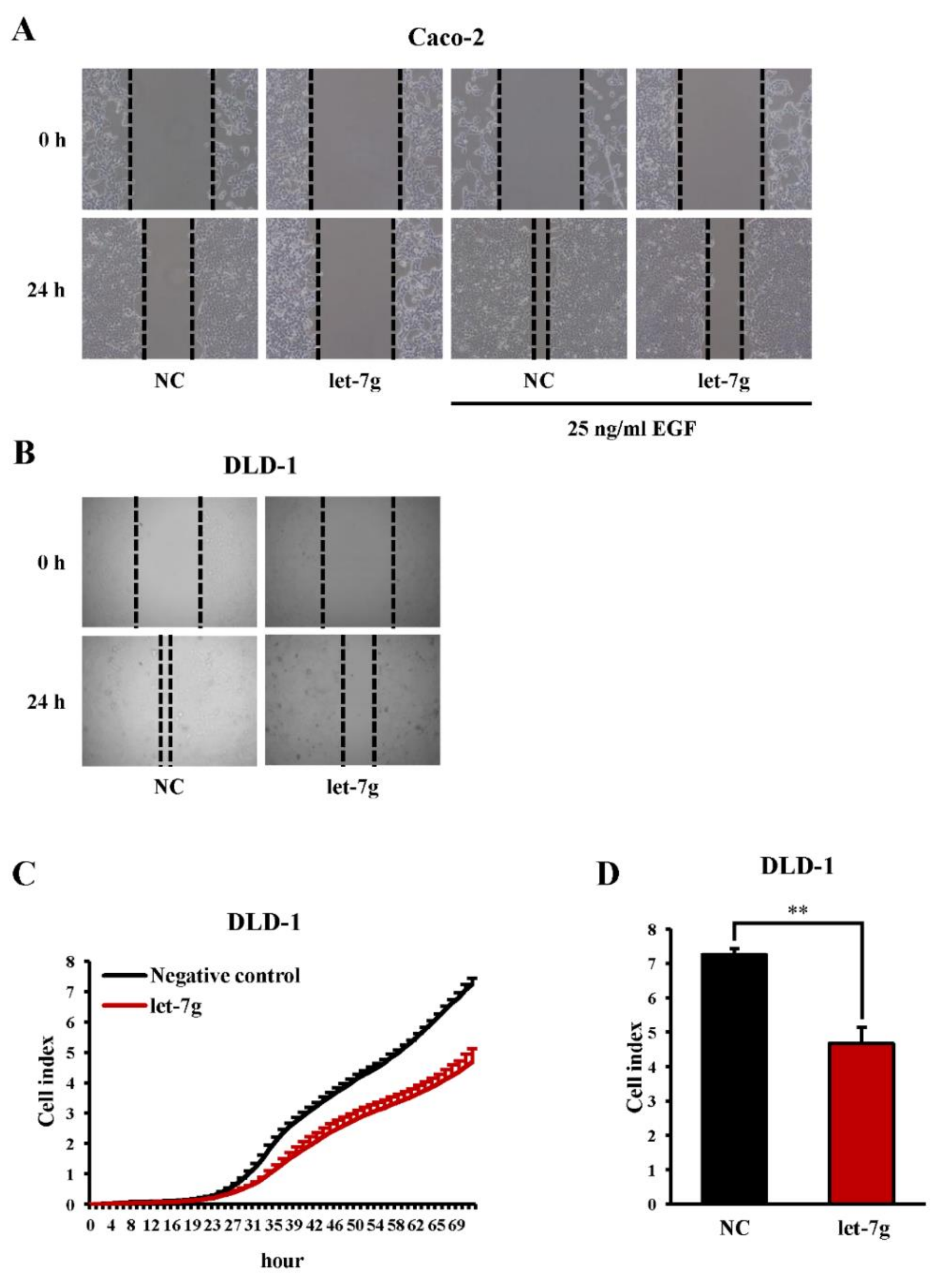
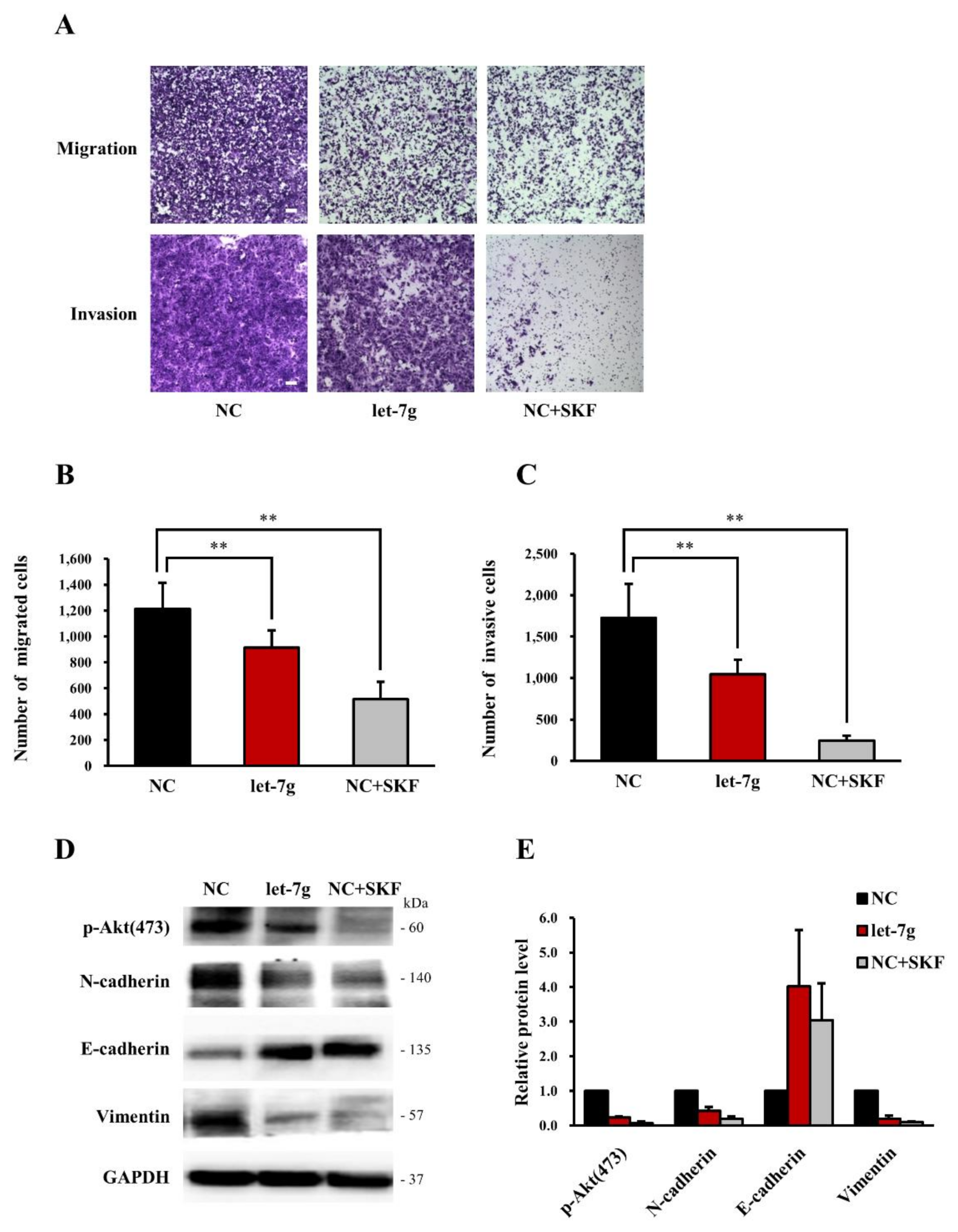
2.6. Let-7g Inhibits CRC Cell Migration in Vitro
2.7. Both let-7g and the Store-Operated Calcium (SOC) Channel Blocker SKF96365 Inhibit CRC Cell Motility in Vitro
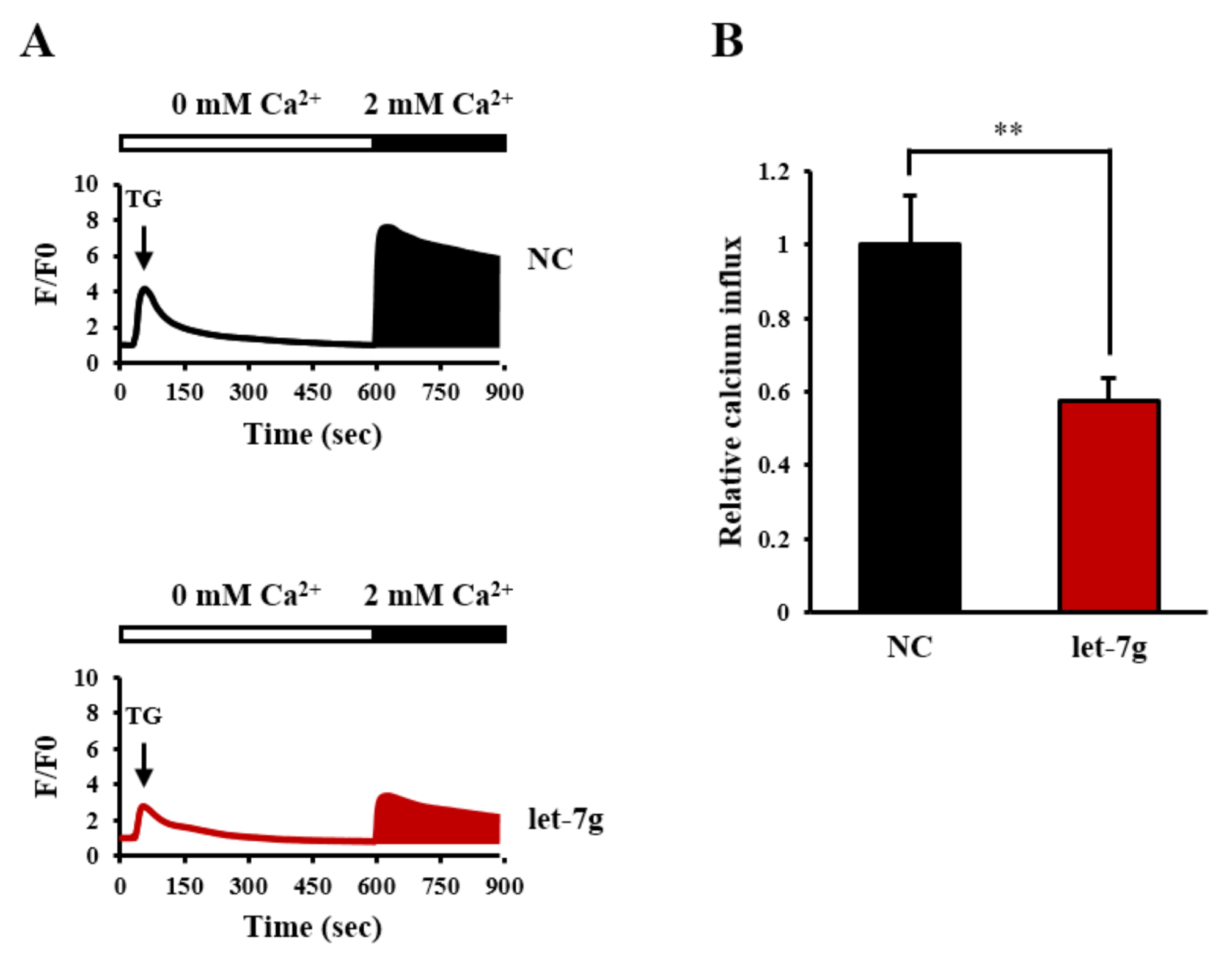
2.8. Let-7g Attenuates SOC Influx in Vitro
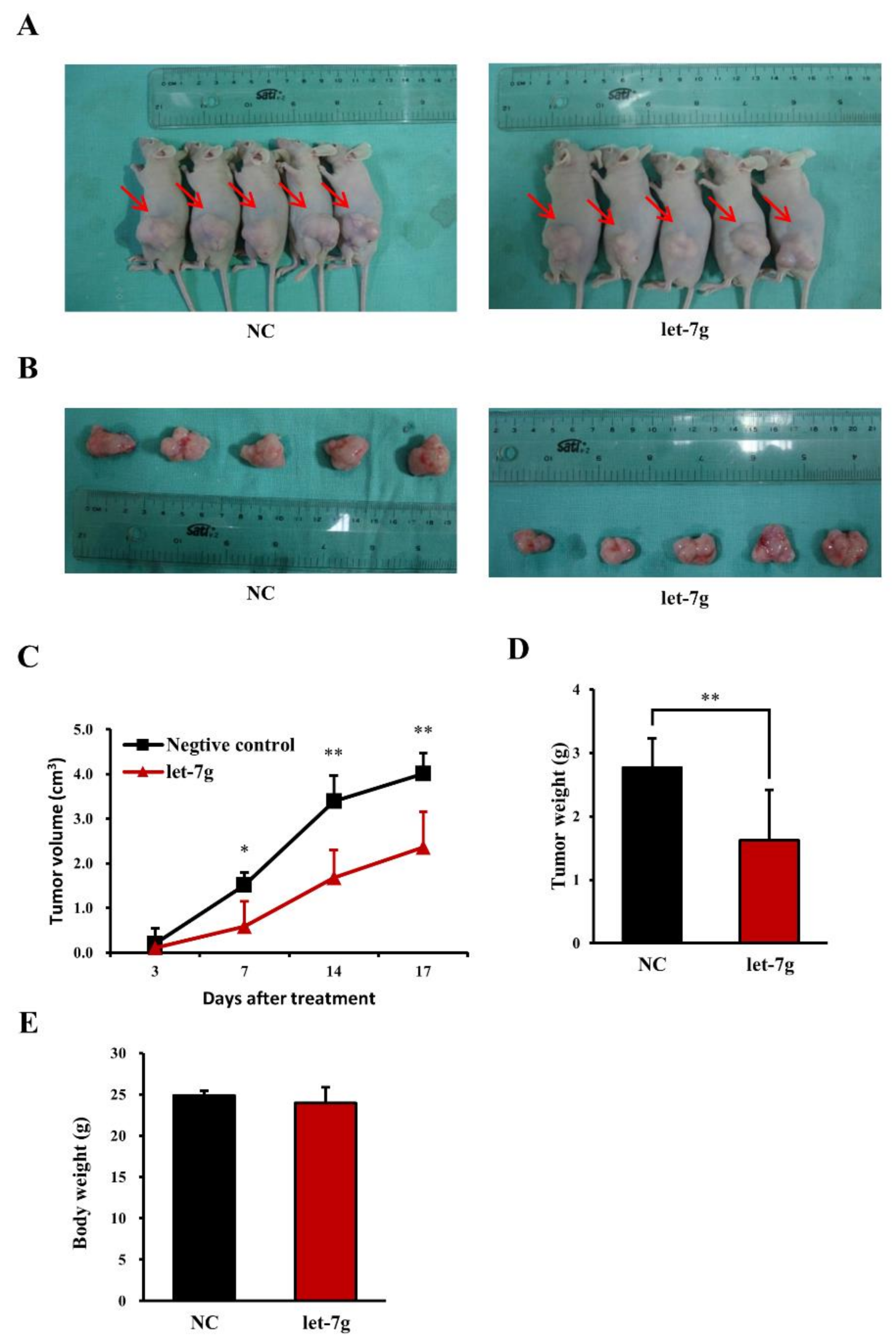
2.9. Let-7g Inhibits Colorectal Tumor Growth in Vivo
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Patient Categorization of the TCGA CRC Cohort
4.2. Analysis of Clinical Demographics in TCGA CRC Samples
4.3. Analysis of Transcript Expression Profiles Based on Sample Categorization
4.4. DIANA-Annotated Targets Gene Set Enrichment Analysis
4.5. Gene Ontology (GO) Enrichment and Pathway Analysis of DEGs in CRC Samples
4.6. Patient Recruitment and Sample Preparation
4.7. Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
4.8. Cell Culture and Transfection
4.9. Cell Migration by a Wound-Healing Assay
4.10. Cell Migration by a Biosensor Approach
4.11. Transwell Cell Migration and Invasion Assays
4.12. Western Blotting
4.13. Calcium Imaging
4.14. Animal Study
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [Green Version]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Liu, B.; Qu, S.; Liang, G.; Luo, W.; Gong, C. MicroRNAs and cancer: Key paradigms in molecular therapy. Oncol. Lett. 2018, 15, 2735–2742. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell. Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef] [Green Version]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Boyerinas, B.; Park, S.M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, G.; Hayashi, K.; Xi, Y.; Kudo, K.; Uchida, K.; Takasaki, K.; Yamamoto, M.; Ju, J. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genom. Proteom. 2006, 3, 317–324. [Google Scholar]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Zhao, L.; Budhu, A.; Forgues, M.; Jia, H.L.; Qin, L.X.; Ye, Q.H.; Yu, J.; Shi, X.; Tang, Z.Y.; et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J. Hepatol. 2010, 52, 690–697. [Google Scholar] [CrossRef]
- Qian, P.; Zuo, Z.; Wu, Z.; Meng, X.; Li, G.; Wu, Z.; Zhang, W.; Tan, S.; Pandey, V.; Yao, Y.; et al. Pivotal role of reduced let-7g expression in breast cancer invasion and metastasis. Cancer Res. 2011, 71, 6463–6474. [Google Scholar] [CrossRef]
- Sathipati, S.Y.; Ho, S.Y. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci. Rep. 2017, 7, 7507. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, H.; Takahashi, H.; Niinuma, T.; Kitajima, H.; Oikawa, R.; Matsumoto, N.; Takeba, Y.; Otsubo, T.; Takagi, M.; Ariizumi, Y.; et al. Dysregulation of miRNA in chronic hepatitis B is associated with hepatocellular carcinoma risk after nucleos(t)ide analogue treatment. Cancer Lett. 2018, 434, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.F.; Wang, H.; Chen, Y.C.; Chan, C.Y.; Ng, S.S.; Li, K.; Xie, D.; He, M.L.; Lin, M.C.; Kung, H.F. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int. J. Cancer 2011, 128, 319–331. [Google Scholar] [CrossRef]
- Hu, X.; Guo, J.; Zheng, L.; Li, C.; Zheng, T.M.; Tanyi, J.L.; Liang, S.; Benedetto, C.; Mitidieri, M.; Katsaros, D.; et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol. Cancer Res. 2013, 11, 240–250. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Kohne, C.H.; Hitre, E.; Zaluski, J.; Chang Chien, C.R.; Makhson, A.; D’Haens, G.; Pinter, T.; Lim, R.; Bodoky, G.; et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 2009, 360, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef] [PubMed]
- Martini, G.; Troiani, T.; Cardone, C.; Vitiello, P.; Sforza, V.; Ciardiello, D.; Napolitano, S.; Della Corte, C.M.; Morgillo, F.; Raucci, A.; et al. Present and future of metastatic colorectal cancer treatment: A review of new candidate targets. World J. Gastroenterol. 2017, 23, 4675–4688. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.K.; Zhao, M.; Yang, M.; Zhong, J.L.; Gu, Y.Y.; Peng, H.; Che, Y.Q.; Huang, C.Z. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS ONE 2014, 9, e87451. [Google Scholar] [CrossRef]
- Wang, J.Y.; Sun, J.; Huang, M.Y.; Wang, Y.S.; Hou, M.F.; Sun, Y.; He, H.; Krishna, N.; Chiu, S.J.; Lin, S.; et al. STIM1 overexpression promotes colorectal cancer progression, cell motility and COX-2 expression. Oncogene 2015, 34, 4358–4367. [Google Scholar] [CrossRef]
- Brabletz, T.; Jung, A.; Reu, S.; Porzner, M.; Hlubek, F.; Kunz-Schughart, L.A.; Knuechel, R.; Kirchner, T. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA 2001, 98, 10356–10361. [Google Scholar] [CrossRef]
- Brabletz, T.; Hlubek, F.; Spaderna, S.; Schmalhofer, O.; Hiendlmeyer, E.; Jung, A.; Kirchner, T. Invasion and metastasis in colorectal cancer: Epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs 2005, 179, 56–65. [Google Scholar] [CrossRef]
- Park, S.; Minai-Tehrani, A.; Xu, C.X.; Chang, S.H.; Woo, M.A.; Noh, M.S.; Lee, E.S.; Lim, H.T.; An, G.H.; Lee, K.H.; et al. Suppression of A549 lung cancer cell migration by precursor let-7g microRNA. Mol. Med. Rep. 2010, 3, 1007–1013. [Google Scholar] [CrossRef]
- Kang, W.; Tong, J.H.; Lung, R.W.; Dong, Y.; Yang, W.; Pan, Y.; Lau, K.M.; Yu, J.; Cheng, A.S.; To, K.F. let-7b/g silencing activates AKT signaling to promote gastric carcinogenesis. J. Transl. Med. 2014, 12, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksen, A.H.; Andersen, R.F.; Pallisgaard, N.; Sorensen, F.B.; Jakobsen, A.; Hansen, T.F. MicroRNA Expression Profiling to Identify and Validate Reference Genes for the Relative Quantification of microRNA in Rectal Cancer. PLoS ONE 2016, 11, e0150593. [Google Scholar] [CrossRef]
- Huang, W.C.; Chai, C.Y.; Chen, W.C.; Hou, M.F.; Wang, Y.S.; Chiu, Y.C.; Lu, S.R.; Chang, W.C.; Juo, S.H.; Wang, J.Y.; et al. Histamine regulates cyclooxygenase 2 gene activation through Orai1-mediated NFkappaB activation in lung cancer cells. Cell Calcium 2011, 50, 27–35. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chen, B.K.; Wang, Y.S.; Tsai, Y.T.; Chen, W.C.; Chang, W.C.; Hou, M.F.; Wu, Y.C.; Chang, W.C. Involvement of store-operated calcium signaling in EGF-mediated COX-2 gene activation in cancer cells. Cell. Signal. 2012, 24, 162–169. [Google Scholar] [CrossRef]
- Mo, P.; Yang, S. The store-operated calcium channels in cancer metastasis: From cell migration, invasion to metastatic colonization. Front. Biosci. 2018, 23, 1241–1256. [Google Scholar]
- Chen, K.J.; Hou, Y.; Wang, K.; Li, J.; Xia, Y.; Yang, X.Y.; Lv, G.; Xing, X.L.; Shen, F. Reexpression of Let-7g microRNA inhibits the proliferation and migration via K-Ras/HMGA2/snail axis in hepatocellular carcinoma. BioMed Res. Int. 2014, 2014, 742417. [Google Scholar] [CrossRef]
- Zhuang, R.; Rao, J.N.; Zou, T.; Liu, L.; Xiao, L.; Cao, S.; Hansraj, N.Z.; Gorospe, M.; Wang, J.Y. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013, 41, 7905–7919. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Liu, X.; Feng, B.; Liu, N.; Wu, Q.; Han, Y.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. STIM1, a direct target of microRNA-185, promotes tumor metastasis and is associated with poor prognosis in colorectal cancer. Oncogene 2015, 34, 4808–4820. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Hebert, C.; Norris, K.; Scheper, M.A.; Nikitakis, N.; Sauk, J.J. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer 2007, 6, 5. [Google Scholar] [CrossRef]
- Shimizu, S.; Takehara, T.; Hikita, H.; Kodama, T.; Miyagi, T.; Hosui, A.; Tatsumi, T.; Ishida, H.; Noda, T.; Nagano, H.; et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J. Hepatol. 2010, 52, 698–704. [Google Scholar] [CrossRef]
- Zhu, Y.; Qiu, P.; Ji, Y. TCGA-assembler: Open-source software for retrieving and processing TCGA data. Nat. Methods 2014, 11, 599–600. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Gaujoux, R.; Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 2010, 11, 367. [Google Scholar] [CrossRef]
- Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, H. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics 2007, 23, 1495–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007, 23, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
| Characteristics | Total Cases | NMF Clusters | p-Value | Covariates Adjusted p a | FDR Adjusted p b | ||
|---|---|---|---|---|---|---|---|
| n | Cluster 1 n (%) | Cluster 2 n (%) | Cluster 3 n (%) | ||||
| Depth of Invasion | |||||||
| T1 + T2 | 28 | 2 (7.1) | 16 (57.1) | 10 (35.8) | 0.015 * | 0.019 * | 0.038 * |
| T3 + T4 | 136 | 38 (27.9) | 45 (33.1) | 53 (39.0) | |||
| Lymphatic Invasion | |||||||
| No | 92 | 17 (18.5) | 37 (40.2) | 38 (41.3) | 0.026 * | 0.044 * | 0.044 * |
| Yes | 49 | 17 (34.7) | 13 (26.5) | 19 (38.8) | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-M.; Wong, H.S.-C.; Huang, C.-Y.; Hsu, W.-L.; Maio, Z.-F.; Chiu, S.-J.; Tsai, Y.-T.; Chen, B.-K.; Wan, Y.-J.Y.; Wang, J.-Y.; et al. Functional Effects of let-7g Expression in Colon Cancer Metastasis. Cancers 2019, 11, 489. https://doi.org/10.3390/cancers11040489
Chang C-M, Wong HS-C, Huang C-Y, Hsu W-L, Maio Z-F, Chiu S-J, Tsai Y-T, Chen B-K, Wan Y-JY, Wang J-Y, et al. Functional Effects of let-7g Expression in Colon Cancer Metastasis. Cancers. 2019; 11(4):489. https://doi.org/10.3390/cancers11040489
Chicago/Turabian StyleChang, Che-Mai, Henry Sung-Ching Wong, Chien-Yu Huang, Wen-Li Hsu, Zhi-Feng Maio, Siou-Jin Chiu, Yao-Ting Tsai, Ben-Kuen Chen, Yu-Jui Yvonne Wan, Jaw-Yuan Wang, and et al. 2019. "Functional Effects of let-7g Expression in Colon Cancer Metastasis" Cancers 11, no. 4: 489. https://doi.org/10.3390/cancers11040489
APA StyleChang, C.-M., Wong, H. S.-C., Huang, C.-Y., Hsu, W.-L., Maio, Z.-F., Chiu, S.-J., Tsai, Y.-T., Chen, B.-K., Wan, Y.-J. Y., Wang, J.-Y., & Chang, W.-C. (2019). Functional Effects of let-7g Expression in Colon Cancer Metastasis. Cancers, 11(4), 489. https://doi.org/10.3390/cancers11040489









