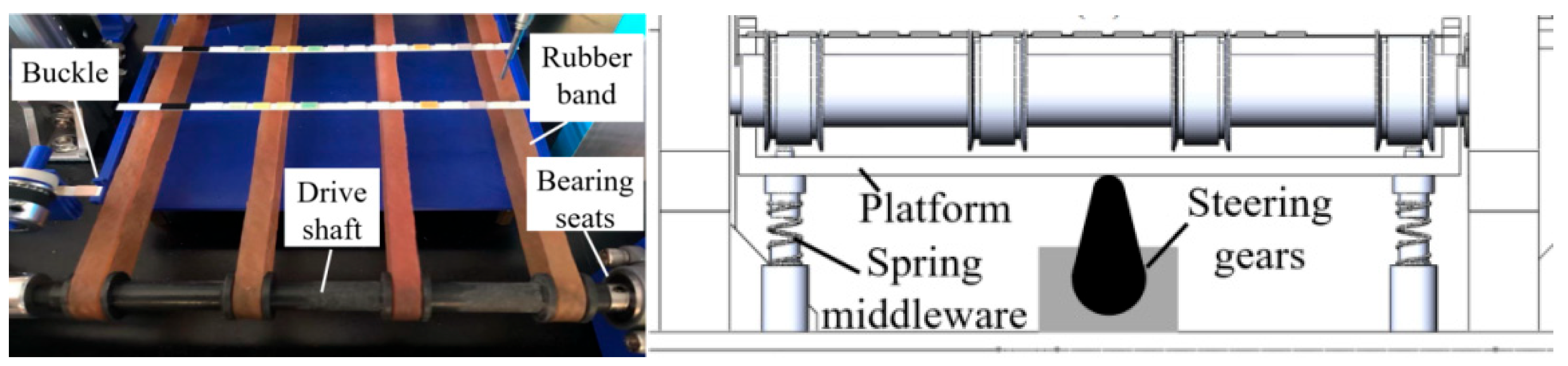
2.1. Principle of Color Recognition
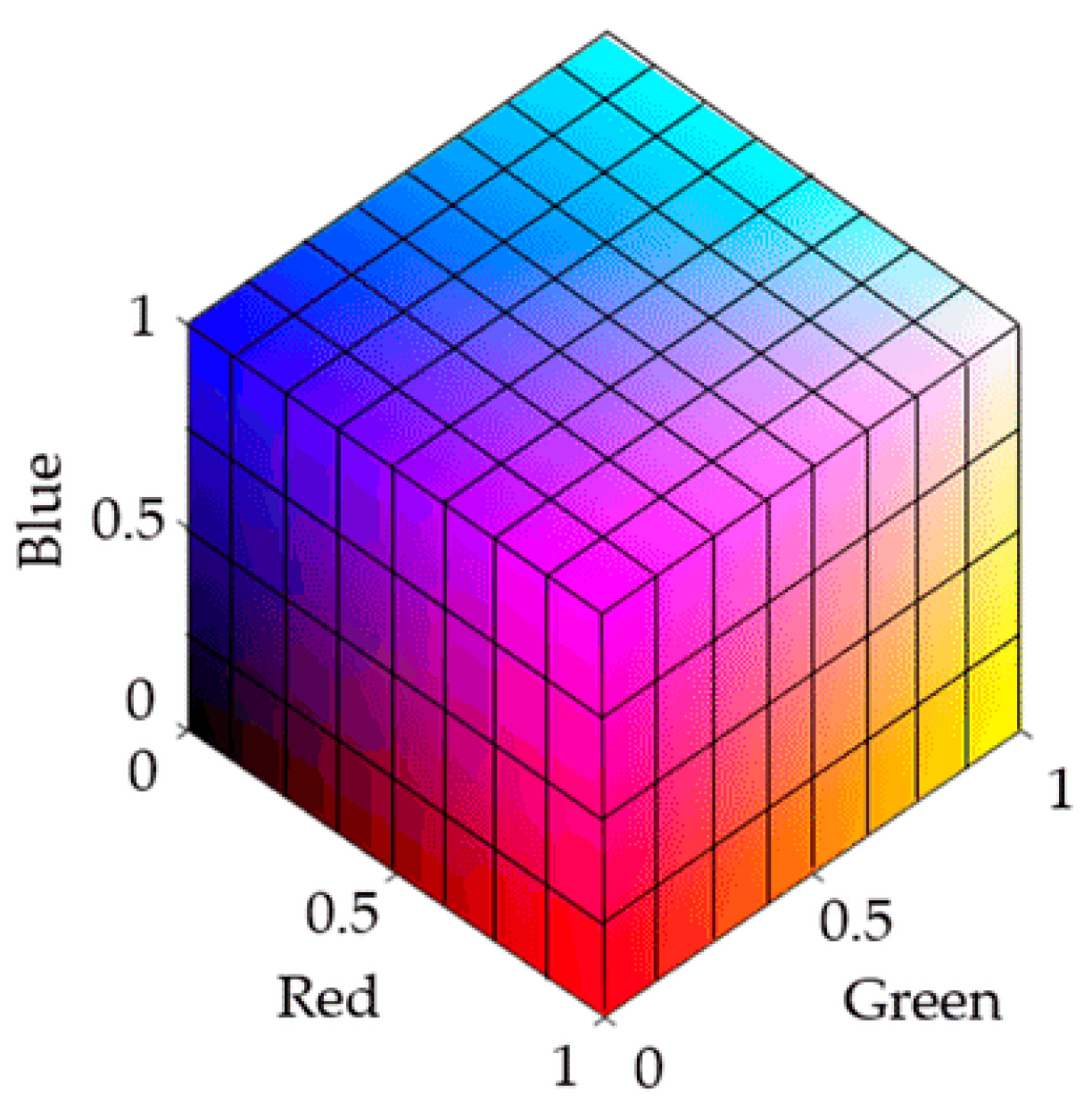
During the test strip detection, color changes are generated by chemical reactions. Different color space models have been used to recognize and analyze these color changes. Common color spaces include the RGB (red, green, blue) and CIELab spaces. RGB is a device-related color model, and the data acquisition system used in this study was a typical RGB input device. As an addition-based color model, RGB represents various colors by combining the intensity values of three primary colors (R, G, B). The intensity of each primary color is usually expressed as an integer value from 0 to 255. Colors range from completely black (0, 0, 0) to completely white (255, 255, 255), between which different intensities of red, green and blue colors are mixed to form other colors. For the RGB color model displayed in
Figure 5, the color range becomes from black (0, 0, 0) to white (1, 1, 1) after normalization [
7].
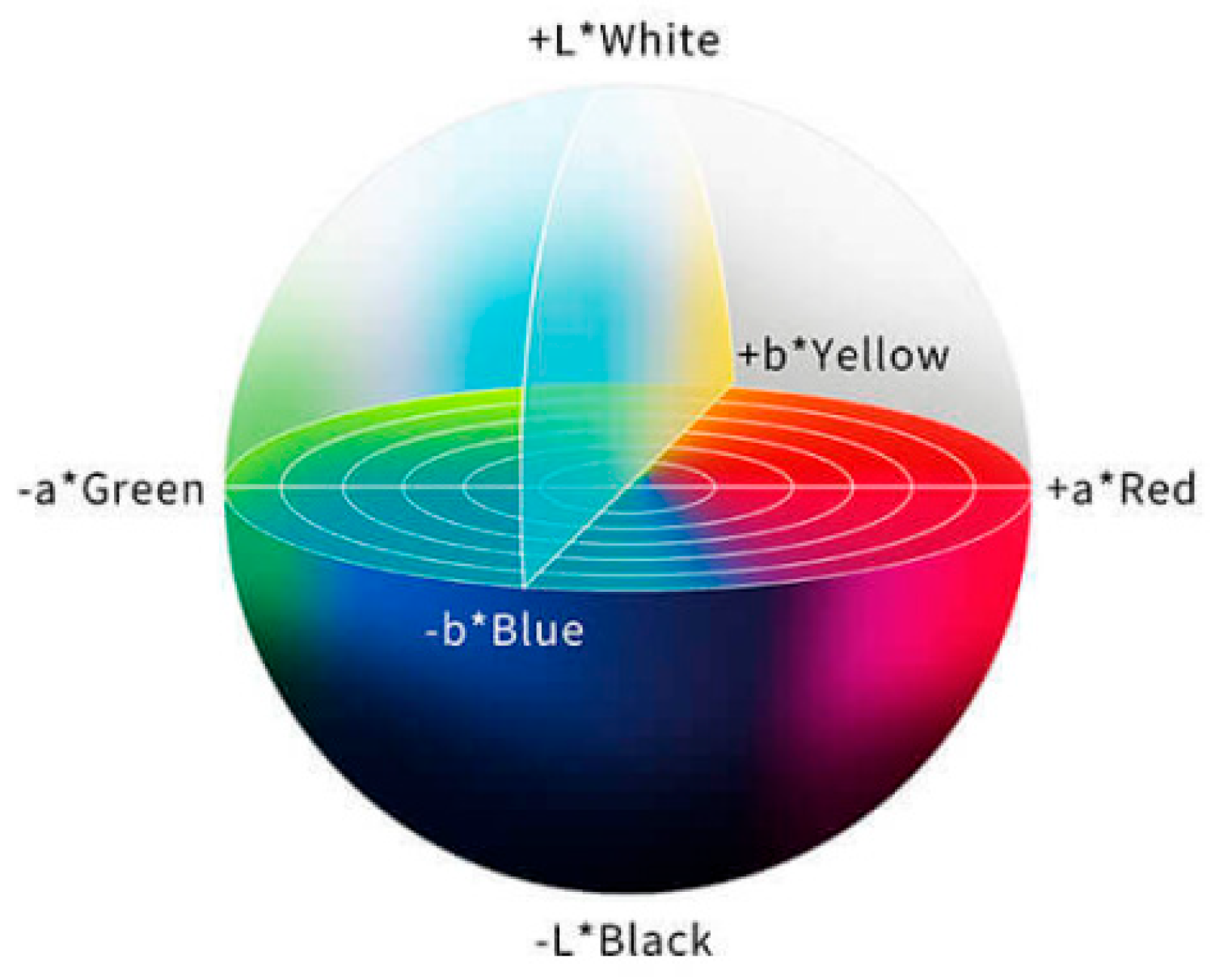
The RGB color space may not accurately represent all colors perceived by the human eye, while the CIELab color space is more in line with the understanding of the human visual system, which makes it easier to operate and facilitates color analysis. As a human visual perception-based color space developed by the International Commission on Illumination, CIELab divides colors into three parts, as shown in
Figure 6: L* (brightness) stands for the brightness of colors, ranging from 0 (black) to 100 (white); a* (green–red) represents the color distribution from green to red, with negative values indicating green and positive values indicating red; and b* (blue–yellow) represents the color distribution from blue to yellow, with negative values indicating blue and positive values indicating yellow. In the CIELAB color space, the asterisk (*) serves as a specific identifier for the parameters of this color model. CIELab is particularly suitable for detecting small changes in color. For example, when the color of test strip changes from light to dark pink, CIELab can accurately reflect the brightness and tonal difference of such change, thereby facilitating more accurate analysis [
8].
The formula for converting a color from RGB to CIELab color spaces is as follows:
Initially, the [R, G, B] value needs to be normalized within the range of [0, 1].
Then, gamma correction is applied to convert this value from nonlinear to linear. For every color channel, the following formula is used:
The same formula applies to and .
The RGB-to-XYZ conversion matrix is employed as:
Subsequently, the XYZ color coordinates are transformed into the CIELab color space.
The normalized X, Y, Z are calculated as:
where X
n, Y
n and Z
n denote the tristimulus values of standard illuminant. In this experiment, X
n = 0.95047, Y
n = 1.0, and Z
n = 1.08883.
xyz is transformed into the CIELab color space using the following function
f(t):
The CIELab L*, a* and b* coordinates are calculated as:
With these formulas, the RGB values can be converted into CIELab coordinates.
2.2. Principle of Concentration Calculation
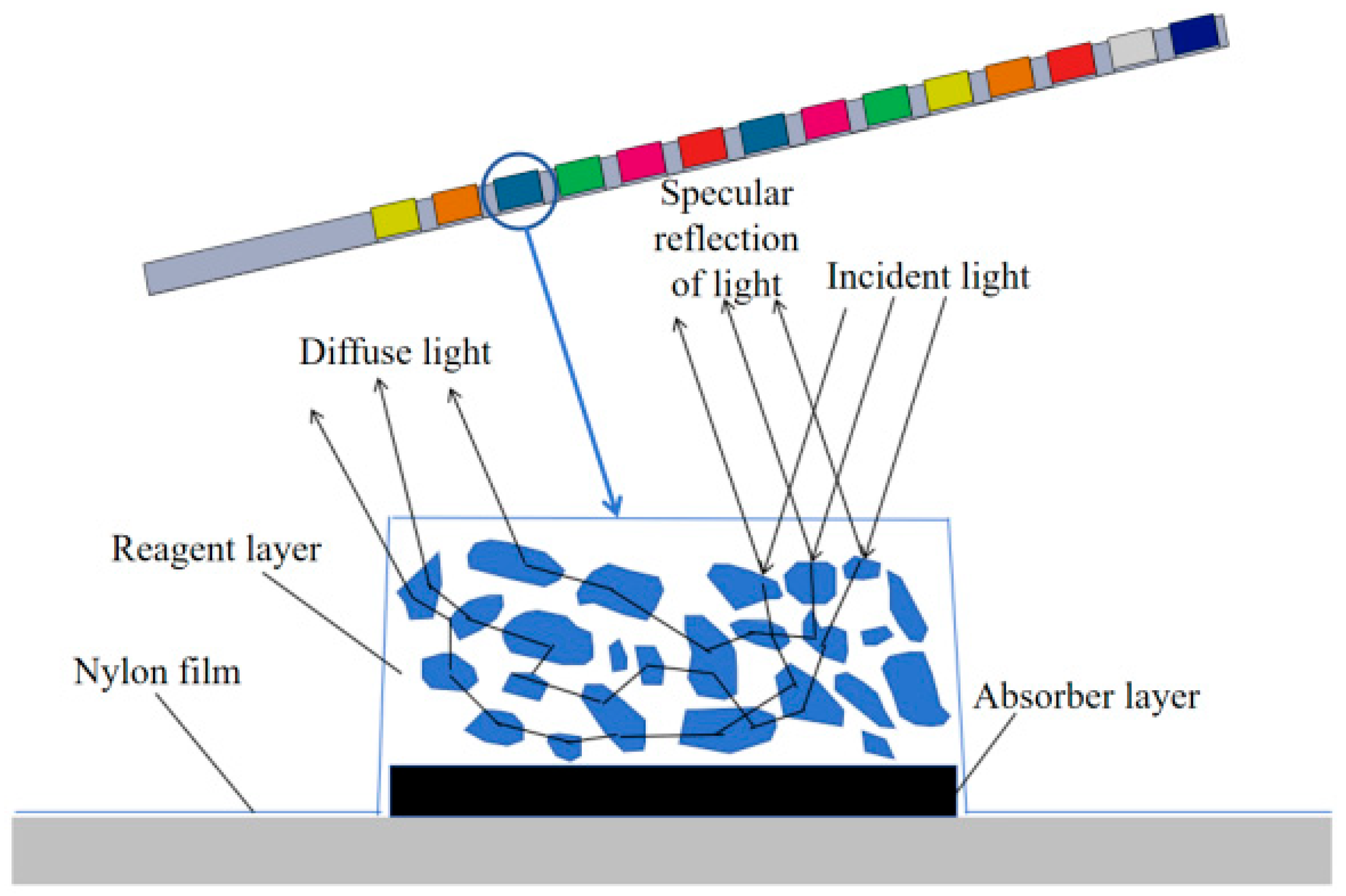
Figure 7 illustrates the structure of reagent blocks. The block surface is covered with a nylon film, which can effectively block the macromolecular entry of the reagent layer, thus protecting the reagents from contamination. At the bottom of the reagent blocks, an absorber layer is designed, whose function is to absorb excess urine and prevent incident light from penetrating the reagent layer. Upon contact of the reagent layer with urine, substantial diffuse reflectors would be formed. It can be observed from
Figure 7 that specular reflection is produced on the surfaces of the detection blocks, while part of the light enters the diffuse reflectors and eventually forms a diffuse reflection after a series of optical processes such as reflection, refraction and diffraction. When the detection zone of the reagent blocks is sufficiently thick, the influence of transmitted light is negligible. By collecting and analyzing the reflected light, concentration-related detection information can be extracted.
According to the Kubelka–Munk theory, the reflectivity of incident light is specifically correlated with the optical absorption coefficient, the scattering coefficient and the degree of diffuse reflected light absorption in the test strip reaction zone. Such correlation can be formulated as:
In the above formula,
R signifies the reflectivity; R
d represents the diffuse reflectance when the test sample thickness is greater than the transmission depth;
K denotes the absorption coefficient of the reagent blocks; and
S represents the scattering coefficient. Through simultaneous Formulas (6) and (7), we can obtain
The scattering coefficient depends mainly on the object material properties. Thus, when the thickness of the reaction zone and the scattering coefficient remain constant, the reflectivity is only correlated with the absorption coefficient. Since the absorption coefficient
K and the substance concentration
C follow the Beer–Lambert law,
where
ε represents the molar absorptivity. The absorption coefficient
K is linearly proportional to the concentration
C of the sample being tested. By measuring the reflectance R of the reagent block, quantitative analysis of the target substance concentration in the urine can be achieved.
In the above relation, ε denotes the molar absorption coefficient. It is thus clear that the absorption coefficient K is directly proportional to the test sample concentration C. Hence, as long as the reflectivity of detection reagent blocks is determined, the urine concentrations of corresponding substances can be calculated.
Combining the Kubelka–Munk theory with the Beer–Lambert law, we can derive that reflectivity is directly proportional to concentration. In the ideal state, if a surface is completely diffuse (i.e., the surface reflects all incident light evenly in all directions), the color value of the surface can be regarded as a direct reflection of reflectivity. However, in practical applications, since cameras and sensors are affected by various interfering factors such as lighting conditions and object surface glossiness, the color values cannot directly reflect the reflectivity. Therefore, a direct relationship between color values and concentrations needs to be established through mathematical modeling.
2.3. Image Processing
A urine test strip image was collected from the image acquisition system, partial functions from the OpenCV library were scheduled in the PyCharm2025.1.1.1 integrated development environment and corresponding program code was written for image processing.
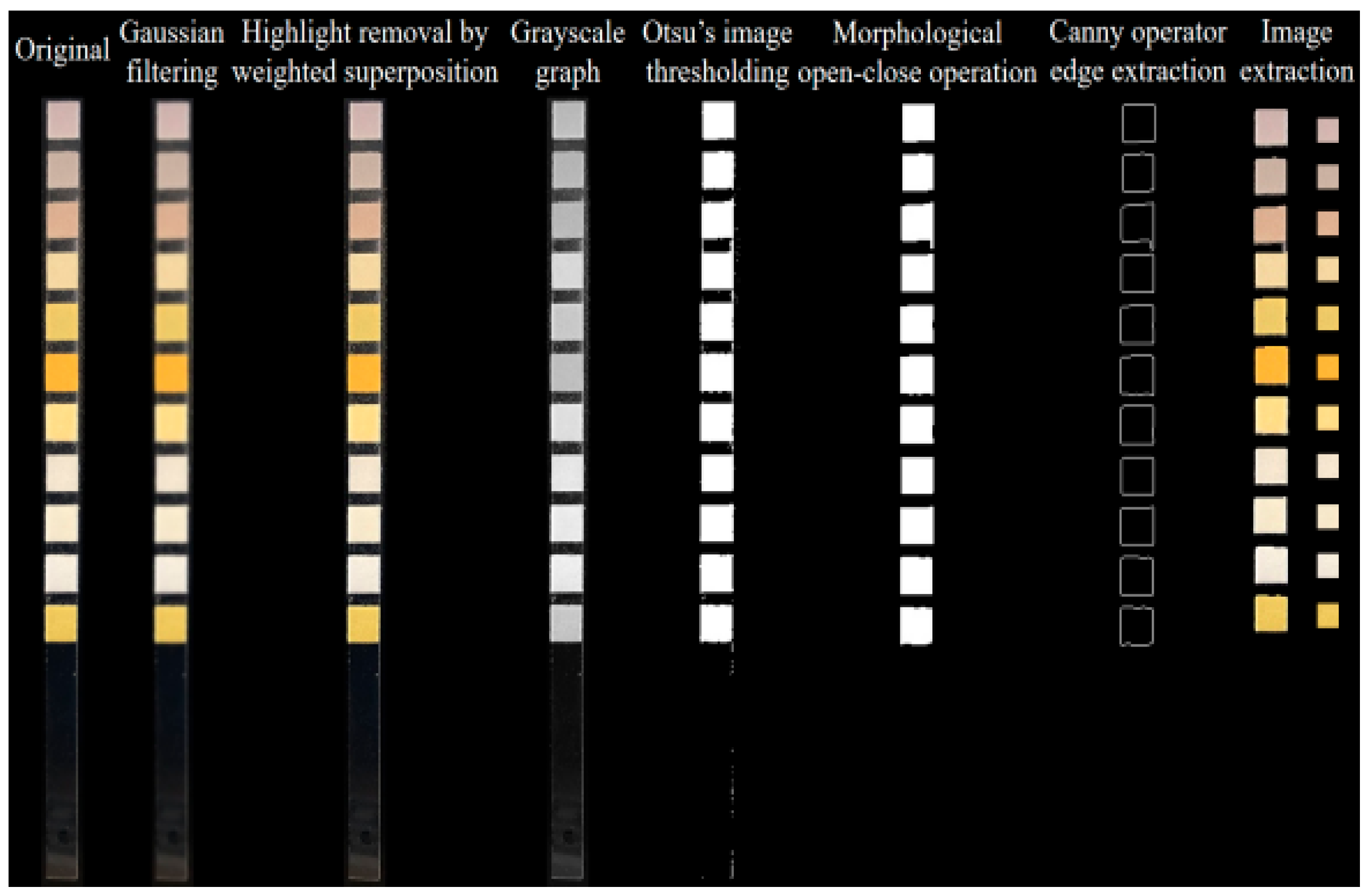
Figure 8 schematizes the processing flow, which includes Gaussian filtering, highlight removal by weighted superposition, Otsu’s image thresholding, morphological open-close operation, Canny operator edge extraction, image extraction, color value extraction and color correction. Considering that during the image acquisition, the camera would introduce some noise (predominantly Gaussian) due to device components and various other factors, the Gaussian smoothing filter was used to accomplish image filtering. The highlight removal reduces the influence of highlight zone through linear weighting of the original image with its smooth version (blurred image). During threshold segmentation, the Otsu’s method was employed to automatically obtain the image thresholds, and the optimal threshold was calculated automatically based on the image gray distribution, thereby separating the background region from the foreground region. The core idea of Otsu’s method is to maximize the variance between classes and find a gray-level threshold T that maximizes the separation between foreground and background pixels. The first step involves calculating the histogram and probability distribution. Assuming the gray level range is from 0 to L − 1, the probability of the i-th gray level is:
For a given threshold T, the Otsu method divides the image into two categories: background pixels (gray levels [0, T]), with probability
and average gray level
; and foreground pixels (gray levels [T, L − 1]), with probability w1 and average gray level
. The inter-class variance is defined as:
By traversing all possible threshold values of T, Ot finds the T that maximizes σb2. This T value is the optimal segmentation threshold.
For morphological processing, the opening operation was performed on the image first to eliminate some small-pixel interfering color blocks. Then, closing operation was performed to fill the small holes in the target color block zones. The primary purpose of morphological processing is to segment the independent elements of the image and reduce the interferences in small and medium-sized regions therein. After the above image operations, the edges of each urine dry chemistry test strip image were sharp and easy to locate. Thus, the Canny operator was directly applied to extract the image edges, obtaining the edge positions of various reagent blocks on the test strip. The image center was determined based on the edges, and by extracting rectangular images with a certain pixel size from the central position, we could obtain respective images of each detection item. Finally, the average RGB value of pixels in the region was calculated, thereby acquiring the representative color information of each reagent block.
In addition to the conventional image processing methods, the YOLOv5 model was also used to train and detect the test strip images. As an advanced object detection model, YOLOv5 has been widely applied in image recognition and localization tasks, and is capable of quickly and accurately identifying objects and their positions in images. Substantial urine test strip images were collected and each detection item color block in the images was accurately annotated with Makesense. These annotated data were utilized to train the YOLOv5 model, allowing it to accurately identify the color block zones of all detection items in the test strip image.
Figure 9 describes the detection effect of the trained model. Regardless of the type of test strip, the model exhibits good detection performance, which can accurately identify the location of each reagent block. During the detection process, the model assigns a confidence value to each detected target zone, which is used for measuring the reliability of detection results. In this study, a detection region with a confidence level of 0.78 or above is regarded as an effective target region and is labelled a “reagent block”. Further processing was carried out on these target regions. Initially, the coordinates of the center point of the detection box were extracted. Then, an image area with a pixel size of 15 × 15 was extracted centering on this central point, which served as the pure color block image of corresponding detection items [
9,
10].
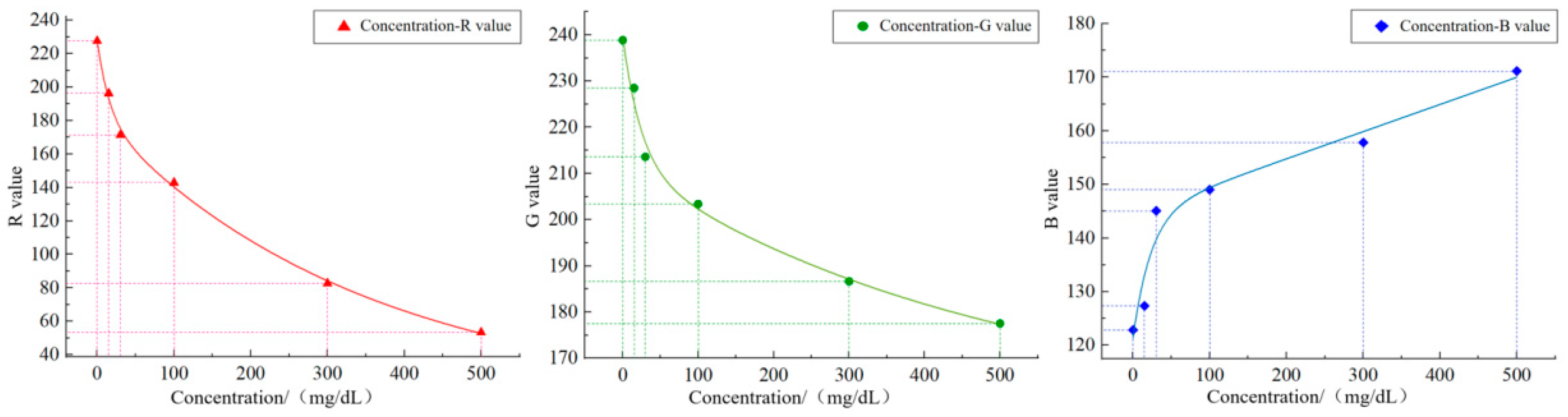
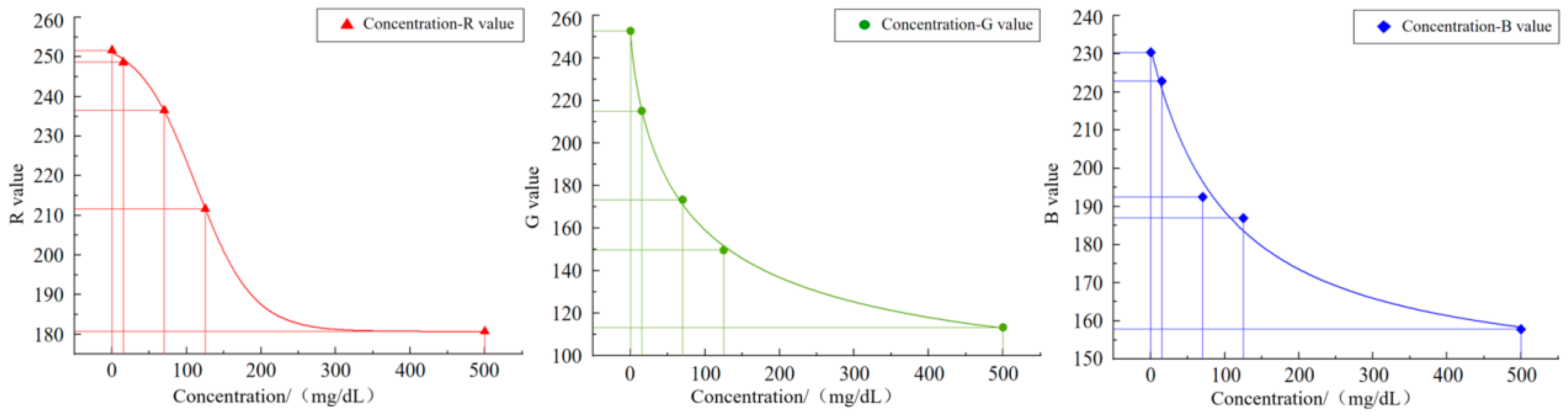
Given the characteristics of the image acquisition system and the influence of environmental factors, the directly extracted color values may have certain deviations. Thus, a final color correction process is required. Through color correction, the extracted color values can be adjusted and corrected, thereby obtaining more accurate color values. The color correction here is specifically the device color correction. By fitting the mapping relationship between the color values captured by the image acquisition system and the known color values on a standard colorimetric card, a polynomial regression model was constructed to correct the color deviations from the device. Initially, it is necessary to photograph the international standard colorimetric card in a fixed lighting environment. All the obtained data were converted from the RGB color space to the CIELab color space with Formulas (11) and (12). The Lab values of the color blocks collected by the system, the known Lab values of standard color blocks, and the corresponding color block images before and after polynomial nonlinear correction are presented in
Figure 10. For every color channel (L, a, b), a polynomial fitting model was built as:
where
xs signifies the standard value of color channel;
a0,
a1, …,
an represent the fitted polynomial coefficient;
xm denotes the measured value of color channel; and
n is the polynomial order. Given the standard colorimetric card dataset, the polynomial coefficients
ai of various channels were fitted by the least squares method as follows:
where
xi represents the device measured value and
m denotes the sample number of standard colorimetric card.
The specific implementation process in the program code is as follows: a NumPy library was used for array operation. The focus was on constructing polynomial features by scheduling the preprocessing function in a sklearn library, thereby extending the input data to polynomial features. For example, when degree = 2, the input data x would be extended to [1, x, x2]. The linear_model function in the sklearn library was applied to fit the polynomial regression equation, and the regression model was trained using the measured extended polynomial feature matrix X_poly and reference target values, thereby obtaining the weight coefficient and intercept of each polynomial feature. The polyfit function code for single-channel data is as follows:
//Polynomial fitting of single-channel data
# Scheduling corresponding function library
from numpy.polynomial.polynomial import Polynomial
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
import numpy as np
# Single-channel fitting function
def fit_polynomial(measured, reference, degree = 2):
# Creating polynomial features
poly = PolynomialFeatures(degree)
X_poly = poly.fit_transform(measured.reshape(−1, 1))
X_poly = poly.fit_transform(measured.reshape(−1, 1)) # Constructing polynomial features
# Fitting polynomial regression model
model = LinearRegression().fit(X_poly, reference)
return model
It is necessary to separately calculate the corresponding polynomial models for the three color channels L, a and b. Substituting the Lab values of the color block images into the model yields corrected Lab color values that are close to the standard.
Figure 10, from left to right, shows the schematic diagram of device color block acquisition, the original image of the standard color chart and the effect diagram of polynomial nonlinear correction.
G
1B
1, R
2G
2B
2, R
3G
3B
3, …, R
iG
iB
i values corresponding to various concentration levels (−, −+, +, ++, +++, ++++) of corresponding items on the standard colorimetric card of urine test strip were separately calculated. The computational formula for the Euclidean distance ΔE
ab is:
Through comparative calculation, all the CIELab distance values ΔEab were obtained. The concentration level on the standard colorimetric card corresponding to the smallest ΔEab was precisely the concentration level of the urine test strip detection item.
2.5. Whale Optimization Algorithm (WOA)
WOA, first proposed by Seyedali Mirjalili et al. in 2016, is an optimization algorithm that simulates whale behavior. The core idea stems from humpback whales’ unique bubble-net feeding strategy. When humpback whales hunt, they blow spiraling circles of bubbles to create a net around their prey, gathering the prey up for easy capture. By simulating this process, WOA searches for the optimal solution in the solution space. The location of each whale corresponds to a potential solution, and the global optimal solution is gradually approached by constantly updating the whale locations. This predation process consists of three stages: the prey encirclement, the bubble-net assaulting and the prey search.
The behavior of encircling prey is modeled by calculating the distance between the whale and the prey (current optimal location) and adjusting the whale location according to this distance [
11,
12,
13]. The specific location update formula is:
The distance vector
D between the current whale location and the optimal location is determined as:
where
C stands for a coefficient vector that adjusts the search range, with
r2 being a random number between [0, 1].
t denotes the number of iterations;
X*(
t) represents the current global optimal location; and
X(
t) represents the current whale location.
The adjustment vector
A is calculated to determine the offset of the whale location relative to the optimal location:
a is a coefficient that decreases linearly from 2 to 0 to control the search convergence process; r is a random number between [0, 1]; and Tmax denotes the maximum number of iterations.
Using the distance vector
D and the adjustment vector
A, the whale location
X(
t + 1) is updated as:
During the bubble-net assaulting behavior, the whale updates its location by calculating its distance from the optimal location and by gradually approaching the optimal location along the spiral path. The specific location update formula is:
The distance vector
D between the current whale location and the optimal location is calculated as follows:
Using the distance vector
D and a spiral path, the whale location
X(
t + 1) is updated as follows:
where
ebl represents an
l-dependent exponential function that simulates the bubble-net
contraction or expansion; cos(2π
l) generates an
l-dependent cosine function to simulate the spiral motion;
b is a constant that determines the spiral shape; and
l is a random number between [−1, 1].
When |A| < 1, the probability parameter p is compared with the preset threshold to identify which of the above behaviors a whale specifically chooses for location updating.
p is a random number between [0, 1]. If p < 0.5, the whale chooses to encircle the prey, which is suitable for the initial stage when a large-area search is required. If p ≥ 0.5, bubble-net assaulting behavior is chosen, which is more suitable for the later stage of the algorithm and enables more accurate approximation when approaching the optimal solution.
When |
A| ≥ 1, under the prey search behavior, the specific location update formula of whale is as follows:
where
Xrand(t) is the location of a whale randomly selected from the whale population.