Microcystis aeruginosa msoT1/msoA1 Locus Displays Features of a Type I Toxin–Antitoxin System
Abstract
1. Introduction
2. Results
2.1. Predicted Type I TA Loci in the Genome of Microcystis aeruginosa PCC 7806SL
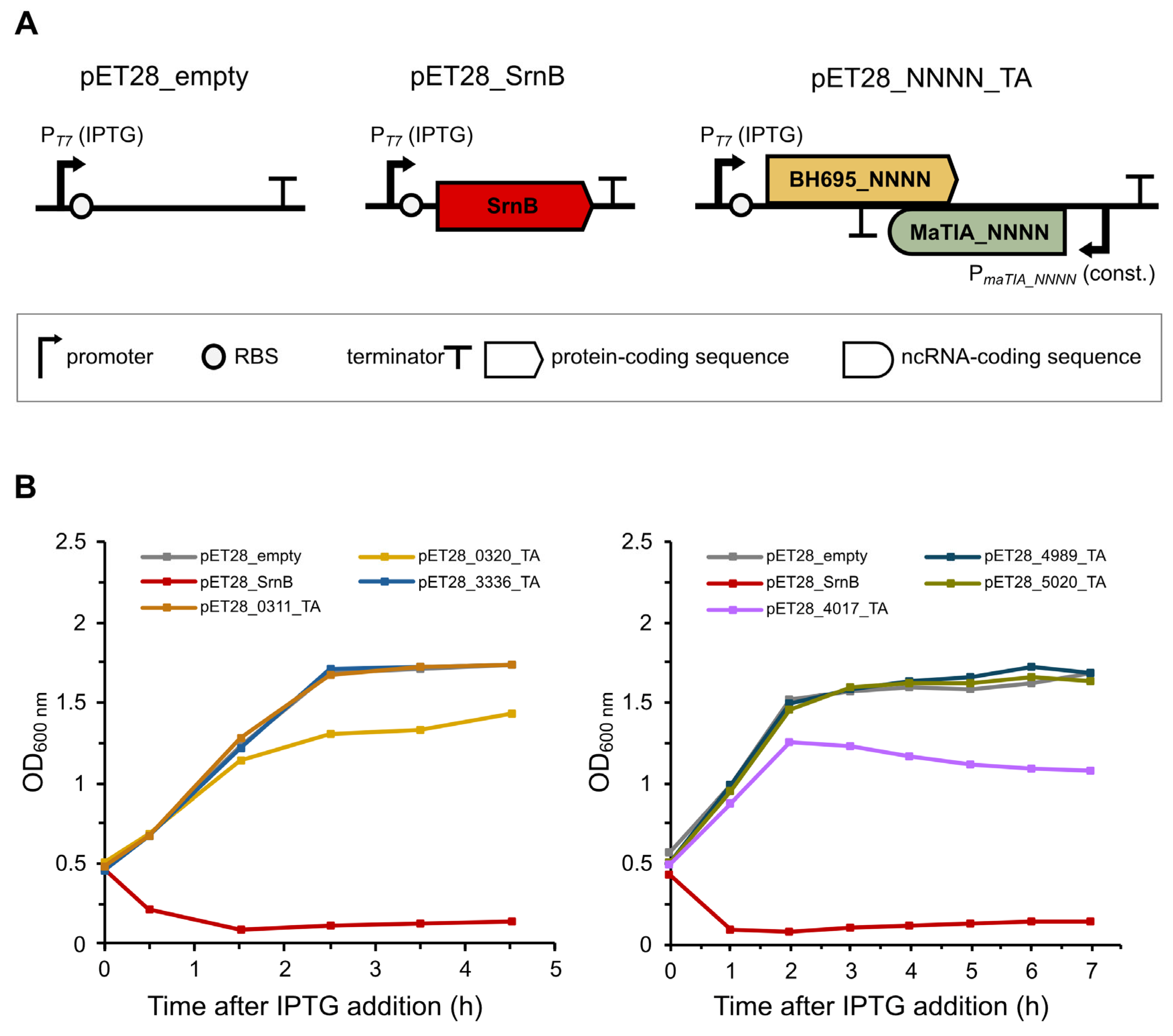
2.2. Overexpression of BH695_0320 and BH695_4017 Negatively Affects Escherichia coli Cell Growth
2.3. Constitutive Expression of MsoA1 Delays Toxic Effect of MsoT1 Expression in Escherichia coli
2.4. The Sequence Upstream of MsoA1 Contains Biologically Active Constitutive Promoters
2.5. Properties of the MsoT1/MsoA1 Toxin–Antitoxin Pair
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bioinformatic Analysis of the Microcystis aeruginosa PCC 7806SL Genome
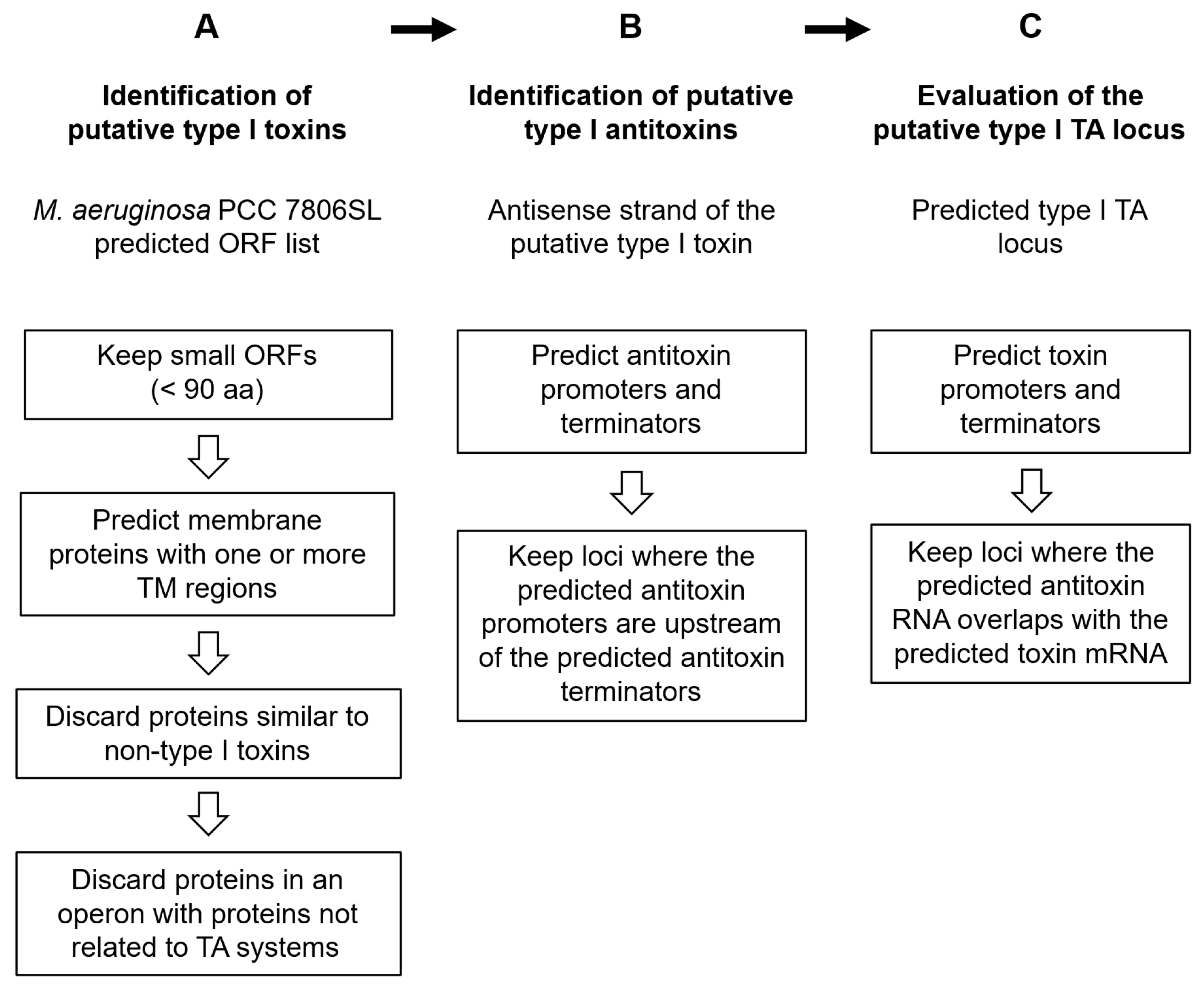
5.1.1. De Novo Search for Novel Putative Type I TA Loci
5.1.2. Nomenclature and Evaluation of the Predicted Putative Loci
5.2. Bacterial Strains
5.2.1. Escherichia coli
5.2.2. Microcystis aeruginosa PCC 7806SL
5.3. Growth Media
5.4. Molecular Techniques
5.5. Plasmids and Their Coding Regions
5.5.1. pET28_empty
5.5.2. pET28_0311_TA, pET28_0320_TA, pET28_3336_TA, pET28_4017_TA (pET28_MsoT1/MsoA1), pET28_4989_TA, and pET28_5020_TA
5.5.3. pET28_4017_T (pET28_MsoT1)
5.5.4. pET28_SrnB
5.5.5. pET28_SrnB/SrnC
5.5.6. pSB1C3_RBS_AmpR
5.5.7. pSB1C3_TT_RBS_AmpR
5.5.8. pSB1C3_PmsoA1_RBS_AmpR
5.5.9. pSB1C3_TT_PmsoA1_RBS_AmpR
5.5.10. pSB1AC3_empty
5.5.11. pSB1C3_K516032
5.5.12. pSB1C3_PmsoA1_K516032
5.5.13. pSB1A2_J23119_K516032
5.6. Cell Toxicity Assay
5.7. Promoter Activity Assays
5.7.1. Antitoxin Promoter Activity Assay with β-Lactamase
5.7.2. Antitoxin Promoter Activity Assay with mRFP
5.8. Western Blotting and Immunodetection
5.9. Erythrosin B Staining of Non-Viable Bacterial Cells
5.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique Type of Plasmid Maintenance Function: Postsegregational Killing of Plasmid-Free Cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef]
- Weaver, K.E.; Clewell, D.B. Construction of Enterococcus Faecalis pAD1 Miniplasmids: Identification of a Minimal Pheromone Response Regulatory Region and Evaluation of a Novel Pheromone-Dependent Growth Inhibition. Plasmid 1989, 22, 106–119. [Google Scholar] [CrossRef]
- Christensen, S.K.; Gerdes, K. RelE Toxins from Bacteria and Archaea Cleave mRNAs on Translating Ribosomes, Which Are Rescued by tmRNA. Mol. Microbiol. 2003, 48, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of Type I Toxin-Antitoxin Systems in Bacteria: Searches for New Candidates and Discovery of Novel Families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef] [PubMed]
- Peltier, J.; Hamiot, A.; Garneau, J.R.; Boudry, P.; Maikova, A.; Hajnsdorf, E.; Fortier, L.C.; Dupuy, B.; Soutourina, O. Type I Toxin-Antitoxin Systems Contribute to the Maintenance of Mobile Genetic Elements in Clostridioides Difficile. Commun. Biol. 2020, 3, 718. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Bech, F.W.; Jørgensen, S.T.; Løbner-Olesen, A.; Rasmussen, P.B.; Atlung, T.; Boe, L.; Karlstrom, O.; Molin, S.; von Meyenburg, K. Mechanism of Postsegregational Killing by the Hok Gene Product of the parB System of Plasmid R1 and Its Homology with the relF Gene Product of the E. coli relB Operon. EMBO J. 1986, 5, 2023–2029. [Google Scholar] [CrossRef]
- Thisted, T.; Gerdes, K. Mechanism of Post-Segregational Killing by the Hok/Sok System of Plasmid R1. J. Mol. Biol. 1992, 223, 41–54. [Google Scholar] [CrossRef]
- Thisted, T.; Nielsen, A.K.; Gerdes, K. Mechanism of Post-Segregational Killing: Translation of Hok, SrnB and Pnd mRNAs of Plasmids R1, F and R483 Is Activated by 3′-End Processing. EMBO J. 1994, 13, 1950–1959. [Google Scholar] [CrossRef]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P.C. The Phage Abortive Infection System, ToxIN, Functions as a Protein–RNA Toxin–Antitoxin Pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef]
- Guegler, C.K.; Laub, M.T. Shutoff of Host Transcription Triggers a Toxin-Antitoxin System to Cleave Phage RNA and Abort Infection. Mol. Cell 2021, 81, 2361–2373. [Google Scholar] [CrossRef] [PubMed]
- De Bast, M.S.; Mine, N.; Van Melderen, L. Chromosomal Toxin-Antitoxin Systems May Act as Antiaddiction Modules. J. Bacteriol. 2008, 190, 4603–4609. [Google Scholar] [CrossRef] [PubMed]
- Korch, S.B.; Henderson, T.A.; Hill, T.M. Characterization of the hipA7 Allele of Escherichia coli and Evidence That High Persistence Is Governed by (p)ppGpp Synthesis. Mol. Microbiol. 2003, 50, 1199–1213. [Google Scholar] [CrossRef]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and Evolution of Bacterial Toxin–Antitoxin Systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef]
- Shore, S.F.H.; Leinberger, F.H.; Fozo, E.M.; Berghoff, B.A. Type I Toxin-Antitoxin Systems in Bacteria: From Regulation to Biological Functions. EcoSal Plus 2024, 12, eesp-0025-2022. [Google Scholar] [CrossRef]
- Kawano, M.; Aravind, L.; Storz, G. An Antisense RNA Controls Synthesis of an SOS-Induced Toxin Evolved from an Antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef]
- Wen, J.; Fozo, E.M. sRNA Antitoxins: More than One Way to Repress a Toxin. Toxins 2014, 6, 2310–2335. [Google Scholar] [CrossRef]
- Darfeuille, F.; Unoson, C.; Vogel, J.; Wagner, E.G.H. An Antisense RNA Inhibits Translation by Competing with Standby Ribosomes. Mol. Cell 2007, 26, 381–392. [Google Scholar] [CrossRef]
- Jahn, N.; Brantl, S. One Antitoxin—Two Functions: SR4 Controls Toxin mRNA Decay and Translation. Nucleic Acids Res. 2013, 41, 9870–9880. [Google Scholar] [CrossRef]
- Durand, S.; Gilet, L.; Condon, C. The Essential Function of B. subtilis RNase III Is to Silence Foreign Toxin Genes. PLoS Genet. 2012, 8, e1003181. [Google Scholar] [CrossRef]
- Reif, C.; Löser, C.; Brantl, S. Bacillus subtilis Type I Antitoxin SR6 Promotes Degradation of Toxin yonT mRNA and Is Required to Prevent Toxic yoyJ Overexpression. Toxins 2018, 10, 74. [Google Scholar] [CrossRef]
- Brielle, R.; Pinel-Marie, M.L.; Felden, B. Linking Bacterial Type I Toxins with Their Actions. Curr. Opin. Microbiol. 2016, 30, 114–121. [Google Scholar] [CrossRef]
- Nonin-Lecomte, S.; Fermon, L.; Felden, B.; Pinel-Marie, M.L. Bacterial Type I Toxins: Folding and Membrane Interactions. Toxins 2021, 13, 490. [Google Scholar] [CrossRef]
- Gurnev, P.A.; Ortenberg, R.; Dörr, T.; Lewis, K.; Bezrukov, S.M. Persister-Promoting Bacterial Toxin TisB Produces Anion-Selective Pores in Planar Lipid Bilayers. FEBS Lett. 2012, 586, 2529–2534. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Bayoumi, M.; Dewachter, L.; Knapen, W.; Mika, J.T.; Hofkens, J.; Dedecker, P.; Maglia, G.; Verstraeten, N.; Michiels, J. The Persistence-Inducing Toxin HokB Forms Dynamic Pores That Cause ATP Leakage. mBio 2018, 9, 1128. [Google Scholar] [CrossRef]
- Kawano, M.; Oshima, T.; Kasai, H.; Mori, H. Molecular Characterization of Long Direct Repeat (LDR) Sequences Expressing a Stable mRNA Encoding for a 35-amino-acid Cell-killing Peptide and a Cis-encoded Small Antisense RNA in Escherichia coli. Mol. Microbiol. 2002, 45, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Weaver, K.E. Addiction Toxin Fst Has Unique Effects on Chromosome Segregation and Cell Division in Enterococcus Faecalis and Bacillus subtilis. J. Bacteriol. 2006, 188, 5374–5384. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M. Divergently Overlapping Cis-Encoded Antisense RNA Regulating Toxin-Antitoxin Systems from E. coli. RNA Biol. 2012, 9, 1520–1527. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Quiroga, C.; Chen, Q.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K.; Wang, X. RalR (a DNase) and RalA (a Small RNA) Form a Type I Toxin-Antitoxin System in Escherichia coli. Nucleic Acids Res. 2014, 42, 6448–6462. [Google Scholar] [CrossRef]
- Tourasse, N.J.; Darfeuille, F. T1TAdb: The Database of Type I Toxin–Antitoxin Systems. RNA 2021, 27, 1471–1481. [Google Scholar] [CrossRef]
- Guan, J.; Chen, Y.; Goh, Y.-X.; Wang, M.; Tai, C.; Deng, Z.; Song, J.; Ou, H.-Y. TADB 3.0: An Updated Database of Bacterial Toxin–Antitoxin Loci and Associated Mobile Genetic Elements. Nucleic Acids Res. 2024, 52, D784–D790. [Google Scholar] [CrossRef] [PubMed]
- Coray, D.S.; Wheeler, N.E.; Heinemann, J.A.; Gardner, P.P. Why so Narrow: Distribution of Anti-Sense Regulated, Type I Toxin-Antitoxin Systems Compared with Type II and Type III Systems. RNA Biol. 2017, 14, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Klemenčič, M.; Halužan Vasle, A.; Dolinar, M. The Cysteine Protease MaOC1, a Prokaryotic Caspase Homolog, Cleaves the Antitoxin of a Type II Toxin-Antitoxin System. Front. Microbiol. 2021, 12, 635684. [Google Scholar] [CrossRef] [PubMed]
- Georg, J.; Voß, B.; Scholz, I.; Mitschke, J.; Wilde, A.; Hess, W.R. Evidence for a Major Role of Antisense RNAs in Cyanobacterial Gene Regulation. Mol. Syst. Biol. 2009, 5, 305. [Google Scholar] [CrossRef]
- Diago-Navarro, E.; Hernández-Arriaga, A.M.; Kubik, S.; Konieczny, I.; Díaz-Orejas, R. Cleavage of the Antitoxin of the parD Toxin–Antitoxin System Is Determined by the ClpAP Protease and Is Modulated by the Relative Ratio of the Toxin and the Antitoxin. Plasmid 2013, 70, 78–85. [Google Scholar] [CrossRef]
- Lehnherr, H.; Yarmolinsky, M.B. Addiction Protein Phd of Plasmid Prophage P1 Is a Substrate of the ClpXP Serine Protease of Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 3274–3277. [Google Scholar] [CrossRef]
- Wilmaerts, D.; De Loose, P.J.; Vercauteren, S.; De Smedt, S.; Verstraeten, N.; Michiels, J. Functional Analysis of Cysteine Residues of the Hok/Gef Type i Toxins in Escherichia coli. FEMS Microbiol. Lett. 2021, 368, fnab069. [Google Scholar] [CrossRef]
- Nitta, T.; Nagamitsu, H.; Murata, M.; Izu, H.; Yamada, M. Function of the ς E Regulon in Dead-Cell Lysis in Stationary-Phase Escherichia coli. J. Bacteriol. 2000, 182, 5231–5237. [Google Scholar] [CrossRef]
- Franke, J.D.; Braverman, A.L.; Cunningham, A.M.; Eberhard, E.E.; Perry, G.A. Erythrosin B: A Versatile Colorimetric and Fluorescent Vital Dye for Bacteria. BioTechniques 2019, 68, 7–13. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Fozo, E.M.; Hemm, M.R.; Storz, G. Small Toxic Proteins and the Antisense RNAs That Repress Them. Microbiol. Mol. Biol. Rev. 2008, 72, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, J. Bacterial Systems. In Production of Membrane Proteins; Robinson, A.S., Ed.; Wiley: New York, NY, USA, 2011; pp. 11–35. [Google Scholar]
- Jahn, N.; Preis, H.; Wiedemann, C.; Brantl, S. BsrG/SR4 from Bacillus subtilis- the First Temperature-Dependent Type I Toxin-Antitoxin System. Mol. Microbiol. 2012, 83, 579–598. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.R.; Muthuramalingam, M.; Murphy, T.; Najar, F.Z.; Bourne, C.R. Expression of Different ParE Toxins Results in Conserved Phenotypes with Distinguishable Classes of Toxicity. MicrobiologyOpen 2019, 8, e902. [Google Scholar] [CrossRef]
- Wada, H.; Murata, N. Membrane Lipids in Cyanobacteria. In Lipids in Photosynthesis: Structure, Function and Genetics; Paul-André, S., Norio, M., Eds.; Springer: Dordrecht, The Netherlands, 1998; Volume 6, pp. 65–81. ISBN 978-0-306-48087-4. [Google Scholar]
- Juteršek, M.; Dolinar, M. A Chimeric Vector for Dual Use in Cyanobacteria and Escherichia coli, Tested with Cystatin, a Nonfluorescent Reporter Protein. PeerJ 2021, 9, e12199. [Google Scholar] [CrossRef]
- Lee, Y.-N.; Chen, L.-K.; Ma, H.-C.; Yang, H.-H.; Li, H.-P.; Lo, S.-Y. Thermal Aggregation of SARS-CoV Membrane Protein. J. Virol. Methods 2005, 129, 152–161. [Google Scholar] [CrossRef]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent Binding Explains Anomalous SDS-PAGE Migration of Membrane Proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef]
- Rawlings, A.E.; Bramble, J.P.; Walker, R.; Bain, J.; Galloway, J.M.; Staniland, S.S. Self-Assembled MmsF Proteinosomes Control Magnetite Nanoparticle Formation in Vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 16094–16099. [Google Scholar] [CrossRef]
- Part: pSB1C3. Available online: https://parts.igem.org/Part:pSB1C3 (accessed on 28 May 2025).
- Škrlj, N.; Vidrih, Z.; Dolinar, M. A Universal Approach for Promoter Strength Evaluation Supported by the Web-Based Tool PromCal. Anal. Biochem. 2010, 396, 83–90. [Google Scholar] [CrossRef]
- Silvaggi, J.M.; Perkins, J.B.; Losick, R. Small Untranslated RNA Antitoxin in Bacillus subtilis. J. Bacteriol. 2005, 187, 6641–6650. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-Mapper: A Web Server for Precise Operon Identification in Bacterial and Archaeal Genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]
- Shahmuradov, I.A.; Mohamad Razali, R.; Bougouffa, S.; Radovanovic, A.; Bajic, V.B. bTSSfinder: A Novel Tool for the Prediction of Promoters in Cyanobacteria and Escherichia coli. Bioinformatics 2017, 33, 334–340. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies; Li, R.W., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Naville, M.; Ghuillot-Gaudeffroy, A.; Marchais, A.; Gautheret, D. ARNold: A Web Tool for the Prediction of Rho-Independent Transcription Terminators. RNA Biol. 2011, 8, 11–13. [Google Scholar] [CrossRef]




| Name | GenBank Accession | Start | End | Strand | Length (aa) | TM Regions |
|---|---|---|---|---|---|---|
| BH695_0311 | ARI79592.1 | 289,680 | 289,489 | −1 | 63 | 1 |
| BH695_0320 | ARI79601.1 | 299,411 | 299,551 | 1 | 46 | 1 |
| BH695_1184 | ARI80465.1 | 1,096,632 | 1,096,486 | −1 | 48 | 1 |
| BH695_1248 | ARI80529.1 | 1,152,754 | 1,152,629 | −1 | 41 | 1 |
| BH695_3336 | ARI82615.1 | 3,177,614 | 3,177,757 | 1 | 47 | 1 |
| BH695_3390 | ARI82669.1 | 3,230,378 | 3,230,259 | −1 | 39 | 1 |
| BH695_4017 | ARI83296.1 | 3,792,667 | 3,792,476 | −1 | 63 | 2 |
| BH695_4253 | ARI83532.1 | 4,016,545 | 4,016,411 | −1 | 44 | 1 |
| BH695_4989 | ARI84268.1 | 4,737,004 | 4,737,249 | 1 | 81 | 2 |
| BH695_5020 | ARI84299.1 | 4,769,107 | 4,768,946 | −1 | 53 | 1 |
| Name | Isoform | Start | End | Strand | Length (nt) | Toxin mRNA Complementarity |
|---|---|---|---|---|---|---|
| MaTIA_0311 | 1 | 289,493 | 289,535 | 1 | 43 | CDS |
| 2 | 289,482 | 289,535 | 1 | 54 | CDS, 3′-UTR | |
| MaTIA_0320 | 1 | 299,493 | 299,423 | −1 | 71 | CDS |
| 2 | 299,517 | 299,423 | −1 | 95 | CDS | |
| 3 | 299,574 | 299,423 | −1 | 152 | CDS, 3′-UTR | |
| MaTIA_1184 | 1 | 1,096,424 | 1,096,466 | 1 | 43 | 3′-UTR |
| 2 | 1,096,327 | 1,096,466 | 1 | 140 | 3′-UTR | |
| MaTIA_1248 | - | 1,152,693 | 1,152,843 | 1 | 151 | CDS, 5′-UTR |
| MaTIA_3336 | 1 | 3,177,829 | 3,177,749 | −1 | 81 | CDS, 3′-UTR |
| 2 | 3,177,945 | 3,177,749 | −1 | 197 | CDS, 3′-UTR | |
| MaT1A_3390 | 1 | 3,230,318 | 3,230,458 | 1 | 141 | CDS, 5′-UTR |
| 2 | 3,230,304 | 3,230,458 | 1 | 155 | CDS, 5′-UTR | |
| MaTIA_4017 | 1 | 3,792,450 | 3,792,505 | 1 | 56 | CDS, 3′-UTR |
| 2 | 3,792,421 | 3,792,505 | 1 | 85 | CDS, 3′-UTR | |
| MaTIA_4253 | - | 4,016,466 | 4,016,717 | 1 | 252 | CDS, 5′-UTR |
| MaTIA_4989 | - | 4,737,319 | 4,737,227 | −1 | 93 | CDS, 3′-UTR |
| MaTIA_5020 | 1 | 4,768,937 | 4,768,986 | 1 | 50 | CDS, 3′-UTR |
| 2 | 4,768,892 | 4,768,986 | 1 | 95 | CDS, 3′-UTR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruparčič, M.; Dolinar, M. Microcystis aeruginosa msoT1/msoA1 Locus Displays Features of a Type I Toxin–Antitoxin System. Toxins 2025, 17, 360. https://doi.org/10.3390/toxins17080360
Ruparčič M, Dolinar M. Microcystis aeruginosa msoT1/msoA1 Locus Displays Features of a Type I Toxin–Antitoxin System. Toxins. 2025; 17(8):360. https://doi.org/10.3390/toxins17080360
Chicago/Turabian StyleRuparčič, Matija, and Marko Dolinar. 2025. "Microcystis aeruginosa msoT1/msoA1 Locus Displays Features of a Type I Toxin–Antitoxin System" Toxins 17, no. 8: 360. https://doi.org/10.3390/toxins17080360
APA StyleRuparčič, M., & Dolinar, M. (2025). Microcystis aeruginosa msoT1/msoA1 Locus Displays Features of a Type I Toxin–Antitoxin System. Toxins, 17(8), 360. https://doi.org/10.3390/toxins17080360





