pH-Triggered Conformational Switching along the Membrane Insertion Pathway of the Diphtheria Toxin T-Domain
Abstract
:1. Introduction
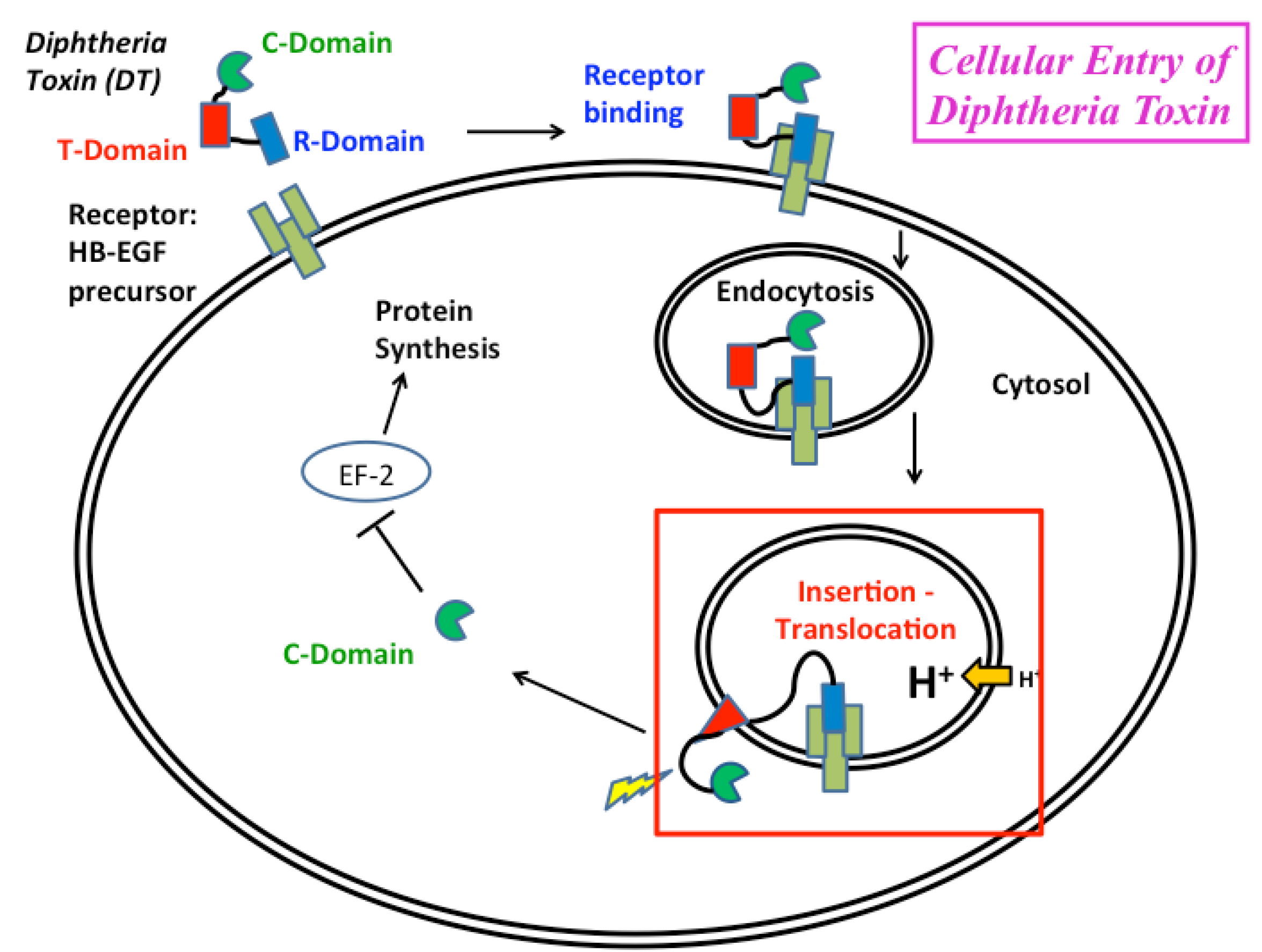
2. Overview of the Insertion Pathway
2.1. Summary of Early Studies

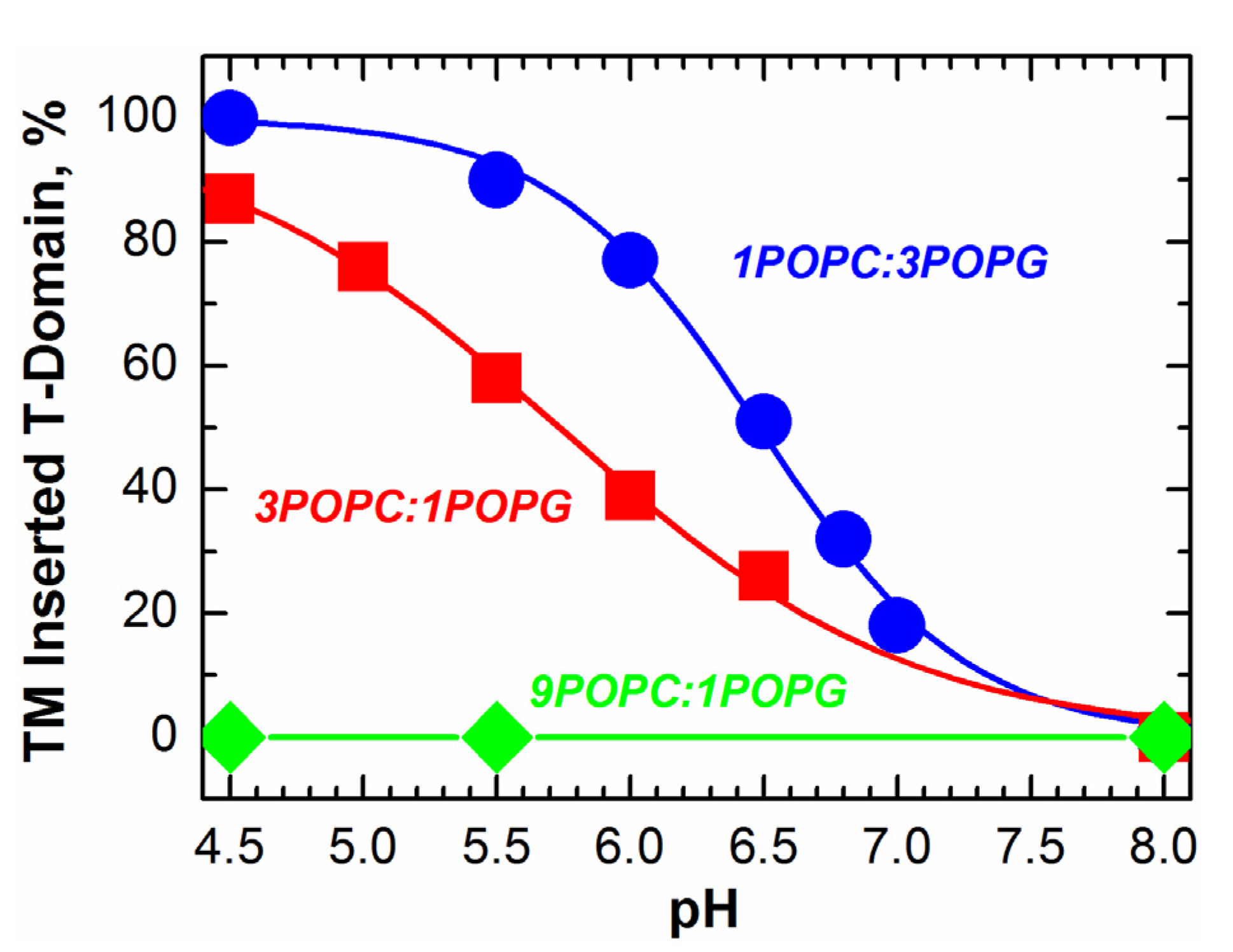
2.2. pH-Dependent Formation of Membrane-Competent Form

2.3. Kinetic Insertion Intermediates
2.4. Insertion Pathway with Two Staggered pH-Dependent Transitions
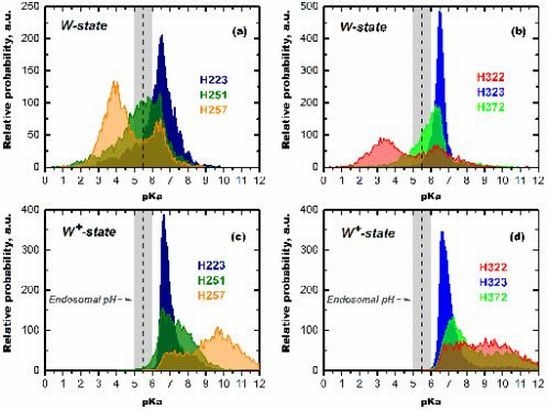
2.5. Multitude of TM-Inserted States Conundrum
3. Role of Histidine Protonation in Conformational Switching
3.1. Mutagenesis Studies
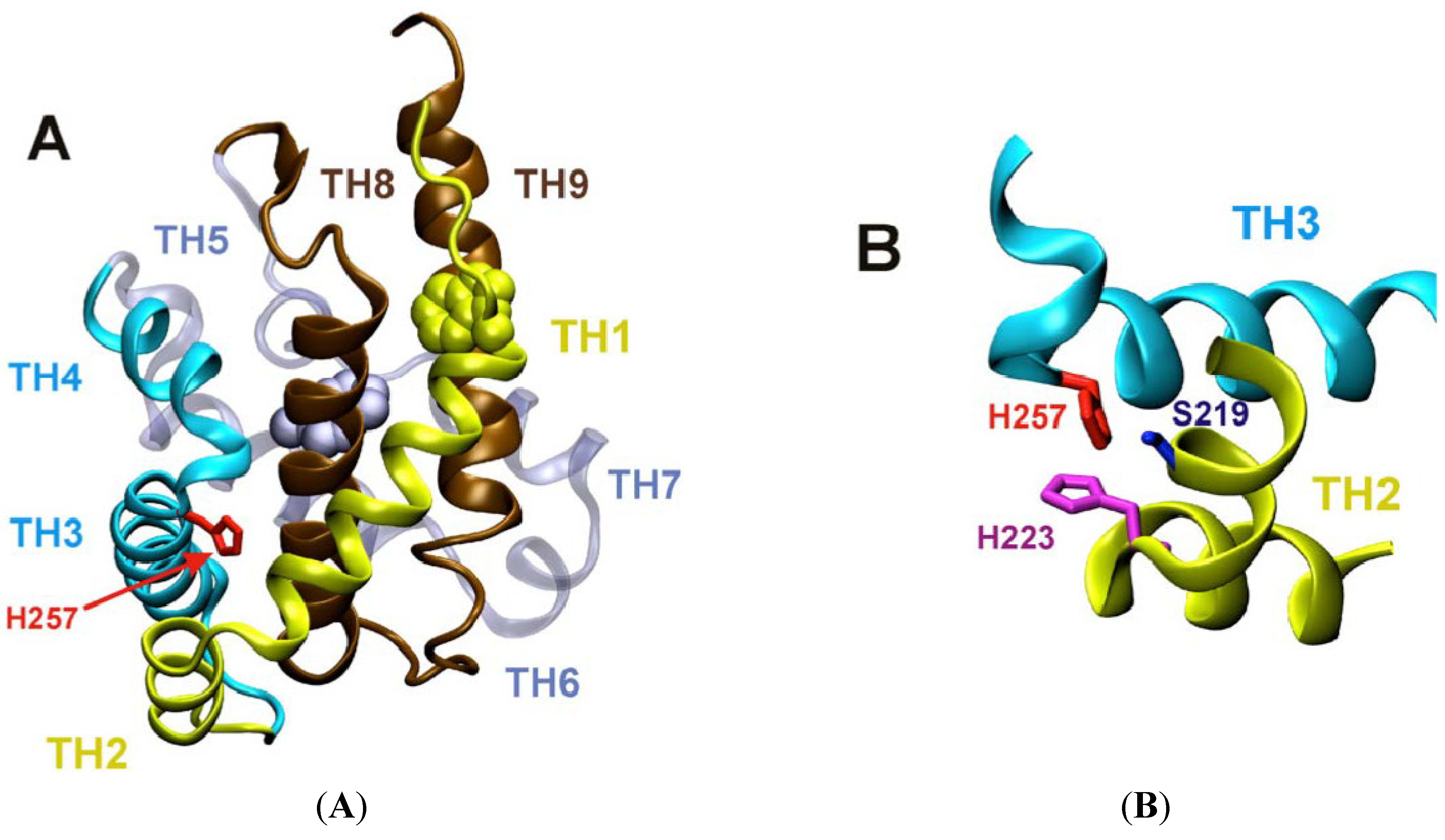
3.1.1. Role of H257 as a Major Component of pH-Dependent Conformational Switch
3.1.2. Role of C-Terminal Histidine Cluster in Membrane Insertion and Translocation
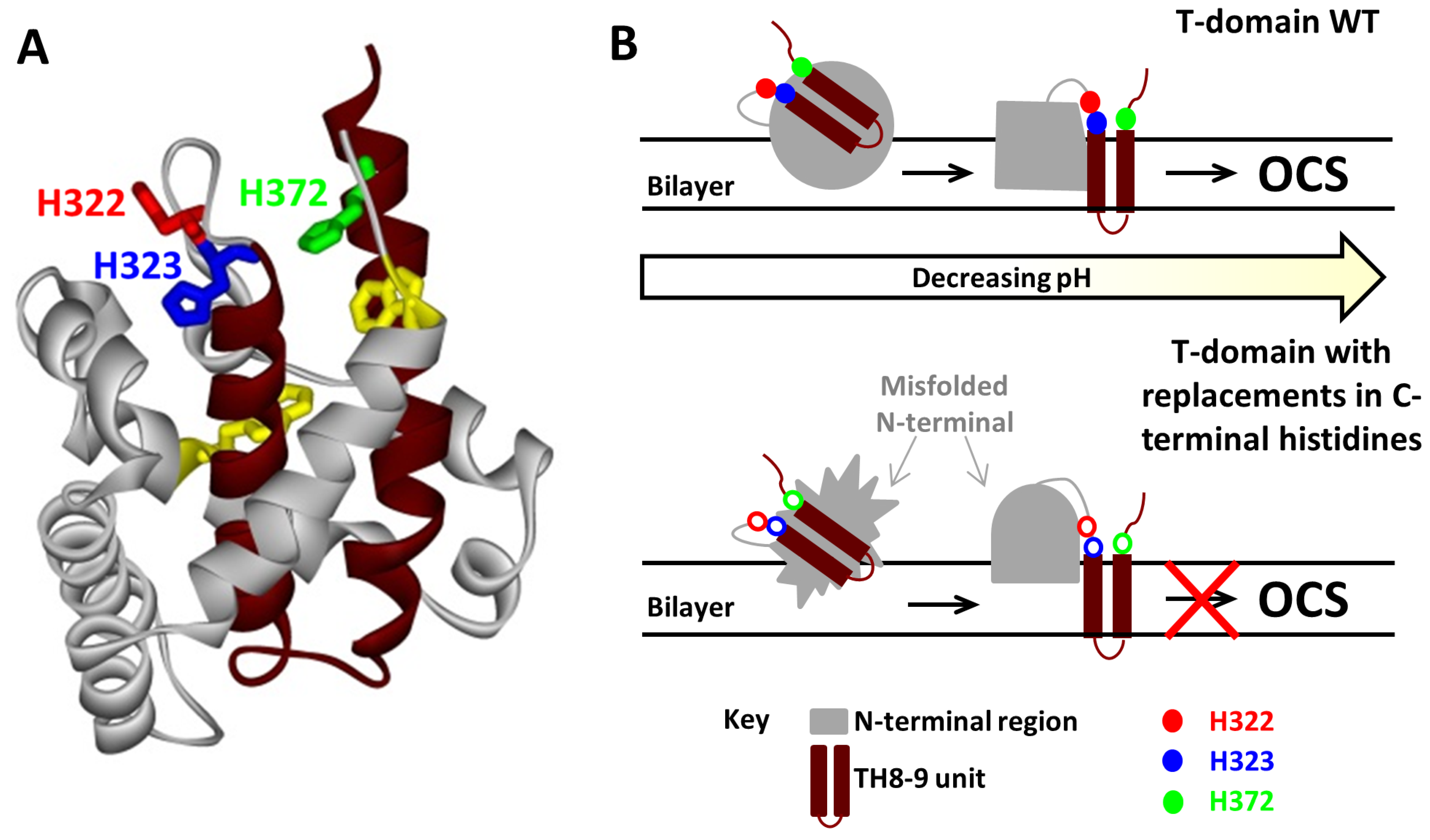
3.2. Computer Simulation Studies
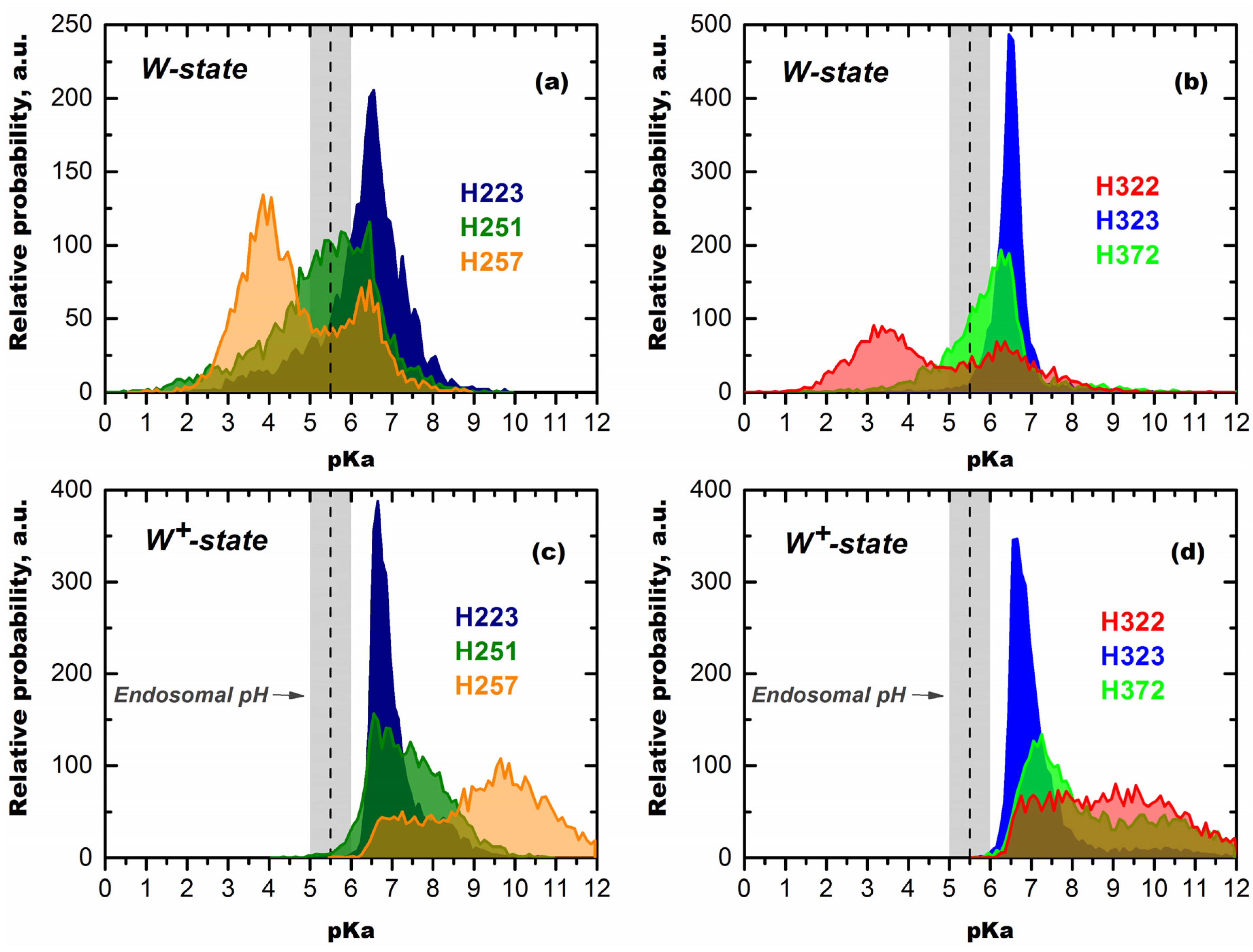
Mechanisms of pH-Trigger and Safety Latch Suggested by MD Simulations
4. Perspectives and Applications
Acknowledgments
Conflict of Interest
References
- Murphy, J.R. Mechanism of diphtheria toxin catalytic domain delivery to the eukaryotic cell cytosol and the cellular factors that directly participate in the process. Toxins 2011, 3, 294–308. [Google Scholar] [CrossRef]
- Hoch, D.H.; Romero-Mira, M.; Ehrlich, B.E.; Finkelstein, A.; DasGupta, B.R.; Simpson, L.L. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: Relevance to translocation of proteins. Proc. Natl. Acad. Sci. USA 1985, 82, 1692–1696. [Google Scholar] [CrossRef]
- Neale, E.A. Moving across membranes. Nat. Struct. Biol. 2003, 10, 2–3. [Google Scholar] [CrossRef]
- Koriazova, L.K.; Montal, M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat. Struct. Biol. 2003, 10, 13–18. [Google Scholar] [CrossRef]
- Collier, R.J.; Young, J.A. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 2003, 19, 45–70. [Google Scholar] [CrossRef]
- Oh, K.J.; Zhan, H.; Cui, C.; Hideg, K.; Collier, R.J.; Hubbell, W.L. Organization of diphtheria toxin T domain in bilayers: A site-directed spin labeling study. Science 1996, 273, 810–812. [Google Scholar]
- Oh, K.J.; Zhan, H.; Cui, C.; Altenbach, C.; Hubbell, W.L.; Collier, R.J. Conformation of the diphtheria toxin t domain in membranes: A site-directed spin-labeling study of the TH8 helix and TL5 loop. Biochemistry 1999, 38, 10336–10343. [Google Scholar] [CrossRef]
- Kachel, K.; Ren, J.H.; Collier, R.J.; London, E. Identifying transmembrane states and defining the membrane insertion boundaries of hydrophobic helices in membrane-inserted diphtheria toxin T domain. J. Biol. Chem. 1998, 273, 22950–22956. [Google Scholar] [CrossRef]
- Senzel, L.; Gordon, M.; Blaustein, R.O.; Oh, K.J.; Collier, R.J.; Finkelstein, A. Topography of diphtheria toxin’s T domain in the open channel state. J. Gen. Physiol. 2000, 115, 421–434. [Google Scholar] [CrossRef]
- Zhao, G.; London, E. Behavior of diphtheria toxin t domain containing substitutions that block normal membrane insertion at Pro345 and Leu307: Control of deep membrane insertion and coupling between deep insertion of hydrophobic subdomains. Biochemistry 2005, 44, 4488–4498. [Google Scholar] [CrossRef]
- Wang, Y.; Malenbaum, S.E.; Kachel, K.; Zhan, H.J.; Collier, R.J.; London, E. Identification of shallow and deep membrane-penetrating forms of diphtheria toxin T domain that are regulated by protein concentration and bilayer width. J. Biol. Chem. 1997, 272, 25091–25098. [Google Scholar]
- Chenal, A.; Savarin, P.; Nizard, P.; Guillain, F.; Gillet, D.; Forge, V. Membrane protein insertion regulated by bringing electrostatic and hydrophobic interactions into play. A case study with the translocation domain of the diphtheria toxin. J. Biol. Chem. 2002, 277, 43425–43432. [Google Scholar]
- Ladokhin, A.S.; Legmann, R.; Collier, R.J.; White, S.H. Reversible refolding of the diphtheria toxin T-domain on lipid membranes. Biochemistry 2004, 43, 7451–7458. [Google Scholar] [CrossRef]
- Palchevskyy, S.S.; Posokhov, Y.O.; Olivier, B.; Popot, J.L.; Pucci, B.; Ladokhin, A.S. Chaperoning of insertion of membrane proteins into lipid bilayers by hemifluorinated surfactants: Application to diphtheria toxin. Biochemistry 2006, 45, 2629–2635. [Google Scholar] [CrossRef]
- Montagner, C.; Perier, A.; Pichard, S.; Vernier, G.; Menez, A.; Gillet, D.; Forge, V.; Chenal, A. Behavior of the N-terminal helices of the diphtheria toxin T domain during the successive steps of membrane interaction. Biochemistry 2007, 46, 1878–1887. [Google Scholar] [CrossRef]
- Perier, A.; Chassaing, A.; Raffestin, S.; Pichard, S.; Masella, M.; Menez, A.; Forge, V.; Chenal, A.; Gillet, D. Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. J. Biol. Chem. 2007, 282, 24239–24245. [Google Scholar] [CrossRef]
- Posokhov, Y.O.; Rodnin, M.V.; Das, S.K.; Pucci, B.; Ladokhin, A.S. FCS study of the thermodynamics of membrane protein insertion into the lipid bilayer chaperoned by fluorinated surfactants. Biophys. J. 2008, 95, L54–L56. [Google Scholar] [CrossRef]
- Bennett, M.J.; Eisenberg, D. Refined structure of monomeric diphtheria toxin at 2.3 å resolution. Protein Sci. 1994, 3, 1464–1475. [Google Scholar] [CrossRef]
- Weiss, M.S.; Blanke, S.R.; Collier, R.J.; Eisenberg, D. Structure of the isolated catalytic domain of diphtheria toxin. Biochemistry 1995, 34, 773–781. [Google Scholar] [CrossRef]
- Oh, K.J.; Senzel, L.; Collier, R.J.; Finkelstein, A. Translocation of the catalytic domain of diphtheria toxin across planar phospholipid bilayers by its own T domain. Proc. Natl. Acad. Sci. USA 1999, 96, 8467–8470. [Google Scholar]
- Senzel, L.; Huynh, P.D.; Jakes, K.S.; Collier, R.J.; Finkelstein, A. The diphtheria toxin channel-forming T domain translocates its own NH2-terminal region across planar bilayers. J. Gen. Physiol. 1998, 112, 317–324. [Google Scholar] [CrossRef]
- Mindell, J.A.; Silverman, J.A.; Collier, R.J.; Finkelstein, A. Structure function relationships in diphtheria toxin channels. II. A residue responsible for the channel's dependence on trans pH. J. Membr. Biol. 1994, 137, 29–44. [Google Scholar]
- Mindell, J.A.; Silverman, J.A.; Collier, R.J.; Finkelstein, A. Structure-function relationships in diphtheria toxin channels. III. Residues which affect the cis pH dependence of channel conductance. J. Membr. Biol. 1994, 137, 45–57. [Google Scholar]
- Huynh, P.D.; Cui, C.; Zhan, H.J.; Oh, K.J.; Collier, R.J.; Finkelstein, A. Probing the structure of the diphtheria toxin channel: Reactivity in planar lipid bilayer membranes of cysteine-substituted mutant channels with methanethiosulfonate derivatives. J. Gen. Physiol. 1997, 110, 229–242. [Google Scholar] [CrossRef]
- Gordon, M.; Finkelstein, A. The number of subunits comprising the channel formed by the T domain of diphtheria toxin. J. Gen. Physiol. 2001, 118, 471–480. [Google Scholar] [CrossRef]
- Kyrychenko, A.; Posokhov, Y.O.; Rodnin, M.V.; Ladokhin, A.S. Kinetic intermediate reveals staggered pH-dependent transitions along the membrane insertion pathway of the diphtheria toxin t-domain. Biochemistry 2009, 48, 7584–7594. [Google Scholar] [CrossRef]
- Rodnin, M.V.; Kyrychenko, A.; Kienker, P.; Sharma, O.; Posokhov, Y.O.; Collier, R.J.; Finkelstein, A.; Ladokhin, A.S. Conformational switching of the diphtheria toxin T domain. J. Mol. Biol. 2010, 402, 1–7. [Google Scholar] [CrossRef]
- Kurnikov, I.V.; Kyrychenko, A.; Flores-Canales, J.C.; Rodnin, M.V.; Simakov, N.; Vargas-Uribe, M.; Posokhov, Y.O.; Kurnikova, M.; Ladokhin, A.S. pH-Triggered conformational switching of the diphtheria toxin t-domain: The roles of N-terminal histidines. J. Mol. Biol. 2013, 425, 2752–2764. [Google Scholar] [CrossRef]
- Vargas-Uribe, M.; Rodnin, M.V.; Kienker, P.; Finkelstein, A.; Ladokhin, A.S. Crucial role of H322 in folding of the diphtheria toxin T-domain into the open-channel state. Biochemistry 2013, 52, 3457–3463. [Google Scholar] [CrossRef]
- Murphy, R.F.; Powers, S.; Cantor, C.R. Endosome ph measured in single cells by dual fluorescence flow cytometry: Rapid acidification of insulin to pH 6. J. Cell Biol. 1984, 98, 1757–1762. [Google Scholar] [CrossRef]
- Yamashiro, D.J.; Maxfield, F.R. Kinetics of endosome acidification in mutant and wild-type chinese hamster ovary cells. J. Cell Biol. 1987, 105, 2713–2721. [Google Scholar] [CrossRef]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef]
- Shaw, D.E.; Bowers, K.J.; Chow, E.; Eastwood, M.P.; Ierardi, D.J.; Klepeis, J.L.; Kuskin, J.S.; Larson, R.H.; Lindorff-Larsen, K.; Maragakis, P.; et al. Millisecond-Scale Molecular Dynamics Simulations on Anton. In Proceedings of the Conference on High Performance Computing NetworkingStorage and Analysis—SC’09, Portland, OR, USA, 14–20 November 2009; ACM Press: New York, NY, USA, 2009; p. 1. [Google Scholar]
- Shaw, D.E.; Chao, J.C.; Eastwood, M.P.; Gagliardo, J.; Grossman, J.P.; Ho, C.R.; Ierardi, D.J.; Kolossváry, I.; Klepeis, J.L.; Layman, T.; et al. ANTON, a special-purpose machine for molecular dynamics simulation. ACM SIGARCH Comput. Archit. News 2007, 35, 1–12. [Google Scholar]
- Ladokhin, A.S.; White, S.H. Interfacial folding and membrane insertion of a designed helical peptide. Biochemistry 2004, 43, 5782–5791. [Google Scholar] [CrossRef]
- Ladokhin, A.S.; White, S.H. Protein chemistry at membrane interfaces: Non-additivity of electrostatic and hydrophobic interactions. J. Mol. Biol. 2001, 309, 543–552. [Google Scholar] [CrossRef]
- Posokhov, Y.O.; Gottlieb, P.A.; Morales, M.J.; Sachs, F.; Ladokhin, A.S. Is lipid bilayer binding a common property of inhibitor cysteine knot ion-channel blockers? Biophys. J. 2007, 93, L20–L22. [Google Scholar] [CrossRef]
- Ladokhin, A.S.; White, S.H. Folding of amphipathic α-helices on membranes: Energetics of helix formation by melittin. J. Mol. Biol. 1999, 285, 1363–1369. [Google Scholar] [CrossRef]
- Fernandez-Vidal, M.; Jayasinghe, S.; Ladokhin, A.S.; White, S.H. Folding amphipathic helices into membranes: Amphiphilicity trumps hydrophobicity. J. Mol. Biol. 2007, 370, 459–470. [Google Scholar] [CrossRef]
- Ladokhin, A.S.; Isas, J.M.; Haigler, H.T.; White, S.H. Determining the membrane topology of proteins: Insertion pathway of a transmembrane helix of annexin 12. Biochemistry 2002, 41, 13617–13626. [Google Scholar] [CrossRef]
- Posokhov, Y.O.; Rodnin, M.V.; Lu, L.; Ladokhin, A.S. Membrane insertion pathway of annexin b12: Thermodynamic and kinetic characterization by fluorescence correlation spectroscopy and fluorescence quenching. Biochemistry 2008, 47, 5078–5087. [Google Scholar] [CrossRef]
- Rodnin, M.V.; Kyrychenko, A.; Kienker, P.; Sharma, O.; Vargas-Uribe, M.; Collier, R.J.; Finkelstein, A.; Ladokhin, A.S. Replacement of C-terminal histidines uncouples membrane insertion and translocation in diphtheria toxin T-domain. Biophys. J. 2011, 101, L41–L43. [Google Scholar] [CrossRef]
- Rodnin, M.V.; Posokhov, Y.O.; Contino-Pepin, C.; Brettmann, J.; Kyrychenko, A.; Palchevskyy, S.S.; Pucci, B.; Ladokhin, A.S. Interactions of fluorinated surfactants with diphtheria toxin T-domain: Testing new media for studies of membrane proteins. Biophys. J. 2008, 94, 4348–4357. [Google Scholar] [CrossRef]
- Posokhov, Y.O.; Ladokhin, A.S. Lifetime fluorescence method for determining membrane topology of proteins. Anal. Biochem. 2006, 348, 87–93. [Google Scholar] [CrossRef]
- Ladokhin, A.S.; Jayasinghe, S.; White, S.H. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 2000, 285, 235–245. [Google Scholar] [CrossRef]
- Chenal, A.; Prongidi-Fix, L.; Perier, A.; Aisenbrey, C.; Vernier, G.; Lambotte, S.; Haertlein, M.; Dauvergne, M.T.; Fragneto, G.; Bechinger, B.; et al. Deciphering membrane insertion of the diphtheria toxin T domain by specular neutron reflectometry and solid-state nmr spectroscopy. J. Mol. Biol. 2009, 391, 872–883. [Google Scholar] [CrossRef]
- Wimalasena, D.S.; Cramer, J.C.; Janowiak, B.E.; Juris, S.J.; Melnyk, R.A.; Anderson, D.E.; Kirk, K.L.; Collier, R.J.; Bann, J.G. Effect of 2-fluorohistidine labeling of the anthrax protective antigen on stability, pore formation, and translocation. Biochemistry 2007, 46, 14928–14936. [Google Scholar] [CrossRef]
- Buckley, J.T.; Wilmsen, H.U.; Lesieur, C.; Schulze, A.; Pattus, F.; Parker, M.W.; van der Goot, F.G. Protonation of histidine-132 promotes oligomerization of the channel-forming toxin aerolysin. Biochemistry 1995, 34, 16450–16455. [Google Scholar] [CrossRef]
- Wimley, W.C.; White, S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996, 3, 842–848. [Google Scholar] [CrossRef]
- Ren, J.; Kachel, K.; Kim, H.; Malenbaum, S.E.; Collier, R.J.; London, E. Interaction of diphtheria toxin t domain with molten globule-like proteins and its implications for translocation. Science 1999, 284, 955–957. [Google Scholar] [CrossRef]
- Kakimoto, S.; Hamada, T.; Komatsu, Y.; Takagi, M.; Tanabe, T.; Azuma, H.; Shinkai, S.; Nagasaki, T. The conjugation of diphtheria toxin T domain to poly(ethylenimine) based vectors for enhanced endosomal escape during gene transfection. Biomaterials 2009, 30, 402–408. [Google Scholar] [CrossRef]
- Zhang, Y.; Schulte, W.; Pink, D.; Phipps, K.; Zijlstra, A.; Lewis, J.D.; Waisman, D.M. Sensitivity of cancer cells to truncated diphtheria toxin. PLoS One 2010, 5, e10498. [Google Scholar]
- Wild, R.; Yokoyama, Y.; Dings, R.P.; Ramakrishnan, S. Vegf-dt385 toxin conjugate inhibits mammary adenocarcinoma development in a transgenic mouse model of spontaneous tumorigenesis. Breast Cancer Res. Treat. 2004, 85, 161–171. [Google Scholar] [CrossRef]
- Urieto, J.O.; Liu, T.; Black, J.H.; Cohen, K.A.; Hall, P.D.; Willingham, M.C.; Pennell, L.K.; Hogge, D.E.; Kreitman, R.J.; Frankel, A.E. Expression and purification of the recombinant diphtheria fusion toxin dt388il3 for phase I clinical trials. Protein Expr. Purif. 2004, 33, 123–133. [Google Scholar] [CrossRef]
- Turturro, F. Denileukin diftitox: A biotherapeutic paradigm shift in the treatment of lymphoid-derived disorders. Expert Rev. Anticancer Ther. 2007, 7, 11–17. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Olson, T.A.; Bautch, V.L.; Mohanraj, D. Vascular endothelial growth factor-toxin conjugate specifically inhibits kdr/flk-1-positive endothelial cell proliferation in vitro and angiogenesis in vivo. Cancer Res. 1996, 56, 1324–1330. [Google Scholar]
- Ramage, J.G.; Vallera, D.A.; Black, J.H.; Aplan, P.D.; Kees, U.R.; Frankel, A.E. The diphtheria toxin/urokinase fusion protein (dtat) is selectively toxic to cd87 expressing leukemic cells. Leuk. Res. 2003, 27, 79–84. [Google Scholar] [CrossRef]
- Murphy, J.R.; Bishai, W.; Borowski, M.; Miyanohara, A.; Boyd, J.; Nagle, S. Genetic construction, expression, and melanoma-selective cytotoxicity of a diphtheria toxin-related alpha-melanocyte-stimulating hormone fusion protein. Proc. Natl. Acad. Sci. USA 1986, 83, 8258–8262. [Google Scholar] [CrossRef]
- Kreitman, R.J. Immunotoxins for targeted cancer therapy. Aaps J. 2006, 8, E532–E551. [Google Scholar] [CrossRef]
- Hogge, D.E.; Yalcintepe, L.; Wong, S.H.; Gerhard, B.; Frankel, A.E. Variant diphtheria toxin-interleukin-3 fusion proteins with increased receptor affinity have enhanced cytotoxicity against acute myeloid leukemia progenitors. Clin. Cancer Res. 2006, 12, 1284–1291. [Google Scholar] [CrossRef]
- Hall, P.D.; Willingham, M.C.; Kreitman, R.J.; Frankel, A.E. Dt388-gm-csf, a novel fusion toxin consisting of a truncated diphtheria toxin fused to human granulocyte-macrophage colony-stimulating factor, prolongs host survival in a scid mouse model of acute myeloid leukemia. Leukemia 1999, 13, 629–633. [Google Scholar] [CrossRef]
- Feuring-Buske, M.; Frankel, A.; Gerhard, B.; Hogge, D. Variable cytotoxicity of diphtheria toxin 388-granulocyte-macrophage colony-stimulating factor fusion protein for acute myelogenous leukemia stem cells. Exp. Hematol. 2000, 28, 1390–1400. [Google Scholar] [CrossRef]
- Duvic, M.; Talpur, R. Optimizing denileukin diftitox (ontak) therapy. Future Oncol. 2008, 4, 457–469. [Google Scholar] [CrossRef]
- Cohen, K.A.; Liu, T.F.; Cline, J.M.; Wagner, J.D.; Hall, P.D.; Frankel, A.E. Toxicology and pharmacokinetics of dt388il3, a fusion toxin consisting of a truncated diphtheria toxin (dt388) linked to human interleukin 3 (il3), in cynomolgus monkeys. Leuk. Lymphoma 2004, 45, 1647–1656. [Google Scholar] [CrossRef]
- Black, J.H.; McCubrey, J.A.; Willingham, M.C.; Ramage, J.; Hogge, D.E.; Frankel, A.E. Diphtheria toxin-interleukin-3 fusion protein (dt(388)il3) prolongs disease-free survival of leukemic immunocompromised mice. Leukemia 2003, 17, 155–159. [Google Scholar] [CrossRef]
- Reshetnyak, Y.K.; Andreev, O.A.; Lehnert, U.; Engelman, D.M. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc. Natl. Acad. Sci. USA 2006, 103, 6460–6465. [Google Scholar] [CrossRef]
- Reshetnyak, Y.K.; Segala, M.; Andreev, O.A.; Engelman, D.M. A monomeric membrane peptide that lives in three worlds: In solution, attached to, and inserted across lipid bilayer. Biophys. J. 2007, 93, 2363–2372. [Google Scholar] [CrossRef]
- Antignani, A.; Youle, R.J. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr. Opin. Cell Biol. 2006, 18, 685–689. [Google Scholar] [CrossRef]
- Youle, R.J.; Strasser, A. The Bcl-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef]
- Minn, A.J.; Velez, P.; Schendel, S.L.; Liang, H.; Muchmore, S.W.; Fesik, S.W.; Fill, M.; Thompson, C.B. Bcl-X(L) forms an ion channel in synthetic lipid membranes. Nature 1997, 385, 353–357. [Google Scholar] [CrossRef]
- Hsu, Y.T.; Wolter, K.G.; Youle, R.J. Cytosol-to-membrane redistribution of bax and Bcl-X(L) during apoptosis. Proc. Natl. Acad. Sci. USA 1997, 94, 3668–3672. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ladokhin, A.S. pH-Triggered Conformational Switching along the Membrane Insertion Pathway of the Diphtheria Toxin T-Domain. Toxins 2013, 5, 1362-1380. https://doi.org/10.3390/toxins5081362
Ladokhin AS. pH-Triggered Conformational Switching along the Membrane Insertion Pathway of the Diphtheria Toxin T-Domain. Toxins. 2013; 5(8):1362-1380. https://doi.org/10.3390/toxins5081362
Chicago/Turabian StyleLadokhin, Alexey S. 2013. "pH-Triggered Conformational Switching along the Membrane Insertion Pathway of the Diphtheria Toxin T-Domain" Toxins 5, no. 8: 1362-1380. https://doi.org/10.3390/toxins5081362
APA StyleLadokhin, A. S. (2013). pH-Triggered Conformational Switching along the Membrane Insertion Pathway of the Diphtheria Toxin T-Domain. Toxins, 5(8), 1362-1380. https://doi.org/10.3390/toxins5081362