Integrated Assessment of Fungi Contamination and Mycotoxins Levels Across the Rice Processing Chain
Abstract
1. Introduction
2. Results and Discussion
2.1. Method Validation
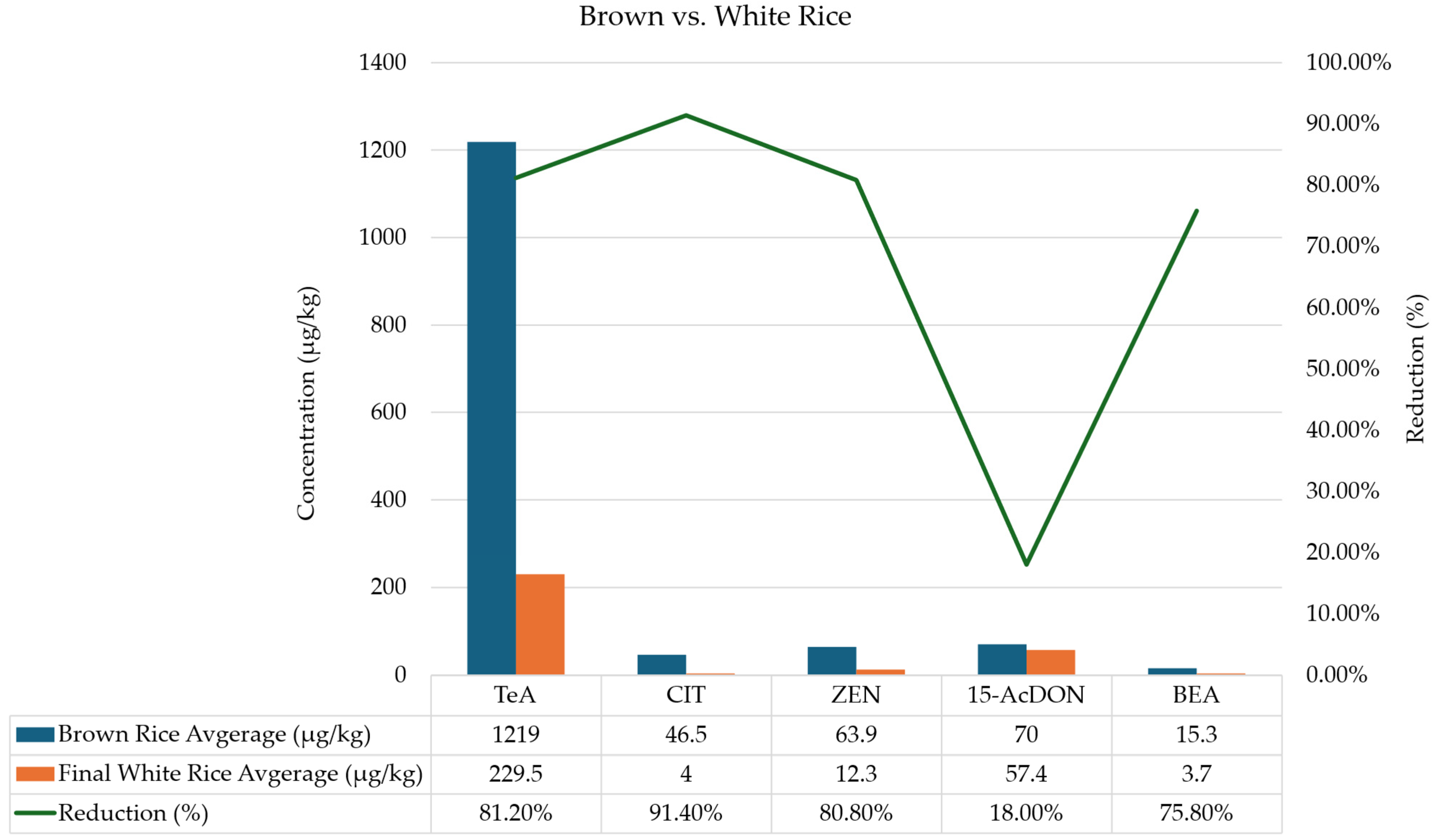
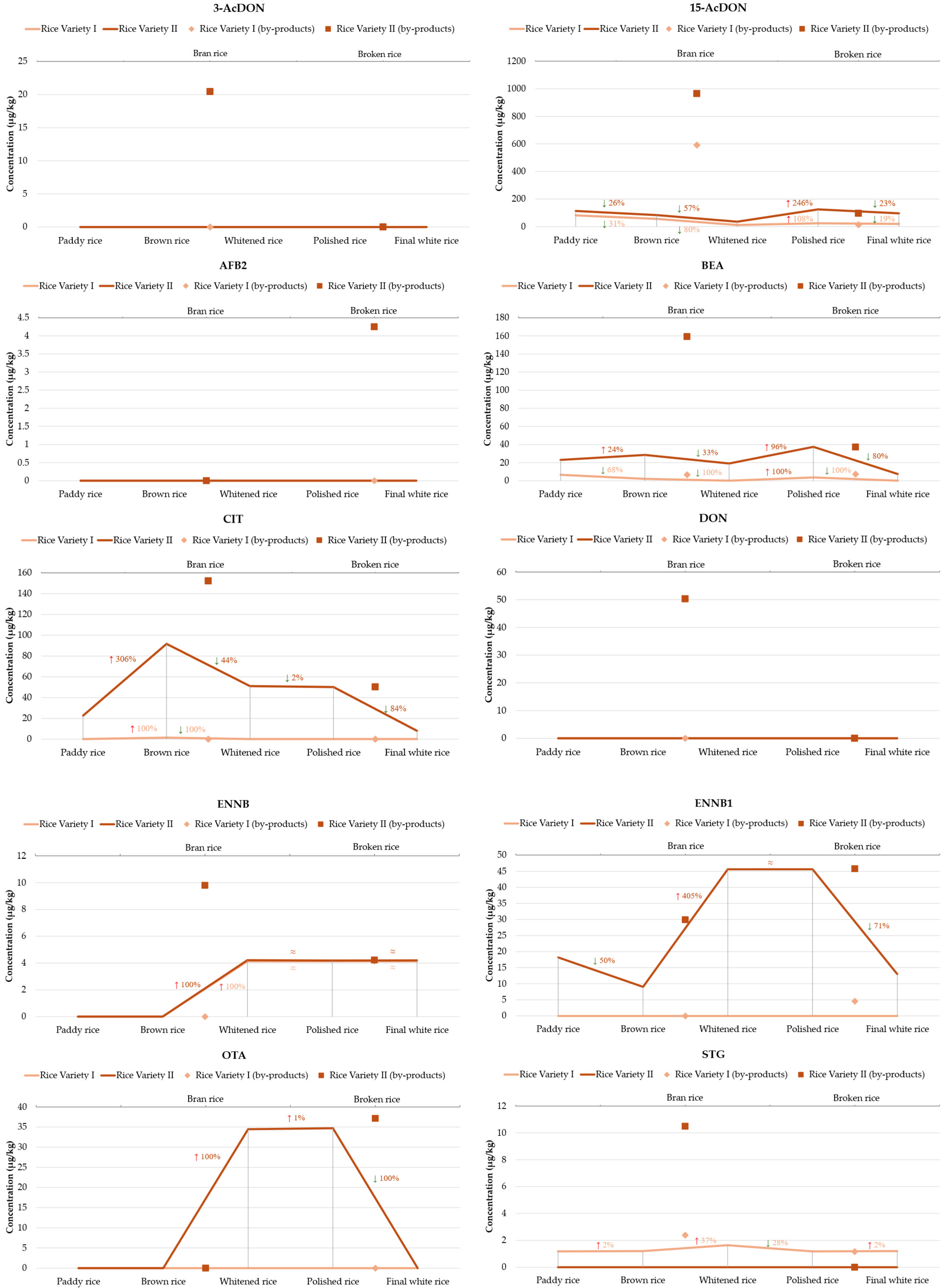
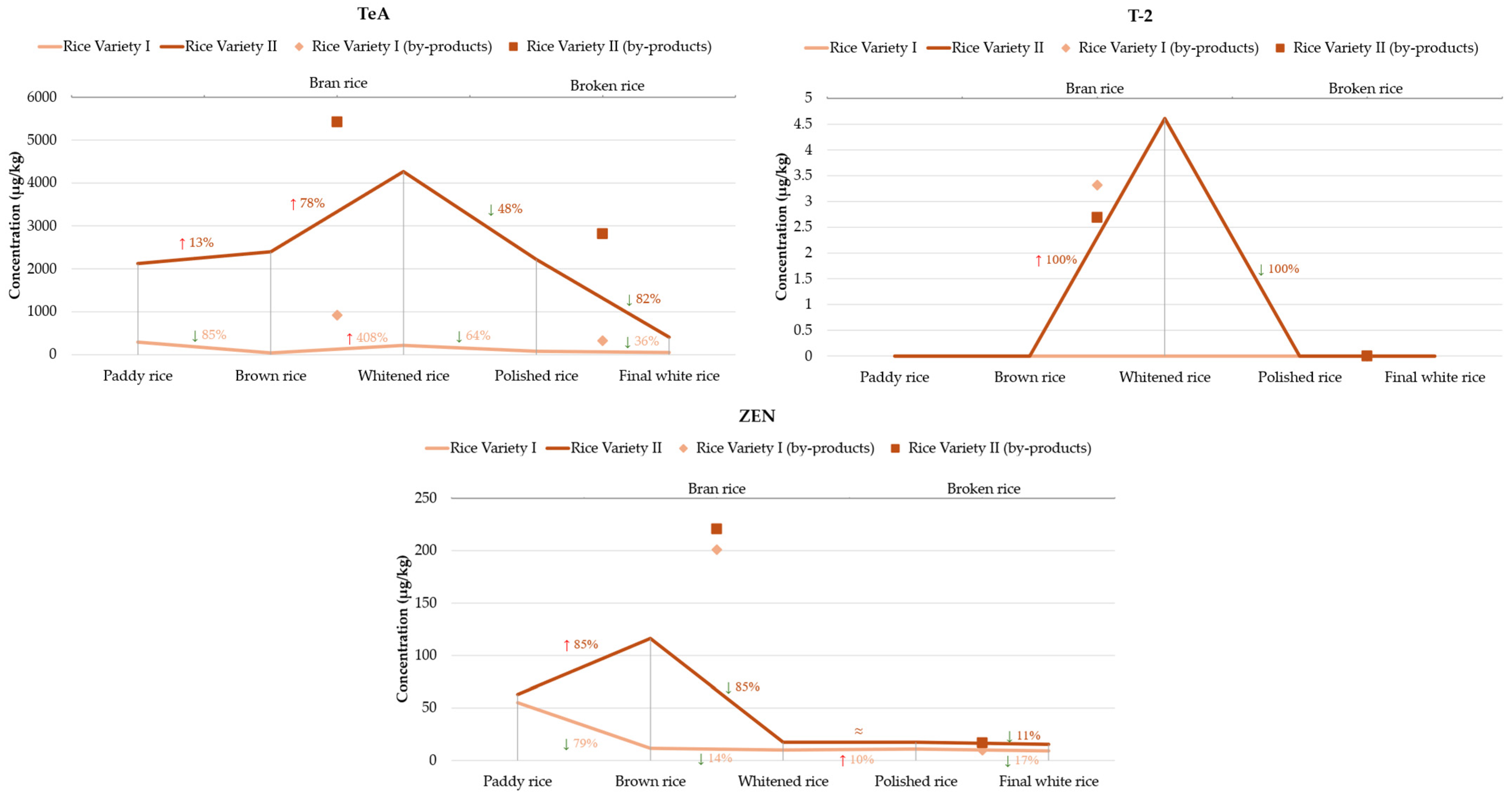
2.2. Occurrence and Distribution of Mycotoxins Along the Rice Processing Chain
2.3. Mycoflora Isolation and Identification
3. Conclusions
4. Materials and Methods
4.1. Reagents and Standard Solutions
4.2. Sampling
4.3. Determination of Mycotoxins by UHPLC-MS/MS
4.3.1. Extraction Protocol
4.3.2. Instrument and Analytical Conditions
4.3.3. Method Validation and Quality Control
4.4. Mycoflora Isolation and Identification
4.4.1. Fugal Isolation
4.4.2. Morphological Identification
4.4.3. Molecular Identification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Rice Market Monitor; Food and Agriculture Organization of the United Nations: Rome, Italy, 2019. [Google Scholar]
- FAO. Crop Prospects and Food Situation—Triannual Global Report; Food and Agriculture Organization of the United Nations: Rome, Italy, 2025. [Google Scholar]
- Atungulu, G.G.; Pan, Z. Rice industrial processing worldwide and impact on macro- and micronutrient content, stability, and retention. Ann. N. Y. Acad. Sci. 2014, 1324, 15–28. [Google Scholar] [CrossRef]
- Gonçalves, A.; Gkrillas, A.; Dorne, J.L.; Dall’Asta, C.; Palumbo, R.; Lima, N.; Battilani, P.; Venâncio, A.; Giorni, P. Pre- and Postharvest Strategies to Minimize Mycotoxin Contamination in the Rice Food Chain. Compr. Rev. Food. Sci. Food Saf. 2019, 18, 441–454. [Google Scholar] [CrossRef]
- Su, Q.; Rohila, J.S.; Ranganathan, S.; Karthikeyan, R. Rice yield and quality in response to daytime and nighttime temperature increase—A meta-analysis perspective. Sci. Total Environ. 2023, 898, 165256. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 4th ed.; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Medina, Á.; Rodríguez, A.; Magan, N. Climate change and mycotoxigenic fungi: Impacts on mycotoxin production. Curr. Opin. Food Sci. 2015, 5, 99–104. [Google Scholar] [CrossRef]
- Ferre, F.S. Worldwide occurrence of mycotoxins in rice. Food Control 2016, 62, 291–298. [Google Scholar] [CrossRef]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25%. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef]
- Santos, A.R.; Carreiró, F.; Freitas, A.; Barros, S.; Brites, C.; Ramos, F.; Sanches Silva, A. Mycotoxins Contamination in Rice: Analytical Methods, Occurrence and Detoxification Strategies. Toxins 2022, 14, 647. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Lee, S.-H.; Lee, S.-H.; Shin, J.Y.; Yun, J.-C.; Lee, Y.-W.; Ryu, J.-G. Occurrence of Fusarium Mycotoxins in Rice and Its Milling By-Products in Korea. J. Food Prot. 2011, 74, 1169–1174. [Google Scholar] [CrossRef]
- Marin, S.; Ramos, A.J.; Cano-Sancho, G.; Sanchis, V. Mycotoxins: Occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. 2013, 60, 218–237. [Google Scholar] [CrossRef]
- Qi, Z.; Zhou, X.; Tian, L.; Zhang, H.; Cai, L.; Tang, F. Distribution of mycotoxin-producing fungi across major rice production areas of China. Food Control 2022, 134, 108572. [Google Scholar] [CrossRef]
- Awuchi, C.G.; Ondari, E.N.; Nwozo, S.; Odongo, G.A.; Eseoghene, I.J.; Twinomuhwezi, H.; Ogbonna, C.U.; Upadhyay, A.K.; Adeleye, A.O.; Okpala, C.O.R. Mycotoxins’ Toxicological Mechanisms Involving Humans, Livestock and Their Associated Health Concerns: A Review. Toxins 2022, 14, 167. [Google Scholar] [CrossRef]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef]
- Dey, D.K.; Kang, J.I.; Bajpai, V.K.; Kim, K.; Lee, H.; Sonwal, S.; Simal-Gandara, J.; Xiao, J.; Ali, S.; Huh, Y.S.; et al. Mycotoxins in food and feed: Toxicity, preventive challenges, and advanced detection techniques for associated diseases. Crit. Rev. Food Sci. Nutr. 2023, 63, 8489–8510. [Google Scholar] [CrossRef] [PubMed]
- The European Commission. COMMISSION REGULATION (EU) 2023/915 on maximum levels for certain contaminants in food and repealing Regulation (EC) No 1881/2006. Off. J. Eur. Union 2023. Available online: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:02006R1881-20230326 (accessed on 1 July 2025).
- Milani, J.; Maleki, G. Effects of processing on mycotoxin stability in cereals. J. Sci. Food Agric. 2014, 94, 2372–2375. [Google Scholar] [CrossRef]
- Müller, A.; Nunes, M.T.; Maldaner, V.; Coradi, P.C.; Moraes, R.S.d.; Martens, S.; Leal, A.F.; Pereira, V.F.; Marin, C.K. Rice Drying, Storage and Processing: Effects of Post-Harvest Operations on Grain Quality. Rice Sci. 2022, 29, 16–30. [Google Scholar] [CrossRef]
- Sawicka, B. Post-Harvest Losses of Agricultural Produce. In Zero Hunger; Leal Filho, W., Azul, A.M., Brandli, L., Özuyar, P.G., Wall, T., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 654–669. [Google Scholar] [CrossRef]
- He, Z.; Zhang, Z.; Valè, G.; San Segundo, B.; Chen, X.; Pasupuleti, J. Editorial: Disease and pest resistance in rice. Front. Plant Sci. 2023, 14, 1333904. [Google Scholar] [CrossRef]
- The European Commission. Community Regulation No. 401_2006 of 23 February 2006—Laying down the methods of sampling and analysis for the official control of the levels of mycotoxins in foodstuffs. Off. J. Eur. Union 2006. Available online: http://data.europa.eu/eli/reg/2006/401/oj (accessed on 1 July 2025).
- The European Commission. COMMISSION REGULATION (EU) 2024/1022 of 8 April 2024 amending Regulation (EU) 2023/915 as regards maximum levels of deoxynivalenol in food. Off. J. Eur. Union 2024. Available online: http://data.europa.eu/eli/reg/2024/1022/oj (accessed on 1 July 2025).
- The European Commission. COMMISSION REGULATION (EU) 2024/1038 of 9 April 2024 amending Regulation (EU) 2023/915 as regards maximum levels of T-2 and HT-2 toxins in food. Off. J. Eur. Union 2024. Available online: http://data.europa.eu/eli/reg/2024/1038/oj (accessed on 1 July 2025).
- The European Commission. COMMISSION REGULATION (EU) 2024/1756 of 25 June 2024 amending and correcting Regulation (EU) 2023/915 on maximum levels for certain contaminants in food. Off. J. Eur. Union 2024. Available online: http://data.europa.eu/eli/reg/2024/1756/oj (accessed on 1 July 2025).
- Ranathunga, A.; Thumanu, K.; Kiatponglarp, W.; Siriwong, S.; Wansuksri, R.; Suwannaporn, P. Image mapping of biological changes and structure-function relationship during rice grain development via Synchrotron FTIR spectroscopy. Food Chem. Adv. 2023, 2, 100290. [Google Scholar] [CrossRef]
- Phan, L.T.K.; Nguyen, T.T.N.; Tran, T.T.T.; De Saeger, S. Diversity of Mycotoxins in Stored Paddy Rice: Contamination Patterns in the Mekong Delta, Vietnam. Toxins 2025, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Chandravarnan, P.; Agyei, D.; Ali, A. The prevalence and concentration of mycotoxins in rice sourced from markets: A global description. Trends Food Sci. Technol. 2024, 146, 104394. [Google Scholar] [CrossRef]
- Majeed, S.; De Boevre, M.; De Saeger, S.; Rauf, W.; Tawab, A.; Rahman, M.; Iqbal, M. Multiple Mycotoxins in Rice: Occurrence and Health Risk Assessment in Children and Adults of Punjab, Pakistan. Toxins 2018, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.Y.; Lee, S.Y.; Jeong, T.K.; Park, S.M.; Auh, J.H.; Shin, H.-S.; Chun, H.S. Natural Occurrence of Alternaria Toxins in Agricultural Products and Processed Foods Marketed in South Korea by LC–MS/MS. Toxins 2022, 14, 824. [Google Scholar] [CrossRef]
- Safavizadeh, V.; Sher, A.; Oliveira, C.A.F.D.; Moore, M.; Ghasemlou, M.; Naderi-Manesh, H.; Nemati, M.; Nokhodchi, A.; Rostami, M.; Tahergorabi, R. The occurrence of tenuazonic acid in food products: A systematic review. Toxin Rev. 2024, 43, 453–462. [Google Scholar] [CrossRef]
- Patriarca, A.; Azcarate, M.P.; Terminiello, L.; Fernández Pinto, V. Mycotoxin production by Alternaria strains isolated from Argentinean wheat. Int. J. Food Microbiol. 2007, 119, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Agriopoulou, S.; Stamatelopoulou, E.; Varzakas, T. Advances in Occurrence, Importance, and Mycotoxin Control Strategies: Prevention and Detoxification in Foods. Foods 2020, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Kodape, A.; Kodape, A.; Desai, R. Rice bran: Nutritional value, health benefits, and global implications for aflatoxin mitigation, cancer, diabetes, and diarrhea prevention. Food Chem. 2025, 464, 141749. [Google Scholar] [CrossRef] [PubMed]
- Mannaa, M.; Kim, K.D. Microbe-mediated control of mycotoxigenic grain fungi in stored rice with focus on aflatoxin biodegradation and biosynthesis inhibition. Mycobiology 2016, 44, 67–78. [Google Scholar] [CrossRef]
- Chandravarnan, P.; Agyei, D.; Ali, A. Green and sustainable technologies for the decontamination of fungi and mycotoxins in rice: A review. Trends Food Sci. Technol. 2022, 124, 278–295. [Google Scholar] [CrossRef]
- Laut, S.; Poapolathep, S.; Piasai, O.; Sommai, S.; Boonyuen, N.; Giorgi, M.; Zhang, Z.; Fink-Gremmels, J.; Poapolathep, A. Storage Fungi and Mycotoxins Associated with Rice Samples Commercialized in Thailand. Foods 2023, 12, 487. [Google Scholar] [CrossRef]
- Tang, E.N.; Ndindeng, S.A.; Onaga, G.; Ortega-Beltran, A.; Falade, T.D.O.; Djouaka, R.; Frei, M. Mycotoxin concentrations in rice are affected by chalkiness, grain shape, processing type, and grain origin. Mycotoxin Res. 2025, 41, 163–177. [Google Scholar] [CrossRef]
- Bertuzzi, T.; Romani, M.; Rastelli, S.; Giorni, P. Mycotoxins and Related Fungi in Italian Paddy Rice During the Growing Season and Storage. Toxins 2019, 11, 151. [Google Scholar] [CrossRef]
- Li, W.; Cui, J.; Li, J.; Guo, J.; Huang, T.; Zhang, J.; Hu, H.; Liu, X. Analysis of the Fungi Community Variation during Rice Storage through High Throughput Sequencing. Processes 2022, 10, 754. [Google Scholar] [CrossRef]
- Cunha, S.C.; Fernandes, J.O. Development and validation of a method based on a QuEChERS procedure and heart-cutting GC-MS for determination of five mycotoxins in cereal products. J. Sep. Sci. 2010, 33, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Cunha, S.C.; Sá, S.V.M.; Fernandes, J.O. Multiple mycotoxin analysis in nut products: Occurrence and risk characterization. Food Chem Toxicol. 2018, 114, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi; Samson, R.A., Ed.; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2010; Volume 2, p. 390. [Google Scholar]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Acharya, T.; Hare, J. Sabouraud Agar and Other Fungal Growth Media. In Laboratory Protocols in Fungal Biology: Current Methods in Fungal Biology; Gupta, V.K., Tuohy, M., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 69–86. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Guarro, J.; Gené, J.; Ahmed, S.A.; Al-Hatmi, A.M.S.; Figueras, M.J.; Vitale, R.G. Atlas of Clinical Fungi, 1st ed.; Westerdijk Institute: Utrecht, The Netherlands, 2019. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; Academic Press: Cambridge, MA, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Hong, S.-B.; Cho, H.-S.; Shin, H.-D.; Frisvad, J.C.; Samson, R.A. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 477–486. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Frandsen, R.J.; Lysøe, E.; Rehner, S.A.; Aoki, T.; et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef]
- O’Donnell, K.; Laraba, I.; Geiser, D.M. Pure Culture and DNA Sequence-Based Identification of Fusarium from Symptomatic Plants and Diverse Substrates. In Fusarium wilt: Methods and Protocols; Coleman, J., Ed.; Springer: New York, NY, USA, 2022; pp. 1–20. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]

| Rice Variety I | ||||||||||
| Mycotoxins | Paddy rice | Brown rice | Rice bran | Whitened rice | Polished rice | Broken rice | Final white rice | Average (µg/kg) | Min (µg/kg) | Max (µg/kg) |
| 3-AcDON | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| 15-AcDON | 81.15 ± 5.60 | 56.21 ± 0.71 | 591.58 ± 4.20 | 11.34 ± 2.20 | 23.54 ± 4.24 | 13.72 ± 2.07 | 19.04 ± 0.33 | 113.80 | 11.34 | 591.58 |
| AFB1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| AFB2 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| AFG1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| AFG2 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| BEA | 6.58 ± 0.30 | 2.10 ± 0.02 | 6.62 ± 0.76 | nd | 3.70 ± 0.01 | 7.37 ± 0.32 | nd | 5.27 | 2.10 | 7.37 |
| CIT | nd | 1.39 ± 0.20 | nd | nd | nd | nd | nd | - | 1.39 | 1.39 |
| CPZ | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| DON | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| ENNA | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| ENNA1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| ENNB | ≤LOQ | ≤LOQ | ≤LOQ | 4.13 ± 0.01 | 4.14 ± 0.01 | 4.15 ± 0.03 | 4.13 ± 0.02 | 4.14 | 4.13 | 4.15 |
| ENNB1 | nd | ≤LOQ | ≤LOQ | nd | nd | 4.57 ± 0.01 | nd | - | 4.57 | 4.57 |
| FB1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| FB2 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| HT-2 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| OTA | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| STG | 1.18 ± 0.03 | 1.20 ± 0.02 | 2.39 ± 0.14 | 1.64 ± 0.01 | 1.18 ± 0.02 | 1.17 ± 0.01 | 1.20 ± 0.01 | 1.42 | 1.17 | 2.39 |
| TeA | 292.10 ± 8.09 | 42.94 ± 9.76 | 917.00 ± 14.65 | 217.99 ± 19.8 | 79.55 ± 9.91 | 323.59 ± 4.07 | 51.30 ± 18.73 | 274.92 | 42.94 | 917.00 |
| T-2 | nd | nd | 3.32 ± 0.52 | nd | nd | nd | nd | - | 3.32 | 3.32 |
| ZEN | 55.14 ± 9.39 | 11.57 ± 1.02 | 200.91 ± 13.09 | 9.92 ± 1.85 | 10.90 ± 1.45 | 9.76 ± 0.38 | 9.10 ± 0.82 | 43.90 | 9.1 | 200.91 |
| Total * | 5 | 6 | 6 | 5 | 6 | 7 | 5 | |||
| Rice Variety II | ||||||||||
| Mycotoxins | Paddy rice | Brown rice | Rice bran | Whitened rice | Polished rice | Broken rice | Final white rice | Average (µg/kg) | Min (µg/kg) | Máx (µg/kg) |
| 3-AcDON | nd | nd | 20.44 ± 2.94 | nd | nd | nd | nd | - | 20.44 | 20.44 |
| 15-AcDON | 113.28 ± 16.23 | 83.75 ± 10.20 | 965.23 ± 14.72 | 35.86 ± 3.91 | 124.25 ± 4.08 | 95.29 ± 11.88 | 95.74 ± 10.09 | 216.20 | 35.86 | 965.23 |
| AFB1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| AFB2 | nd | nd | nd | nd | nd | 4.25 ± 0.82 | nd | - | 4.25 | 4.25 |
| AFG1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| AFG2 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| BEA | 22.99 ± 0.05 | 28.52 ± 0.57 | 159.19 ± 0.84 | 19.16 ± 1.92 | 37.57 ± 0.43 | 37.30 ± 0.23 | 7.41 ± 0.03 | 44.59 | 7.41 | 159.19 |
| CIT | 22.57 ± 1.28 | 91.61 ± 0.40 | 152.23 ± 0.99 | 50.95 ± 0.92 | 50.10 ± 0.05 | 50.21 ± 0.04 | 7.99 ± 0.07 | 60.81 | 7.99 | 152.23 |
| CPZ | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| DON | nd | nd | 50.32 ± 5.57 | nd | nd | nd | nd | - | 50.32 | 50.32 |
| ENNA | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| ENNA1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| ENNB | ≤LOQ | ≤LOQ | 9.80 ± 0.08 | 4.21 ± 0.09 | 4.19 ± 0.02 | 4.22 ± 0.04 | 4.20 ± 0.06 | 5.32 | 4.19 | 9.80 |
| ENNB1 | 18.21 ± 0.88 | 9.03 ± 0.02 | 29.90 ± 0.08 | 45.61 ± 0.02 | 45.62 ± 0.01 | 45.78 ± 0.33 | 13.01 ± 1.94 | 29.59 | 9.03 | 45.78 |
| FB1 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| FB2 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| HT-2 | nd | nd | nd | nd | nd | nd | nd | - | - | - |
| OTA | nd | nd | nd | 34.44 ± 0.09 | 34.68 ± 0.09 | 37.11 ± 0.06 | nd | 35.41 | 34.44 | 37.11 |
| STG | nd | nd | 10.49 ± 0.14 | nd | nd | nd | nd | - | 10.49 | 10.49 |
| TeA | 2121.37 ± 5.10 | 2395.11 ± 3.51 | 5419.88 ± 4.74 | 4264.81 ± 6.42 | 2219.83 ± 4.41 | 2812.20 ± 13.05 | 407.76 ± 14.92 | 2805.85 | 407.76 | 5419.88 |
| T-2 | nd | nd | 2.69 ± 0.38 | 4.61 ± 0.55 | nd | nd | nd | 3.65 | 2.69 | 4.61 |
| ZEN | 62.98 ± 0.56 | 116.31 ± 15.97 | 220.45 ± 7.91 | 17.41 ± 2.93 | 17.35 ± 3.11 | 16.64 ± 0.39 | 15.44 ± 0.29 | 66.62 | 15.44 | 220.45 |
| Total * | 6 | 6 | 11 | 9 | 8 | 9 | 7 | |||
| Rice Variety I | ||||||
| Samples | ||||||
| Morphological ID | Molecular ID | Paddy rice | Brown rice | Whitened rice | Final white rice | Total n (%) |
| Aspergillus spp. | 1 | 5 | 3 | 1 | 10 (20.8) | |
| A. fumigatus | 1 | |||||
| A. chevalieri | 2 | |||||
| A. porosus | 2 | |||||
| A. pseudoglaucus | 1 | |||||
| A. clavatus | 2 | |||||
| A. montevidensis | 1 | |||||
| A. flavus | 1 | |||||
| Penicillium spp. | 4 | 7 | 8 | 1 | 20 (41.7) | |
| P. charlesii | 1 | 2 | 7 | |||
| P. viridicatum | 2 | 1 | ||||
| P. citrinum | 1 | |||||
| P. polonicum | 1 | |||||
| P. sajarovii | 2 | |||||
| P. echinulonalgiovense | 2 | |||||
| P. brevicompactum | 1 | |||||
| Talaromyces spp. | 0 | 0 | 1 | 2 | 3 (6.3) | |
| T. wortmannii | 1 | |||||
| T. rugulosus | 2 | |||||
| Fusarium sp. | 1 | 0 | 0 | 1 | 2 (4.2) | |
| F. fujikuroi | 1 | 1 | ||||
| Alternaria spp. | 6 | 2 | 4 | 1 | 13 (27.1) | |
| A. resedae | 1 | 1 | 1 | |||
| A. alternata | 3 | 1 | 1 | |||
| A. destruens | 1 | |||||
| A. jacinthicola | 1 | |||||
| A. tenuissima | 1 | |||||
| A. arborescens | 1 | 1 | ||||
| Total n (%) | 12 (25.0) | 14 (29.2) | 16 (33.3) | 6 (12.5) | 48 | |
| Rice Variety II | ||||||
| Samples | ||||||
| Morphological ID | Molecular ID | Paddy rice | Brown rice | Whitened rice | Final white rice | Total n (%) |
| Aspergillus spp. | 2 | 0 | 2 | 1 | 5 (18.5) | |
| A. fumigatus | 2 | |||||
| A. chevalieri | 1 | |||||
| A. neotritici | 1 | 1 | ||||
| Penicillium sp. | 2 | 0 | 0 | 0 | 2 (7.4) | |
| P. viridicatum | 2 | |||||
| Talaromyces sp. | 1 | 0 | 0 | 0 | 1 (3.7) | |
| T. rugulosus | 1 | |||||
| Fusarium sp. | 1 | 1 | 0 | 0 | 2 (7.4) | |
| F. tanahbumbuense | 1 | 1 | ||||
| Alternaria spp. | 4 | 9 | 4 | 0 | 17 (63.0) | |
| A. resedae | 1 | |||||
| A. alternata | 3 | 4 | 3 | |||
| A. destruens | 4 | |||||
| A. longipes | 1 | |||||
| A. arborescens | 1 | |||||
| Total n (%) | 10 (37.0) | 10 (37.0) | 6 (22.2) | 1 (3.7) | 27 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro, C.S.; Pinto, E.; López-Ruiz, R.; Marín-Sáez, J.; Frenich, A.G.; Faria, M.A.; Cunha, S.C. Integrated Assessment of Fungi Contamination and Mycotoxins Levels Across the Rice Processing Chain. Toxins 2025, 17, 468. https://doi.org/10.3390/toxins17090468
Monteiro CS, Pinto E, López-Ruiz R, Marín-Sáez J, Frenich AG, Faria MA, Cunha SC. Integrated Assessment of Fungi Contamination and Mycotoxins Levels Across the Rice Processing Chain. Toxins. 2025; 17(9):468. https://doi.org/10.3390/toxins17090468
Chicago/Turabian StyleMonteiro, Carolina Sousa, Eugénia Pinto, Rosalía López-Ruiz, Jesús Marín-Sáez, Antonia Garrido Frenich, Miguel A. Faria, and Sara C. Cunha. 2025. "Integrated Assessment of Fungi Contamination and Mycotoxins Levels Across the Rice Processing Chain" Toxins 17, no. 9: 468. https://doi.org/10.3390/toxins17090468
APA StyleMonteiro, C. S., Pinto, E., López-Ruiz, R., Marín-Sáez, J., Frenich, A. G., Faria, M. A., & Cunha, S. C. (2025). Integrated Assessment of Fungi Contamination and Mycotoxins Levels Across the Rice Processing Chain. Toxins, 17(9), 468. https://doi.org/10.3390/toxins17090468






