High Throughput Identification of Antimicrobial Peptides from Fish Gastrointestinal Microbiota
Abstract
:1. Introduction
2. Results
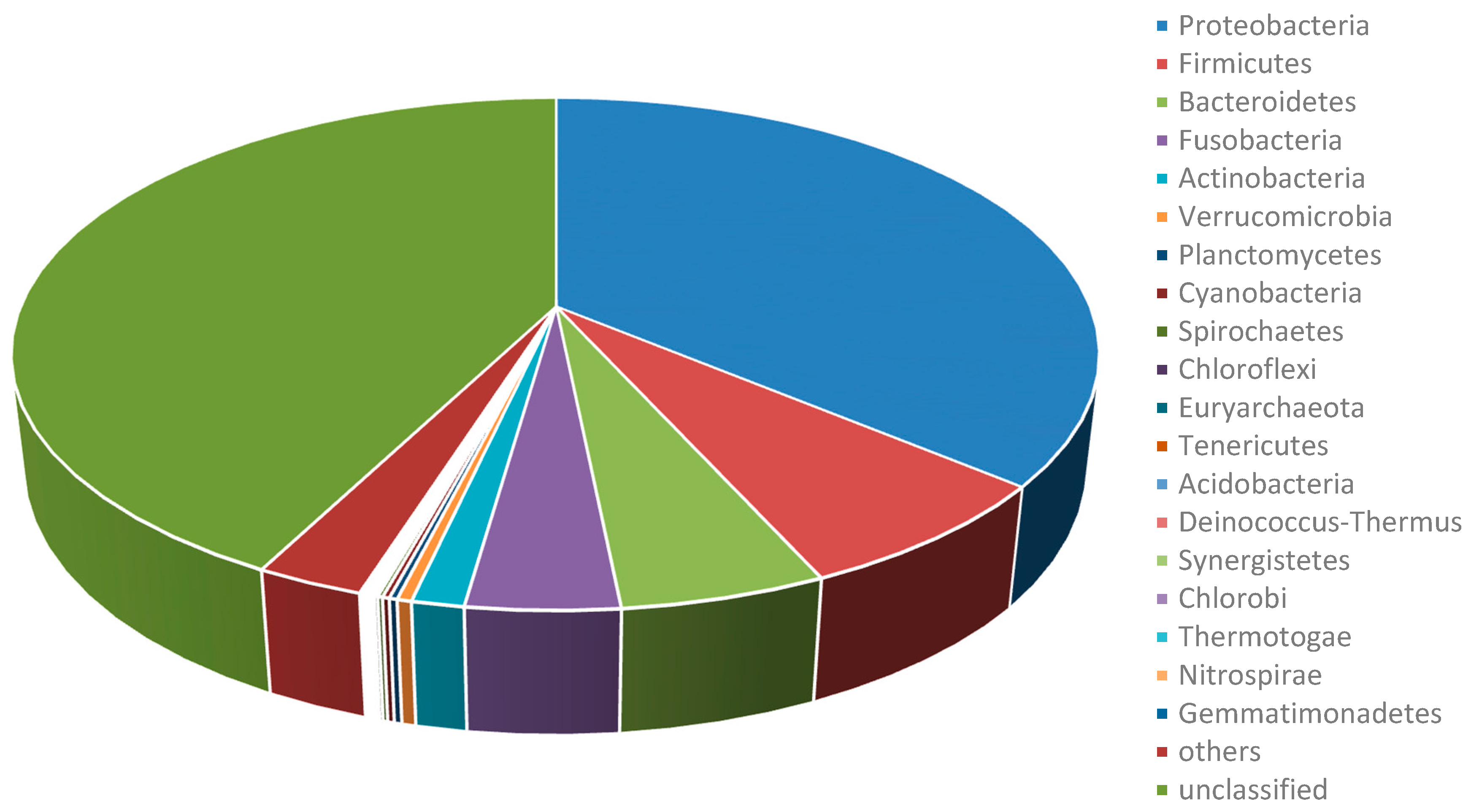
2.1. Summary of Achieved Metagenome Datasets
2.2. AMPs Identified in the Metagenome
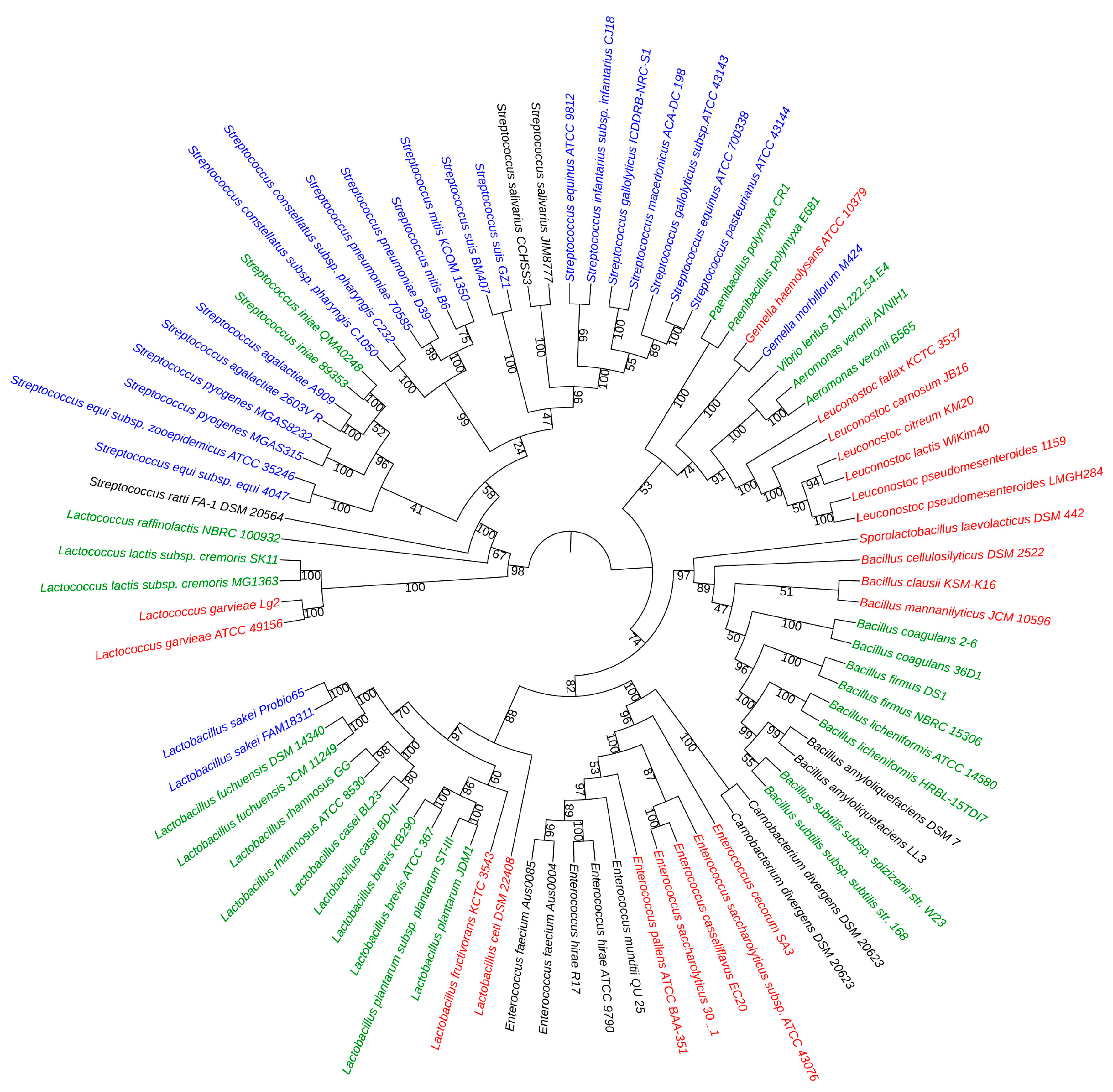
2.3. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Assembly and Annotation of the Metagenome
4.3. Prediction and Identification of AMPs
4.4. Alignment and Homology of AMPs
4.5. Construction of the Phylogenetic Tree
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rodney, H.P.; Takeshi, Z.; Kenji, S. Novel bacteriocins from lactic acid bacteria (LAB): Various structures and applications. Microb. Cell Fact. 2014, 13 (Suppl. 1), S3. [Google Scholar]
- Zacharof, M.P.; Lovitt, R.W. Bacteriocins produced by lactic acid bacteria a review article. APCBEE Procedia 2012, 2, 50–56. [Google Scholar] [CrossRef]
- Field, D.; Cotter, P.D.; Ross, R.P.; Hill, C. Bioengineering of the model lantibiotic nisin. Bioengineered 2015, 6, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Harris, H.M.; McCann, A.; Guo, C.; Argimon, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M.; Anastasiadou, S. Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications. Microb. Cell Fact. 2009, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Zhang, X.H.; Boon, N.; Bossier, P. Probiotics in aquaculture of China—Current state, problems and prospect. Aquaculture 2009, 290, 15–21. [Google Scholar] [CrossRef]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037-15. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, G.; Angert, E.R.; Wang, W.; Li, W.; Zou, H. Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS ONE 2012, 7, e30440. [Google Scholar] [CrossRef] [PubMed]
- Dischinger, J.; Basi, C.S.; Bierbaum, G. Lantibiotics: Promising candidates for future applications in health care. Int. J. Med. Microbiol. 2014, 304, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Vaillancourt, K.; LeBel, G.; Frenette, M.; Fittipaldi, N.; Gottschalk, M.; Grenier, D. Purification and characterization of Suicin 65, a novel class I type B lantibiotic produced by Streptococcus suis. PLoS ONE 2015, 10, e0145854. [Google Scholar] [CrossRef] [PubMed]
- Drider, D.; Fimland, G.; Hechard, Y.; McMullen, L.M.; Prevost, H. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa bacteriocins: Diversity and new developments. Int. J. Mol. Sci. 2012, 13, 16668–16707. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Suárez, J.E.; Rodríguez, A. Lactococcin 972: A hornodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 1996, 142, 2393–2398. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Bottiger, T.; Schneider, T.; Rodriguez, A.; Sahl, H.G.; Wiedemann, I. Specific interaction of the unmodified bacteriocin Lactococcin 972 with the cell wall precursor lipid II. Appl. Environ. Microbiol. 2008, 74, 4666–4670. [Google Scholar] [CrossRef] [PubMed]
- Parveen, R.R.; Anandharaj, M.; Hema, S.; Deepika, R.; David, R.A. Purification of antilisterial peptide (Subtilosin A) from novel Bacillus tequilensis FR9 and demonstrate their pathogen invasion protection ability using human carcinoma cell line. Front. Microbiol. 2016, 7, 1910. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.A.; Pohl, R.; Beck-Sickinger, A.G.; Selmer, T.; Pierik, A.J.; Bastos, M.F.; Sahl, H.G. Biochemical characterisation and genetic analysis of aureocin A53, a new, a typical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 2002, 319, 745–756. [Google Scholar] [CrossRef]
- Netz, D.A.; Bastos, M.C.; Sahl, H.G. Mode of action of the antimicrobial peptide aureocin A53 from Staphylococcus aureus. Appl. Environ. Microbiol. 2002, 68, 5274–5280. [Google Scholar] [CrossRef] [PubMed]
- Vendrell, D.; Balcázar, J.L.; Ruiz, Z.I.; De, B.I.; Gironés, O.; Múzquiz, J.L. Lactococcus garvieae in fish: A review. Comp. Immunol. Microbiol. Infect. Dis. 2006, 29, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Delpech, P.; Rifa, E.; Ball, G.; Nidelet, S.; Dubois, E.; Gagne, G.; Montel, M.C.; Delbes, C.; Bornes, S. New insights into the anti-pathogenic potential of Lactococcus garvieae against Staphylococcus aureus based on RNA sequencing profiling. Front. Microbiol. 2017, 8, 359. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, Y.; Hagi, T.; Hoshino, T. Correlation between in vitro mucus adhesion and the in vivo colonization ability of lactic acid bacteria: Screening of new candidate carp probiotics. Biosci. Biotechnol. Biochem. 2011, 75, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Hagi, T.; Hoshino, T. Screening and characterization of potential probiotic lactic acid bacteria from cultured common carp intestine. Biosci. Biotechnol. Biochem. 2009, 73, 1479–1483. [Google Scholar] [CrossRef] [PubMed]
- Pandiyan, P.; Balaraman, D.; Thirunavukkarasu, R.; George, E.J.; Subaramaniyan, K.; Manikkam, S.; Sadayappan, B. Probiotics in aquaculture. Drug Invent. Today 2013, 5, 55–59. [Google Scholar] [CrossRef]
- Merrifield, D.L.; Dimitroglou, A.; Foey, A.; Davies, S.J.; Baker, R.M.; Bøgwald, J.; Castex, M.; Ringø, E. The current status and future focus of probiotic and prebiotic applications for salmonids. Aquaculture 2010, 302, 1–18. [Google Scholar] [CrossRef]
- Sugita, H.; Hirose, Y.; Matsuo, N.; Deguchi, Y. Production of the antibacterial substance by Bacillus sp. strain NM 12, an intestinal bacterium of Japanese coastal fish. Aquaculture 1998, 165, 269–280. [Google Scholar] [CrossRef]
- Giri, S.S.; Sukumaran, V.; Sen, S.S.; Vinumonia, J.; Banu, B.N.; Jena, P.K. Antagonistic activity of cellular components of potential probiotic bacteria, isolated from the gut of Labeo rohita, against Aeromonas hydrophila. Probiotics Antimicrob. Proteins 2011, 3, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, M.; Jami, M.; Domig, K.J.; Kneifel, W. Seafood biopreservation by lactic acid bacteria—A review. LWT Food Sci. Technol. 2013, 54, 315–324. [Google Scholar] [CrossRef]
- Offret, C.; Desriac, F.; Le, C.P.; Mounier, J.; Jegou, C.; Fleury, Y. Spotlight on antimicrobial metabolites from the marine bacteria Pseudoalteromonas: Chemodiversity and ecological significance. Mar. Drugs 2016, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Harikrishnan, R.; Balasundaram, C.; Heo, M.S. Potential use of probiotic- and triherbal extract-enriched diets to control Aeromonas hydrophila infection in carp. Dis. Aquat. Org. 2010, 92, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Yan, Q.; Yu, Y.; Zhang, T. Factors influencing the grass carp gut microbiome and its effect on metabolism. FEMS Microbiol. Ecol. 2014, 87, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Zendo, T.; Nakayama, J.; Fujita, K.; Sonomoto, K. Bacteriocin detection by liquid chromatography/mass spectrometry for rapid identification. J. Appl. Environ. Microbiol. 2008, 104, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.T.; Freed, S.D.; Lee, S.W.; Friedberg, I. A large scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC Bioinform. 2015, 16, 381. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.J.; Guinane, C.M.; O’Toole, P.W.; Cotter, P.D. A Profile Hidden Markov Model to investigate the distribution and frequency of LanB-encoding lantibiotic modification genes in the human oral and gut microbiome. PeerJ 2017, 5, e3254. [Google Scholar] [CrossRef] [PubMed]
- De Jong, A.; van Heel, A.J.; Kok, J.; Kuipers, O.P. BAGEL2: Mining for bacteriocins in genomic data. Nucleic Acids Res. 2010, 38, W647–W651. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, R.J.; Colmer, S.; Tynan, H.; Demain, A.L.; Gullo, V.P. Antimicrobials, drug discovery, and genome mining. Appl. Microbiol. Biotechnol. 2013, 97, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, R.; Liu, C.M.; Leung, C.M.; Ting, H.F.; Sadakane, K.; Yamashita, H.; Lam, T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Beitz, E. TeXshade: Shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
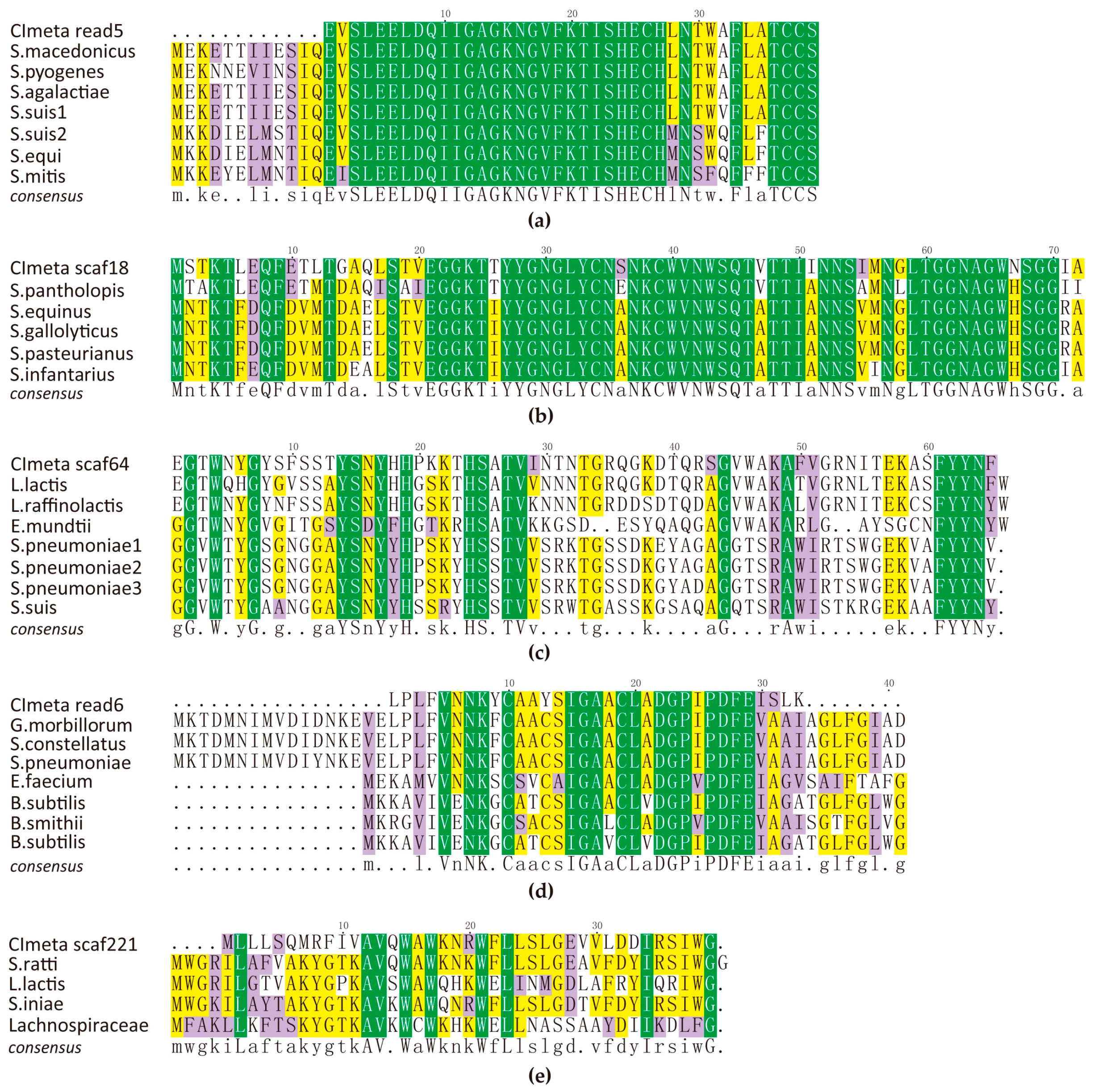
| Category | Identified AMPs 1 | NCBI AMPs | GenBank Accession No. | NCBI AMP-Producing Bacterial Species |
|---|---|---|---|---|
| Class I (Lantibiotic) | a | McdA | ABI30227.1 | Streptococcus macedonicus |
| Streptococcin A-FF22 | P36501.1 | Streptococcus pyogenes | ||
| Nukacin | CZT39525.1 | Streptococcus agalactiae | ||
| Macedocin | CZA89639.1 | Streptococcus suis | ||
| type-A lantibiotic | EEF65507.1 | Streptococcus suis 89/1591 | ||
| type A2 lantipeptide | WP_041790396.1 | Streptococcus equi | ||
| type A2 lantipeptide | WP_033685037.1 | Streptococcus mitis | ||
| lantibiotic nukacin | KEO43205.1 | Streptococcus salivarius | ||
| Class IIa (Pediocin-like) | b | Hypothetical | AND78905.1 | Streptococcus pantholopis |
| piscicolin-126 | EFM26697.1 | Streptococcus equinus | ||
| Bacteriocin | KUE92317.1 | Streptococcus gallolyticus | ||
| putative piscicolin-126 | KXI11412.1 | Streptococcus pasteurianus | ||
| infantaricin E | AHW46171.1 | Streptococcus infantarius | ||
| MundKS | ACI25616.1 | Enterococcus mundtii | ||
| leucocin C | AEY55410.1 | Leuconostoc carnosum | ||
| SakX | AAP44569.1 | Lactobacillus sakei | ||
| Class IIc | c | lactococcin 972 | CAA05247.1 | Lactococcus lactis |
| lactococcin 972 family | WP_061775386.1 | Lactococcus raffinolactis | ||
| lactococcin 972 family | WP_065096983.1 | Enterococcus mundtii | ||
| lactococcin 972 | CTL98394.1 | Streptococcus pneumoniae | ||
| lactococcin 972 family | SNP59245.1 | Streptococcus pneumoniae | ||
| lactococcin 972 family | CWJ26067.1 | Streptococcus pneumoniae | ||
| lactococcin 972 family | CYW17154.1 | Streptococcus suis | ||
| d | sboA protein | EFV34710.1 | Gemella morbillorum M424 | |
| subtilosin A | EGV07582.1 | Streptococcus constellatus | ||
| putative subtilosin A | CVX48913.1 | Streptococcus pneumoniae | ||
| Hypothetical | ELB10075.1 | Enterococcus faecium | ||
| subtilosin A | CAD23198.1 | Bacillus subtilis | ||
| subtilosin A | AKP46487.1 | Bacillus smithii | ||
| subtilosin A | WP_087992738.1 | Bacillus subtilis | ||
| Class IId | e | Mutacin BhtB | AAZ76605.1 | Streptococcus ratti |
| lactolisterin BU | SDR48784.1 | Lactococcus lactis | ||
| Hypothetical | WP_081348647.1 | Streptococcus iniae | ||
| Aureocin-like | SFG15527.1 | Lachnospiraceae C7 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, B.; Yi, Y.; Liang, L.; Shi, Q. High Throughput Identification of Antimicrobial Peptides from Fish Gastrointestinal Microbiota. Toxins 2017, 9, 266. https://doi.org/10.3390/toxins9090266
Dong B, Yi Y, Liang L, Shi Q. High Throughput Identification of Antimicrobial Peptides from Fish Gastrointestinal Microbiota. Toxins. 2017; 9(9):266. https://doi.org/10.3390/toxins9090266
Chicago/Turabian StyleDong, Bo, Yunhai Yi, Lifeng Liang, and Qiong Shi. 2017. "High Throughput Identification of Antimicrobial Peptides from Fish Gastrointestinal Microbiota" Toxins 9, no. 9: 266. https://doi.org/10.3390/toxins9090266






