Mechanisms for Differential Protein Production in Toxin–Antitoxin Systems
Abstract
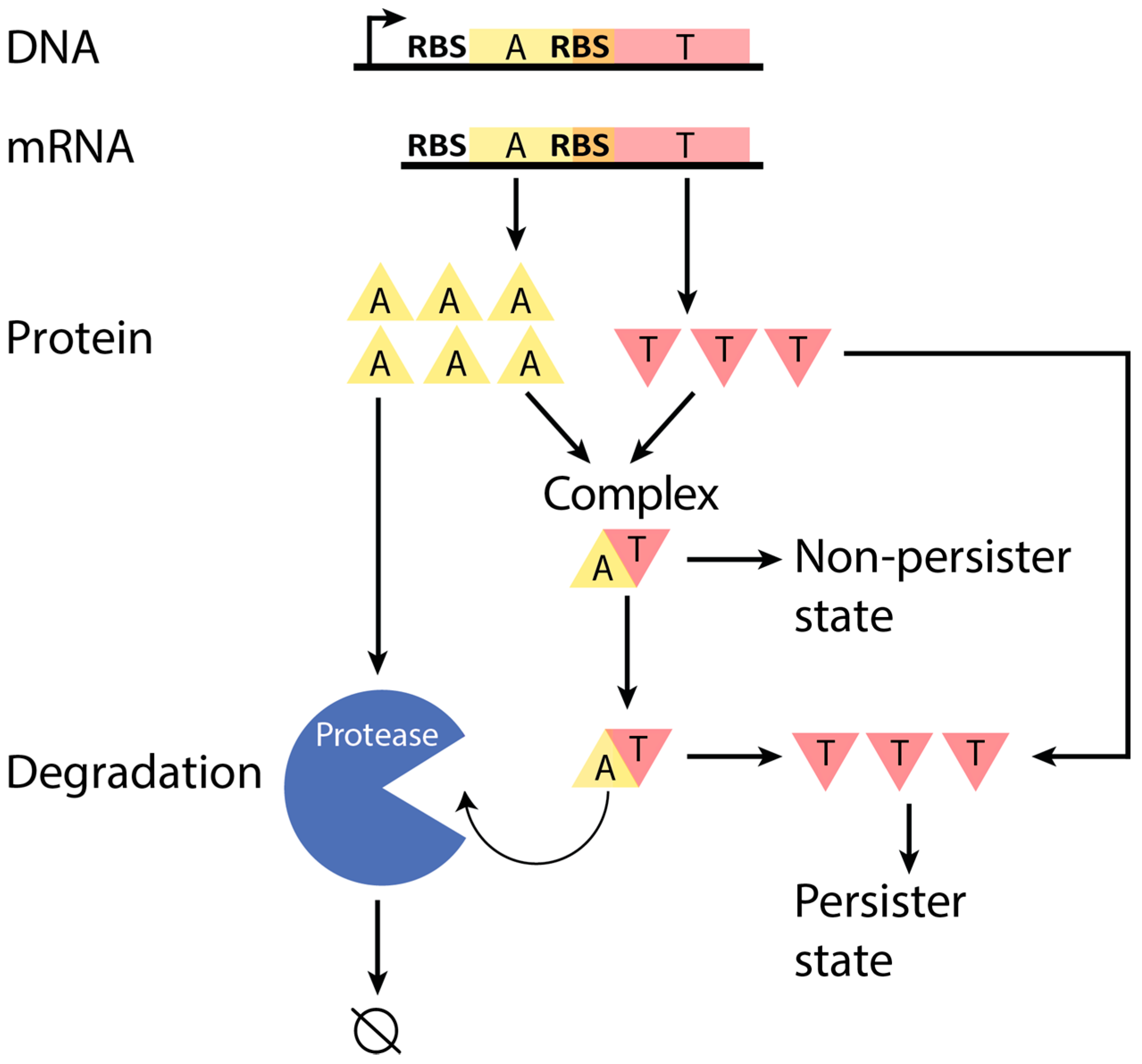
:1. Introduction
2. Results and Discussion
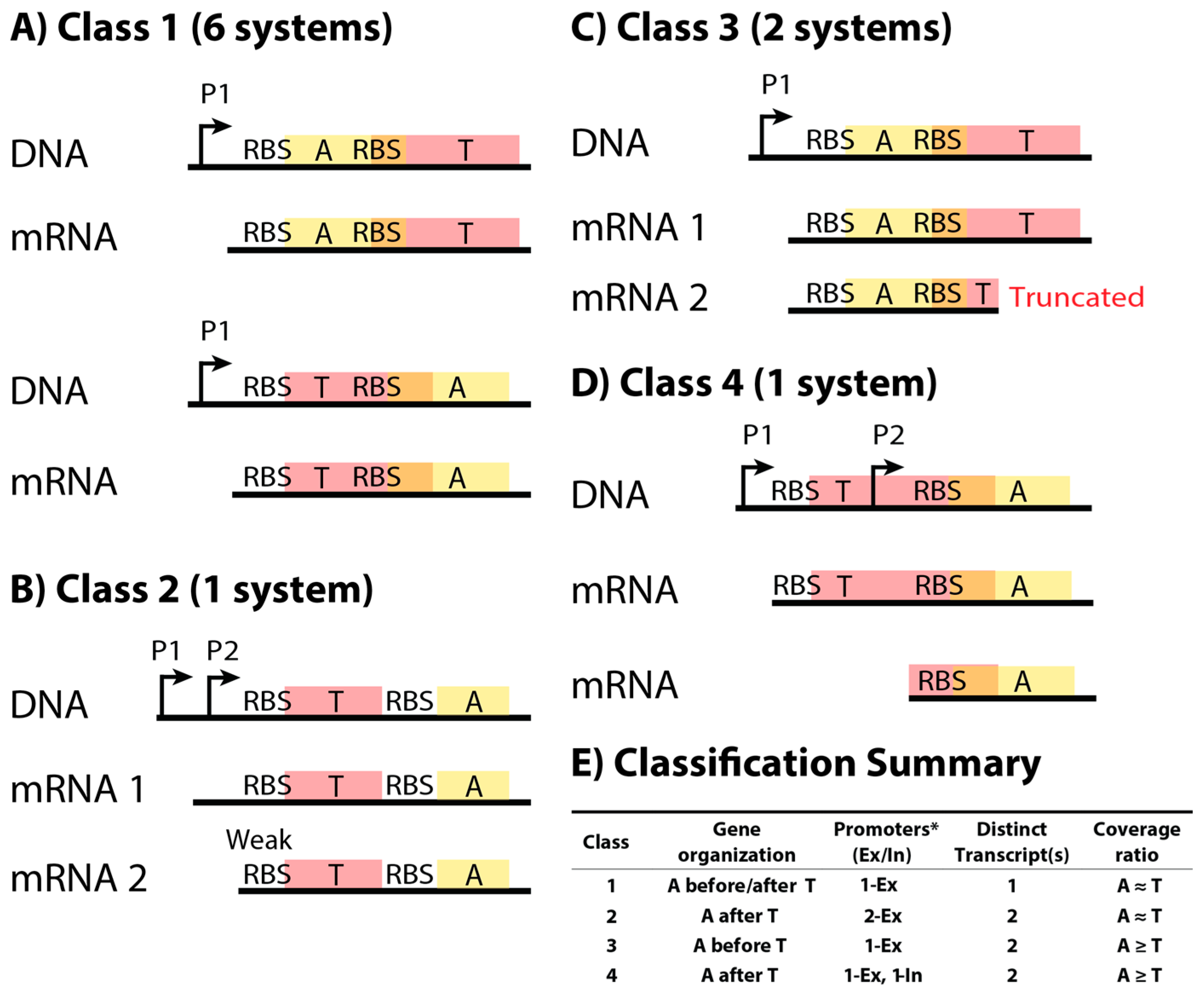
2.1. Classification Scheme for Type II Toxin–Antitoxin Systems Based on DNA Sequence and mRNA Products
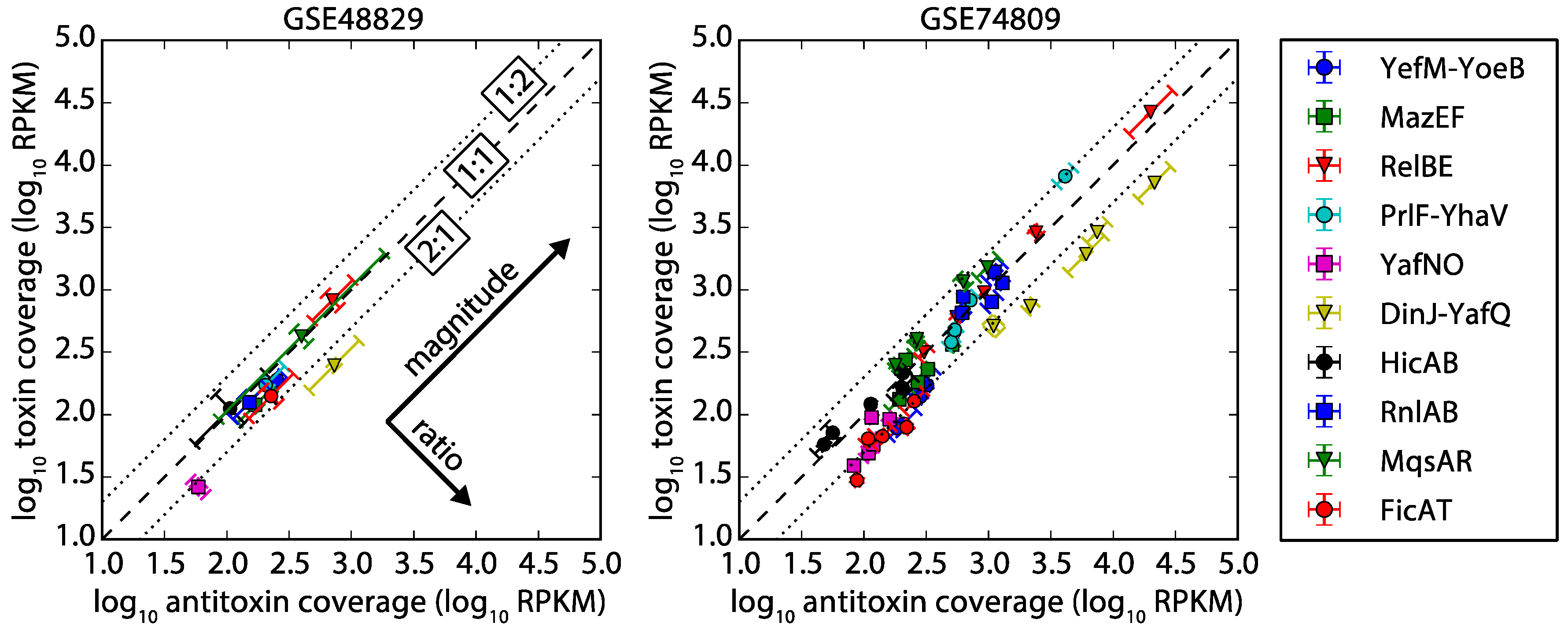
2.2. Antitoxin and Toxin mRNA Coverage by RNA-seq
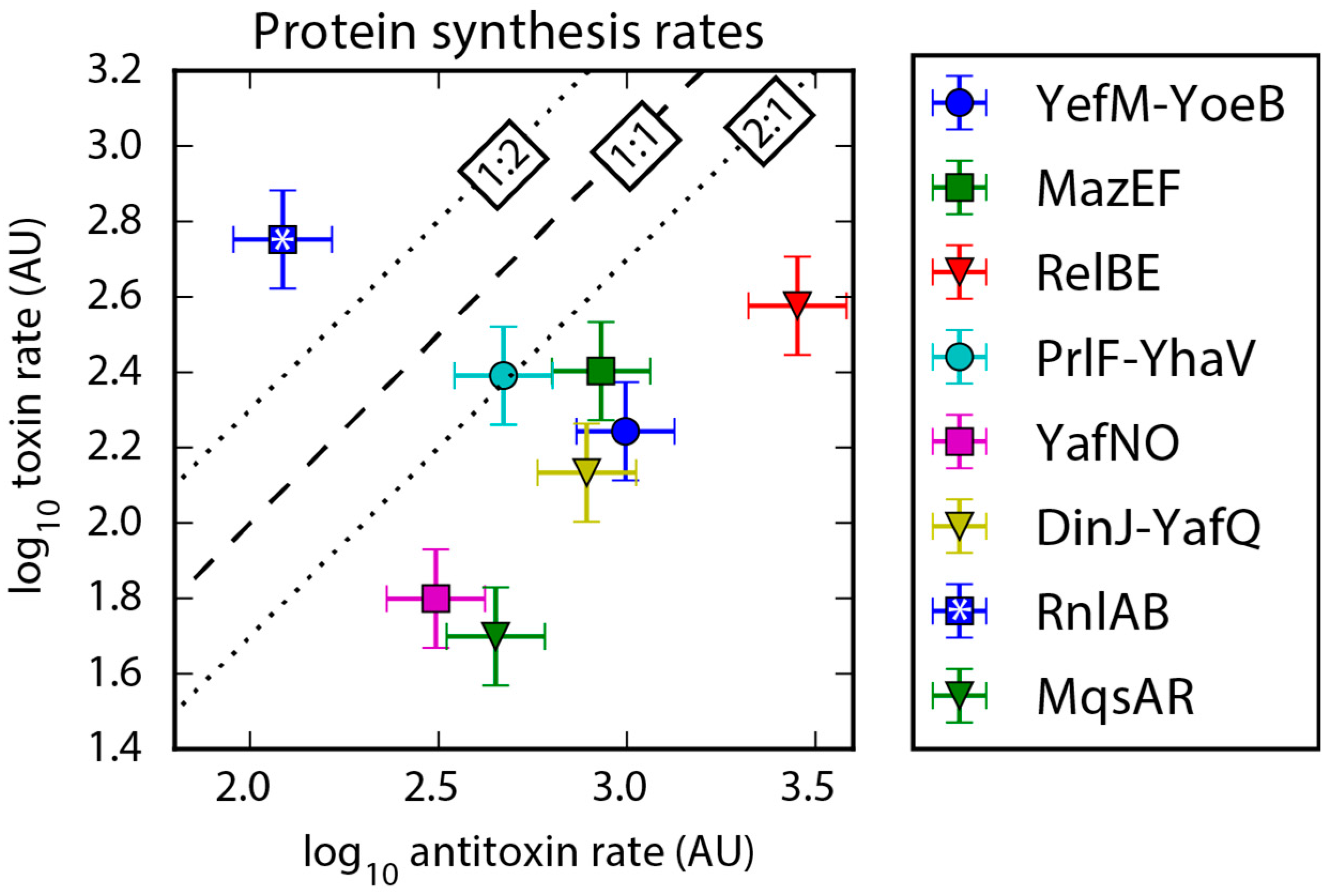
2.3. Protein Synthesis Rates Determined by Ribo-Seq
2.4. Analysis of Differential Protein Expression Using Translation Initiation Calculators (TIRs)
2.5. Summary and Discussion of Major Trends and Exceptions for TA System Classes
2.6. Incorporation of Our Results into Current Models
3. Conclusions
4. Materials and Methods
4.1. DNA Sequence and mRNA Sequence Analysis
4.2. RNA-seq Analysis
4.3. Protein Synthesis Rates Based on Ribosome Profiling (Ribo-Seq)
4.4. Translation Initation Rate (TIR) Calculators
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Michiels, J.E.; van den Bergh, B.; Verstraeten, N.; Michiels, J. Molecular mechanisms and clinical implications of bacterial persistence. Drug Resist. Updates 2016, 29, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Rowe, S.E.; Conlon, B.P.; Keren, I.; Lewis, K. Persisters: Methods for isolation and identifying contributing factors—A review. Methods Mol. Biol. 2016, 1333, 17–28. [Google Scholar] [PubMed]
- Korch, S.B.; Hill, T.M. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: Effects on macromolecular synthesis and persister formation. J. Bacteriol. 2006, 188, 3826–3836. [Google Scholar] [CrossRef] [PubMed]
- Fasani, R.A.; Savageau, M.A. Unrelated toxin-antitoxin systems cooperate to induce persistence. J. R. Soc. Interface 2015, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Shakespeare, L.J.; Jørgensen, M.G.; Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 13206–13211. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Maisonneuve, E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012, 66, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Goeders, N.; van Melderen, L. Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel) 2014, 6, 304–324. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Inouye, M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011, 9, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Leplae, R.; Geeraerts, D.; Hallez, R.; Guglielmini, J.; Dreze, P.; van Melderen, L. Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011, 39, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Rocker, A.; Meinhart, A. Type II toxin: Antitoxin systems. More than small selfish entities? Curr. Genet. 2016, 62, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.T.; Espinosa, M.; Yeo, C.C. Keeping the wolves at bay: Antitoxins of prokaryotic type II toxin-antitoxin systems. Front. Mol. Biosci. 2016, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, M.; Borch, J.; Gerdes, K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J. Mol. Biol. 2009, 394, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Vandervelde, A.; Drobnak, I.; Hadzi, S.; Sterckx, Y.G.; Welte, T.; de Greve, H.; Charlier, D.; Efremov, R.; Loris, R.; Lah, J. Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon. Nucleic Acids Res. 2017, 45, 2937–2950. [Google Scholar] [CrossRef] [PubMed]
- Hayes, F.; Kedzierska, B. Regulating toxin-antitoxin expression: Controlled detonation of intracellular molecular timebombs. Toxins (Basel) 2014, 6, 337–358. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elem. 2013, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cataudella, I.; Trusina, A.; Sneppen, K.; Gerdes, K.; Mitarai, N. Conditional cooperativity in toxin-antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Res. 2012, 40, 6424–6434. [Google Scholar] [CrossRef] [PubMed]
- Gelens, L.; Hill, L.; Vandervelde, A.; Danckaert, J.; Loris, R. A general model for toxin-antitoxin module dynamics can explain persister cell formation in E. coli. PLoS Comput. Biol. 2013, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Miki, K.; Koga, M.; Katayama, N.; Morimoto, W.; Takahashi, Y.; Yonesaki, T. IscR regulates RNase LS activity by repressing rnlA transcription. Genetics 2010, 185, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Ogle, C.T.; Mather, W.H. Proteolytically coordinated activation of toin-antitoxin modules. bioRxiv 2017, 1–24. [Google Scholar] [CrossRef]
- Quax, T.E.; Wolf, Y.I.; Koehorst, J.J.; Wurtzel, O.; van der Oost, R.; Ran, W.; Blombach, F.; Makarova, K.S.; Brouns, S.J.; Forster, A.C.; et al. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013, 4, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Rex, G.; Surin, B.; Besse, G.; Schneppe, B.; McCarthy, J.E. The mechanism of translational coupling in Escherichia coli. Higher order structure in the atpHA mRNA acts as a conformational switch regulating the access of de novo initiating ribosomes. J. Biol. Chem. 1994, 269, 18118–18127. [Google Scholar] [PubMed]
- Karp, P.D.; Weaver, D.; Paley, S.; Fulcher, C.; Kubo, A.; Kothari, A.; Krummenacker, M.; Subhraveti, P.; Weerasinghe, D.; Gama-Castro, S.; et al. The EcoCyc Database. EcoSal Plus 2014, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, D.H.; Rouskin, S.; Zhang, Y.; Li, G.W.; Weissman, J.S.; Gross, C.A. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife 2017, 6, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Armalyte, J.; Jurenaite, M.; Beinoraviciute, G.; Teiserskas, J.; Suziedeliene, E. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J. Bacteriol. 2012, 194, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Stanger, F.V.; Harms, A.; Dehio, C.; Schirmer, T. Crystal structure of the Escherichia coli Fic toxin-like protein in complex with its cognate antitoxin. PLoS ONE 2016, 11, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.G.; Pandey, D.P.; Jaskolska, M.; Gerdes, K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009, 191, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Inouye, M. Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins (Basel) 2017, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Hara, H.; Kato, I.; Inouye, M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005, 280, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Grigoriu, S.; Kim, Y.; Arruda, J.M.; Davenport, A.; Wood, T.K.; Peti, W.; Page, R. Three dimensional structure of the MqsR:MqsA complex: A novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.; Schuenemann, V.J.; Hand, N.J.; Silhavy, T.J.; Martin, J.; Lupas, A.N.; Djuranovic, S. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 2007, 372, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Zavialov, A.V.; Pavlov, M.Y.; Elf, J.; Gerdes, K.; Ehrenberg, M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003, 112, 131–140. [Google Scholar] [CrossRef]
- Zhang, Y.; Yamaguchi, Y.; Inouye, M. Characterization of YafO, an Escherichia coli toxin. J. Biol. Chem. 2009, 284, 25522–25531. [Google Scholar] [CrossRef] [PubMed]
- Grady, R.; Hayes, F. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 2003, 47, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, K.J.; Gerdes, K. HicA toxin of Escherichia coli derepresses hicAB transcription to selectively produce HicB antitoxin. Mol. Microbiol. 2017, 104, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Li, G.W.; Burkhardt, D.; Gross, C.; Weissman, J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 2014, 157, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T. Ribosome profiling: New views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014, 15, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Espah Borujeni, A.; Channarasappa, A.S.; Salis, H.M. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014, 42, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.W.; Yang, J.S.; Kim, I.; Yang, J.; Min, B.E.; Kim, S.; Jung, G.Y. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013, 15, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Barrick, D.; Villanueba, K.; Childs, J.; Kalil, R.; Schneider, T.D.; Lawrence, C.E.; Gold, L.; Stormo, G.D. Quantitative analysis of ribosome binding sites in E. coli. Nucleic Acids Res. 1994, 22, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Naka, K.; Koga, M.; Yonesaki, T.; Otsuka, Y. RNase HI stimulates the activity of RnlA toxin in Escherichia coli. Mol. Microbiol. 2014, 91, 596–605. [Google Scholar] [CrossRef] [PubMed]
- Naka, K.; Qi, D.; Yonesaki, T.; Otsuka, Y. RnlB antitoxin of the Escherichia coli RnlA-RnlB toxin-antitoxin module requires RNase HI for inhibition of RnlA toxin activity. Toxins (Basel) 2017, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Zhang, Y.; Inouye, M.; Ikura, M. Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J. Mol. Biol. 2008, 380, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, M.; Borch, J.; Jorgensen, M.G.; Gerdes, K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 2008, 69, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Ruangprasert, A.; Maehigashi, T.; Miles, S.J.; Giridharan, N.; Liu, J.X.; Dunham, C.M. Mechanisms of toxin inhibition and transcriptional repression by Escherichia coli DinJ-YafQ. J. Biol. Chem. 2014, 289, 20559–20569. [Google Scholar] [CrossRef] [PubMed]
- Meysman, P.; Sonego, P.; Bianco, L.; Fu, Q.; Ledezma-Tejeida, D.; Gama-Castro, S.; Liebens, V.; Michiels, J.; Laukens, K.; Marchal, K.; et al. COLOMBOS v2.0: An ever expanding collection of bacterial expression compendia. Nucleic Acids Res. 2014, 42, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Krishna, S.; Seshasayee, A.S.N. Regulation of global transcription in E. coli by Rsd and 6S RNA. bioRxiv 2016, 1–55. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Van der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy Array: A structure for efficient numerical computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar] [CrossRef]
| TA System | Toxin Function | Toxin Family |
|---|---|---|
| DinJ-YafQ | Endoribonuclease that act 5′ to adenine between the codon second and third nucleotides [28] | RelE [13] |
| FicAT | Mediates post-translational protein modification [29] | Unknown |
| HicAB | mRNase [30] | Unknown [31] |
| MazEF | mRNA interferase that cleaves mRNA at ACA sites [32] | CcdB/MazF [13] |
| MqsAR | Ribosome-independent RNase [33] | RelE [31] |
| PrlF-YhaV | Ribonuclease [34] | RelE [31] |
| RelBE | mRNA interferase that cleaves mRNA in the ribosome A site [35] | RelE [13] |
| RnlAB | RNase [22] | Unknown [31] |
| YafNO | Ribosome-dependent mRNA interferase [36] | YafO [13] |
| YefM-YoeB | Ribosome-dependent mRNase [37] | RelE [13] |
| TA System | A/T | St. Dev | Class |
|---|---|---|---|
| FicAT * | 1.99 | 0.53 | 1 |
| YefM-YoeB * | 1.89 | 0.42 | 1 |
| MazEF | 1.42 | 0.25 | 1 |
| PrlF-YhaV | 1.11 | 0.27 | 1 |
| MqsAR | 0.69 | 0.16 | 1 |
| RelBE | 0.95 | 0.10 | 1 |
| HicAB | 0.93 | 0.20 | 2 |
| DinJ-YafQ | 2.89 | 0.35 | 3 |
| YafNO | 2.08 | 0.39 | 3 |
| RnlAB ** | 1.01 | 0.25 | 4 |
| TA System | Class | mRNA Ratio | RBS Calculator | UTR Designer | Barrick Calculator | Synthesis Rates |
|---|---|---|---|---|---|---|
| FicAT | 1 | <2 * | + | + | + | LC |
| MazEF | 1 | <2 | + | + | + | + |
| MqsAR | 1 | <2 | − | + | + | + |
| PrlF-YhaV | 1 | <2 | + | + | + | + |
| RelBE | 1 | <2 | + | + | + | + |
| YefM-YoeB | 1 | <2 * | + | + | + | + |
| HicAB P1 | 2 | <2 | − | − | + | LC |
| HicAB P2 | + | − | + | |||
| DinJ-YafQ | 3 | >2 | + | − | − | + |
| YafNO | 3 | >2 | + | − | − | + |
| RnlAB | 4 | <2 ** | + | + | − | − |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deter, H.S.; Jensen, R.V.; Mather, W.H.; Butzin, N.C. Mechanisms for Differential Protein Production in Toxin–Antitoxin Systems. Toxins 2017, 9, 211. https://doi.org/10.3390/toxins9070211
Deter HS, Jensen RV, Mather WH, Butzin NC. Mechanisms for Differential Protein Production in Toxin–Antitoxin Systems. Toxins. 2017; 9(7):211. https://doi.org/10.3390/toxins9070211
Chicago/Turabian StyleDeter, Heather S., Roderick V. Jensen, William H. Mather, and Nicholas C. Butzin. 2017. "Mechanisms for Differential Protein Production in Toxin–Antitoxin Systems" Toxins 9, no. 7: 211. https://doi.org/10.3390/toxins9070211
APA StyleDeter, H. S., Jensen, R. V., Mather, W. H., & Butzin, N. C. (2017). Mechanisms for Differential Protein Production in Toxin–Antitoxin Systems. Toxins, 9(7), 211. https://doi.org/10.3390/toxins9070211





