Uniaxial Cyclic Cell Stretching Device for Accelerating Cellular Studies
Abstract
1. Introduction
2. Materials and Methods
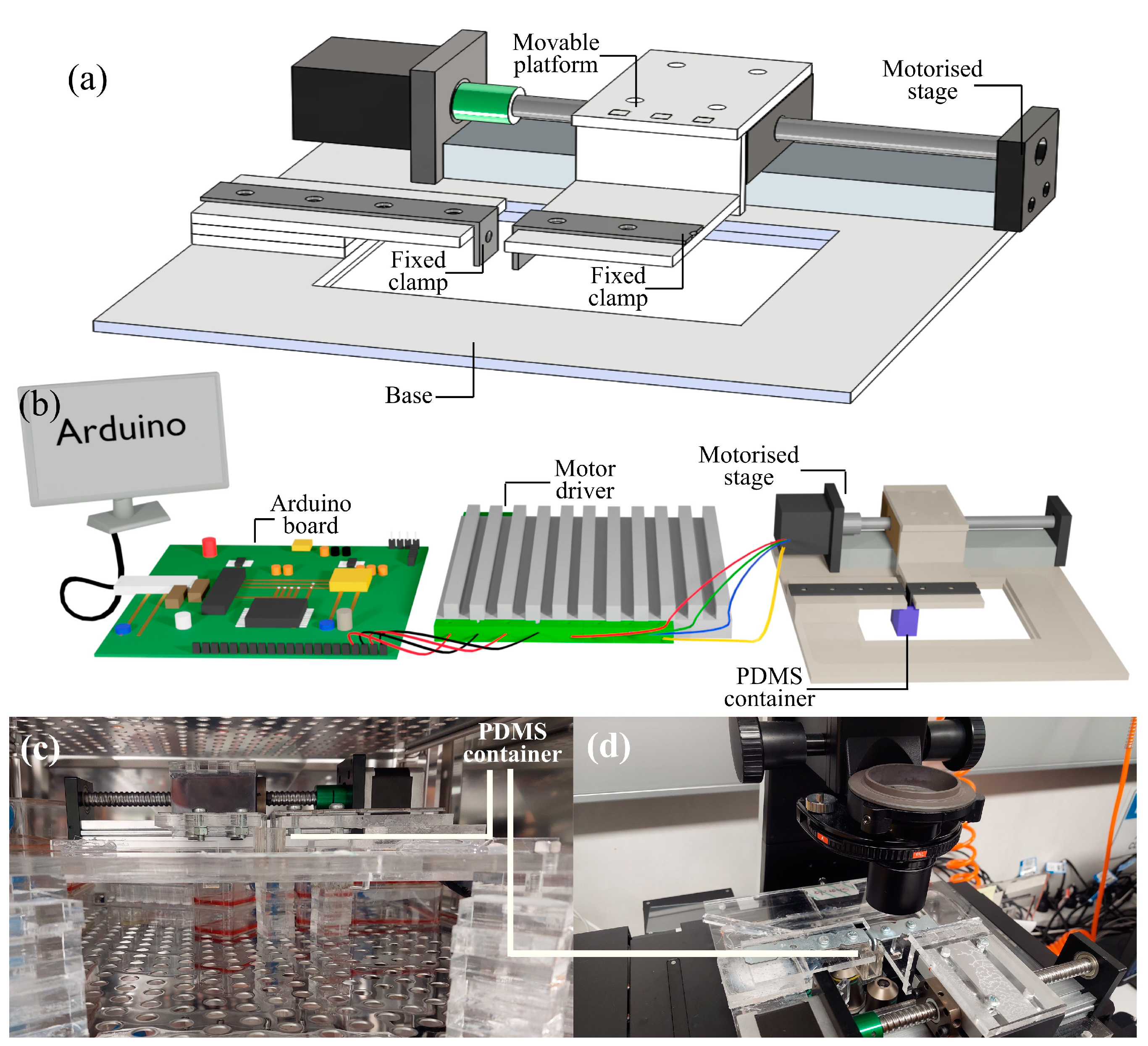
2.1. Cell Stretching Platform
2.2. Motorized Stage and Controller
2.3. PDMS Container
2.4. Cell Culture, Maintenance, and Co-Culture Using the Cell Stretching Device
2.5. Operation of the Cell Stretching Device
2.6. Immunofluorescence Staining
2.7. Image Analysis of the Stretched Cells
2.8. Statistical Analysis
3. Results
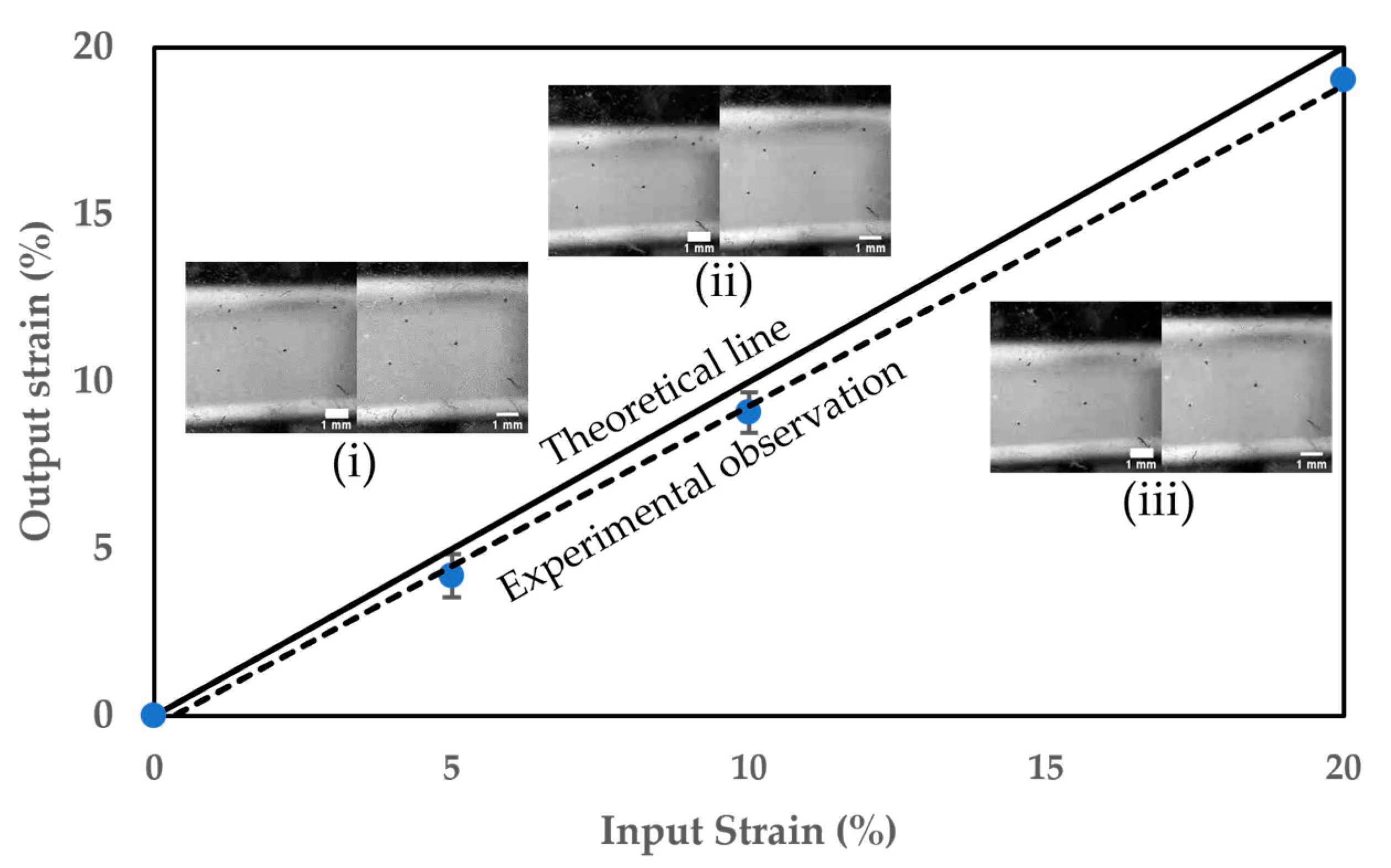
3.1. Performance of the Cell Stretcher
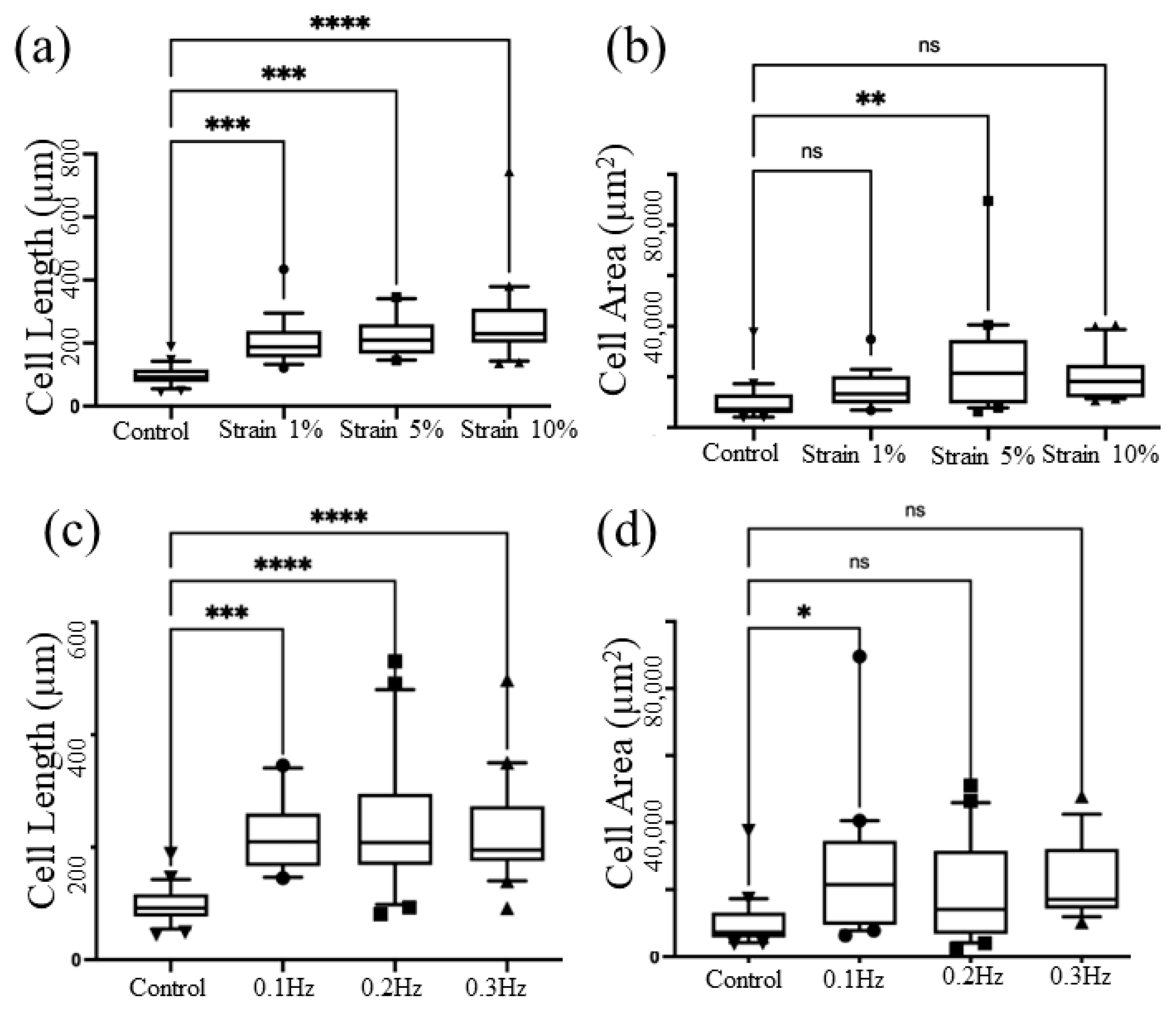
3.2. Parameter Optimization
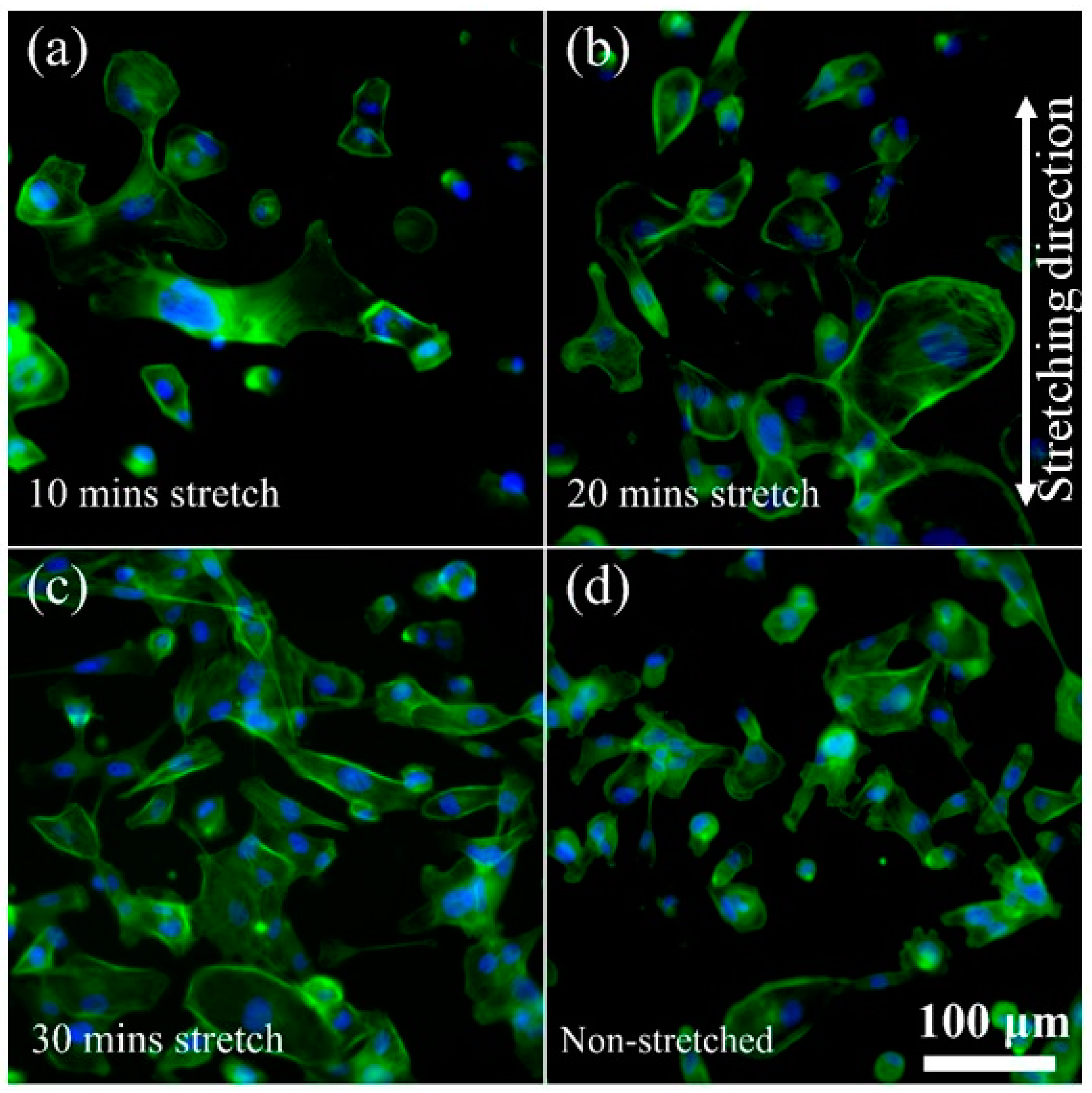
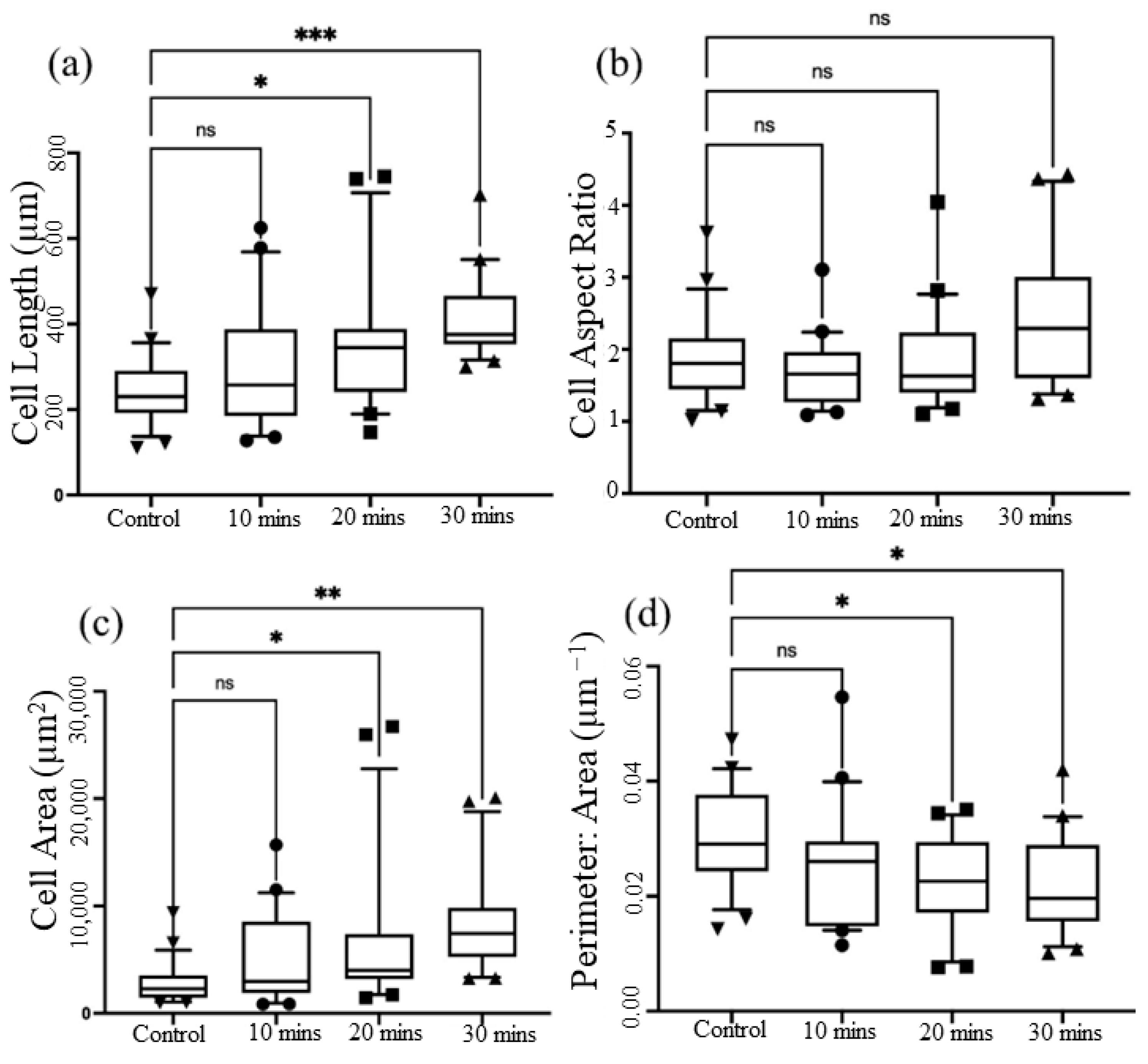
3.3. Cellular Rearrangement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, C.S. Mechanotransduction–a field pulling together? J. Cell Sci. 2008, 121, 3285–3292. [Google Scholar] [CrossRef]
- Orr, A.W.; Helmke, B.P.; Blackman, B.R.; Schwartz, M.A. Mechanisms of Mechanotransduction. Dev. Cell 2006, 10, 11–20. [Google Scholar] [CrossRef]
- Wang, N. Review of cellular mechanotransduction. J. Phys. D Appl. Phys. 2017, 50, 233002. [Google Scholar] [CrossRef]
- Martino, F.; Perestrelo, A.R.; Vinarsky, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Dufort, C.C.; Paszek, M.J.; Weaver, V.M. Balancing forces: Architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 2011, 12, 308–319. [Google Scholar] [CrossRef]
- Schwartz, M.A. Integrins and Extracellular Matrix in Mechanotransduction. Cold Spring Harb. Perspect. Biol. 2010, 2, a005066. [Google Scholar] [CrossRef]
- Jaalouk, D.E.; Lammerding, J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009, 10, 63–73. [Google Scholar] [CrossRef]
- Müller-Marschhausen, K.; Waschke, J.; Drenckhahn, D. Physiological hydrostatic pressure protects endothelial monolayer integrity. Am. J. Physiol. Physiol. 2008, 294, C324–C332. [Google Scholar] [CrossRef]
- Charbonier, F.W.; Zamani, M.; Huang, N.F. Endothelial Cell Mechanotransduction in the Dynamic Vascular Environment. Adv. Biosyst. 2019, 3, e1800252. [Google Scholar] [CrossRef]
- Collinsworth, A.M.; Torgan, C.E.; Nagda, S.N.; Rajalingam, R.J.; Kraus, W.E.; Truskey, G.A. Orientation and length of mammalian skeletal myocytes in response to a unidirectional stretch. Cell Tissue Res. 2000, 302, 243–251. [Google Scholar] [CrossRef]
- Garoffolo, G.; Pesce, M. Mechanotransduction in the Cardiovascular System: From Developmental Origins to Homeostasis and Pathology. Cells 2019, 8, 1607. [Google Scholar] [CrossRef]
- Makale, M. Cellular mechanobiology and cancer metastasis. Birth Defects Res. 2008, 81, 329–343. [Google Scholar] [CrossRef]
- Moghadas, H.; Saidi, M.S.; Kashaninejad, N.; Nguyen, N.-T. A high-performance polydimethylsiloxane electrospun membrane for cell culture in lab-on-a-chip. Biomicrofluidics 2018, 12, 024117. [Google Scholar] [CrossRef]
- Shourabi, A.Y.; Kashaninejad, N.; Saidi, M.S. An integrated microfluidic concentration gradient generator for mechanical stimulation and drug delivery. J. Sci. Adv. Mater. Devices 2021, 6, 280–290. [Google Scholar] [CrossRef]
- Møller, A.B.; Vendelbo, M.H.; Rahbek, S.K.; Clasen, B.F.; Schjerling, P.; Vissing, K.; Jessen, N. Resistance exercise, but not endurance exercise, induces ikkβ phosphorylation in human skeletal muscle of training-accustomed individuals. Pflügers Arch. Eur. J. Physiol. 2013, 465, 1785–1795. [Google Scholar] [CrossRef]
- Swaminathan, V.; Gloerich, M. Decoding mechanical cues by molecular mechanotransduction. Curr. Opin. Cell Biol. 2021, 72, 72–80. [Google Scholar] [CrossRef]
- Du, H.; Bartleson, J.M.; Butenko, S.; Alonso, V.; Liu, W.F.; Winer, D.A.; Butte, M.J. Tuning immunity through tissue mechanotransduction. Nat. Rev. Immunol. 2023, 23, 174–188. [Google Scholar] [CrossRef]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef]
- Li, L.; Eyckmans, J.; Chen, C.S. Designer biomaterials for mechanobiology. Nat. Mater. 2017, 16, 1164–1168. [Google Scholar] [CrossRef]
- Thompson, M.; Epari, D.; Bieler, F.; Duda, G. In vitro models for bone mechanobiology: Applications in bone regeneration and tissue engineering. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2010, 224, 1533–1541. [Google Scholar] [CrossRef]
- Zhao, J.; Meng, F.; Qian, J.; Huang, Y.; Fan, Y. In vitro cell stretching devices and their applications: From cardiomyogenic differentiation to tissue engineering. Med. Nov. Technol. Devices 2023, 18, 100220. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 2020, 584, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Gracey, E.; Burssens, A.; Cambré, I.; Schett, G.; Lories, R.; McInnes, I.B.; Asahara, H.; Elewaut, D. Tendon and ligament mechanical loading in the pathogenesis of inflammatory arthritis. Nat. Rev. Rheumatol. 2020, 16, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Li, B. Mechanics rules cell biology. BMC Sports Sci. Med. Rehabil. 2010, 2, 16. [Google Scholar] [CrossRef]
- Seriani, S.; Del Favero, G.; Mahaffey, J.; Marko, D.; Gallina, P.; Long, C.S.; Mestroni, L.; Sbaizero, O. The cell-stretcher: A novel device for the mechanical stimulation of cell populations. Rev. Sci. Instrum. 2016, 87, 084301. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Parekh, S.H.; Lam, W.A.; Fletcher, D.A. Combined atomic force microscopy and side-view optical imaging for mechanical studies of cells. Nat. Methods 2009, 6, 383–387. [Google Scholar] [CrossRef]
- Kamble, H.; Barton, M.J.; Jun, M.; Park, S.; Nguyen, N.-T. Cell stretching devices as research tools: Engineering and biological considerations. Lab Chip 2016, 16, 3193–3203. [Google Scholar] [CrossRef]
- Al-Maslamani, N.A.; Khilan, A.A.; Horn, H.F. Design of a 3D printed, motorized, uniaxial cell stretcher for microscopic and biochemical analysis of mechanotransduction. Biol. Open 2021, 10, bio057778. [Google Scholar] [CrossRef]
- Boulter, E.; Tissot, F.S.; Dilly, J.; Pisano, S.; Féral, C.C. Cyclic uniaxial mechanical stretching of cells using a LEGO® parts-based mechanical stretcher system. J. Cell Sci. 2020, 133, jcs234666. [Google Scholar] [CrossRef]
- Bonakdar, N. Cell Mechanics in Response to Large Forces and Deformations. Ph.D. Thesis, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 2014. [Google Scholar]
- Matsugaki, A.; Fujiwara, N.; Nakano, T. Conditions for Osteoblast Arrangement Induced under Long-Term Cyclic Stretching. Mater. Trans. 2013, 54, 1195–1199. [Google Scholar] [CrossRef]
- Huang, Y.; Nguyen, N.-T. A polymeric cell stretching device for real-time imaging with optical microscopy. Biomed. Microdevices 2013, 15, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Yost, M.J.; Simpson, D.; Wrona, K.; Ridley, S.; Ploehn, H.J.; Borg, T.K.; Terracio, L. Design and construction of a uniaxial cell stretcher. Am. J. Physiol. Heart Circ. Physiol. 2000, 279, H3124–H3130. [Google Scholar] [CrossRef]
- Deng, Y.; Davis, S.P.; Yang, F.; Paulsen, K.S.; Kumar, M.; Sinnott DeVaux, R.; Wang, X.; Conklin, D.S.; Oberai, A.; Herschkowitz, J.I. Inertial microfluidic cell stretcher (imcs): Fully automated, high-throughput, and near real-time cell mechanotyping. Small 2017, 13, 1700705. [Google Scholar] [CrossRef]
- Kamble, H.; Vadivelu, R.; Barton, M.; Boriachek, K.; Munaz, A.; Park, S.; Shiddiky, M.J.; Nguyen, N.-T. An electromagnetically actuated double-sided cell-stretching device for mechanobiology research. Micromachines 2017, 8, 256. [Google Scholar] [CrossRef]
- Kamotani, Y.; Bersano-Begey, T.; Kato, N.; Tung, Y.-C.; Huh, D.; Song, J.W.; Takayama, S. Individually programmable cell stretching microwell arrays actuated by a Braille display. Biomaterials 2008, 29, 2646–2655. [Google Scholar] [CrossRef]
- Huang, Y.; Nguyen, N.-T.; Lok, K.S.; Lee, P.P.F.; Su, M.; Wu, M.; Kocgozlu, L.; Ladoux, B.; Chua, C.K.; Tan, L.P.; et al. Multiarray cell stretching platform for high-magnification real-time imaging. Nanomedicine 2013, 8, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, D.; Chagnon-Lessard, S.; Mirzaei, M.; Pelling, A.E.; Godin, M. A microscale anisotropic biaxial cell stretching device for applications in mechanobiology. Biotechnol. Lett. 2014, 36, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Constantinou, I.; Bastounis, E.E. Cell-stretching devices: Advances and challenges in biomedical research and live-cell imaging. Trends Biotechnol. 2023, 41, 939–950. [Google Scholar] [CrossRef]
- Atcha, H.; Davis, C.T.; Sullivan, N.R.; Smith, T.D.; Anis, S.; Dahbour, W.Z.; Robinson, Z.R.; Grosberg, A.; Liu, W.F. A Low-Cost Mechanical Stretching Device for Uniaxial Strain of Cells: A Platform for Pedagogy in Mechanobiology. J. Biomech. Eng. 2018, 140, 081005. [Google Scholar] [CrossRef]
- Yadav, S.; Vadivelu, R.; Ahmed, M.; Barton, M.; Nguyen, N.-T. Stretching cells–an approach for early cancer diagnosis. Exp. Cell Res. 2019, 378, 191–197. [Google Scholar] [CrossRef]
- Yadav, S.; Barton, M.; Nguyen, N.T. Stretching induces overexpression of rhoa and rac1 gtpases in breast cancer cells. Adv. Biosyst. 2020, 4, 1900222. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Kashaninejad, N.; Nguyen, N.-T. RhoA and Rac1 in Liver Cancer Cells: Induction of Overexpression Using Mechanical Stimulation. Micromachines 2020, 11, 729. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, S.; Singha, P.; Nguyen, N.-K.; Ooi, C.H.; Kashaninejad, N.; Nguyen, N.-T. Uniaxial Cyclic Cell Stretching Device for Accelerating Cellular Studies. Micromachines 2023, 14, 1537. https://doi.org/10.3390/mi14081537
Yadav S, Singha P, Nguyen N-K, Ooi CH, Kashaninejad N, Nguyen N-T. Uniaxial Cyclic Cell Stretching Device for Accelerating Cellular Studies. Micromachines. 2023; 14(8):1537. https://doi.org/10.3390/mi14081537
Chicago/Turabian StyleYadav, Sharda, Pradip Singha, Nhat-Khuong Nguyen, Chin Hong Ooi, Navid Kashaninejad, and Nam-Trung Nguyen. 2023. "Uniaxial Cyclic Cell Stretching Device for Accelerating Cellular Studies" Micromachines 14, no. 8: 1537. https://doi.org/10.3390/mi14081537
APA StyleYadav, S., Singha, P., Nguyen, N.-K., Ooi, C. H., Kashaninejad, N., & Nguyen, N.-T. (2023). Uniaxial Cyclic Cell Stretching Device for Accelerating Cellular Studies. Micromachines, 14(8), 1537. https://doi.org/10.3390/mi14081537








