Plant-Derived UDP-Glycosyltransferases for Glycosylation-Mediated Detoxification of Deoxynivalenol: Enzyme Discovery, Characterization, and In Vivo Resistance Assessment
Abstract
1. Introduction
2. Results and Discussion
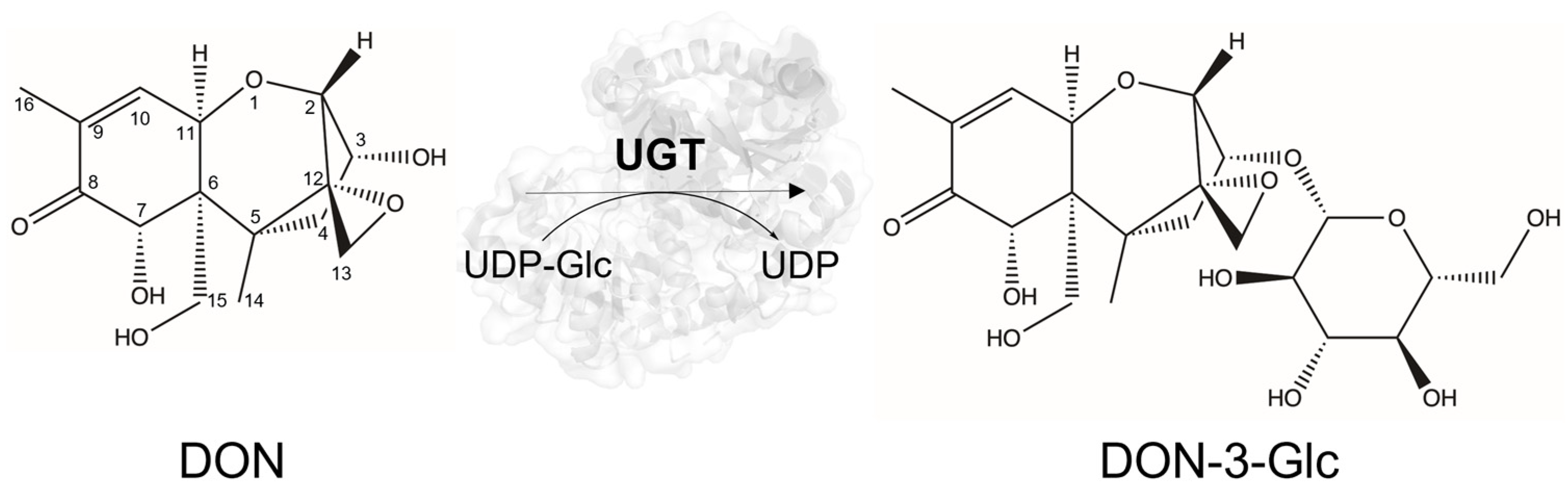
2.1. Screening of the GLY-it Library
2.2. Characterization of the Novel DON UGTs
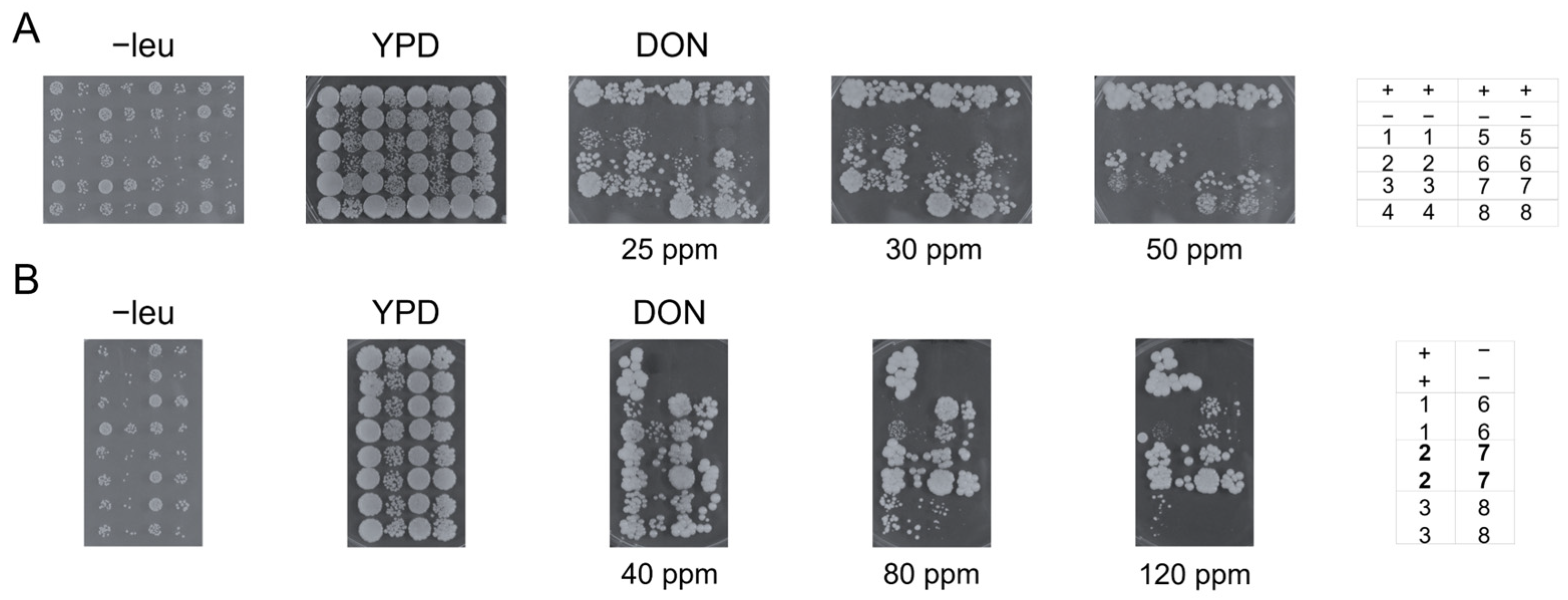
2.3. UGT-Mediated Resistance to DON in Yeast
3. Conclusions
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Preliminary Screening of the GLY-it Library
4.3. In Silico Sequence Analysis
4.4. Protein Expression in E. coli and Purification
4.5. Time-Course Activity Assay with DON
4.6. Biochemical and Kinetic Characterization
4.7. LC–MS/MS and HPLC Analysis
4.8. Cloning of UGTs in Yeast
4.9. Data Analysis and Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The United Nations. Global Issues: Population; The United Nations: New York, NY, USA, 2023. [Google Scholar]
- FAO. Towards a Waste-Free Future; FAO: Rome, Italy, 2023. [Google Scholar]
- Singh, B.K.; Delgado-Baquerizo, M.; Egidi, E.; Guirado, E.; Leach, J.E.; Liu, H.; Trivedi, P. Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 2023, 21, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Nnadi, N.E.; Carter, D.A. Climate change and the emergence of fungal pathogens. PLoS Pathog. 2021, 17, e1009503. [Google Scholar] [CrossRef]
- Reddy, G.K.K.; Padmavathi, A.R.; Nancharaiah, Y.V. Fungal infections: Pathogenesis, antifungals and alternate treatment approaches. Curr. Res. Microb. Sci. 2022, 3, 100137. [Google Scholar] [CrossRef] [PubMed]
- Stukenbrock, E.; Gurr, S. Address the growing urgency of fungal disease in crops. Nature 2023, 617, 31–34. [Google Scholar] [CrossRef]
- Steinberg, G.; Gurr, S.J. Fungi, fungicide discovery and global food security. Fungal Genet. Biol. 2020, 144, 103476. [Google Scholar] [CrossRef]
- FAO. OECD-FAO Agricultural Outlook 2023–2032; FAO: Rome, Italy, 2023; pp. 143–157. [Google Scholar]
- Pecoraro, F.; Giannini, M.; Beccari, G.; Covarelli, L.; Filippini, G.; Pisi, A.; Nipoti, P.; Prodi, A. Comparative studies about fungal colonization and deoxynivalenol translocation in barley plants inoculated at the base with Fusarium graminearum, Fusarium culmorum and Fusarium pseudograminearum. AFSci 2018, 27, 74–83. [Google Scholar] [CrossRef]
- N Matny, O. Fusarium Head Blight and Crown Rot on Wheat & Barley: Losses and Health Risks. Adv. Plants Agric. Res. 2015, 2, 103476. [Google Scholar]
- EFSA. Mycotoxins; EFSA: Parma, Italy, 2024.
- Sudakin, D.L. Trichothecenes in the environment: Relevance to human health. Toxicol. Lett. 2003, 143, 97–107. [Google Scholar] [CrossRef]
- Perochon, A.; Doohan, F.M. Trichothecenes and Fumonisins: Key Players in Fusarium–Cereal Ecosystem Interactions. Toxins 2024, 16, 90. [Google Scholar] [CrossRef]
- Karlovsky, P. Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl. Microbiol. Biotechnol. 2011, 91, 491–504. [Google Scholar] [CrossRef]
- Pierron, A.; Mimoun, S.; Murate, L.S.; Loiseau, N.; Lippi, Y.; Bracarense, A.P.F.L.; Schatzmayr, G.; He, J.W.; Zhou, T.; Moll, W.D.; et al. Microbial biotransformation of DON: Molecular basis for reduced toxicity. Sci. Rep. 2016, 6, 29105. [Google Scholar]
- Wang, X.; Fan, M.; Chu, X.; Zhang, Y.; Rahman, S.U.; Jiang, Y.; Chen, X.; Zhu, D.; Feng, S.; Li, Y.; et al. Deoxynivalenol induces toxicity and apoptosis in piglet hippocampal nerve cells via the MAPK signaling pathway. Toxicon 2018, 155, 1–8. [Google Scholar] [PubMed]
- Sobrova, P.; Adam, V.; Vasatkova, A.; Beklova, M.; Zeman, L.; Kizek, R. Deoxynivalenol and its toxicity. Interdiscip. Toxicol. 2010, 3, 94–99. [Google Scholar]
- Pestka, J.J. Deoxynivalenol: Toxicity, mechanisms and animal health risks. Anim. Feed Sci. Technol. 2007, 137, 283–298. [Google Scholar] [CrossRef]
- Pinton, P.; Oswald, I.P. Effect of deoxynivalenol and other type B trichothecenes on the intestine: A review. Toxins 2014, 6, 1615–1643. [Google Scholar] [CrossRef]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25%. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar]
- Johns, L.E.; Bebber, D.P.; Gurr, S.J.; Brown, N.A. Emerging health threat and cost of Fusarium mycotoxins in European wheat. Nat. Food 2022, 3, 1014–1019. [Google Scholar]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Campbell, J.A.; Davies, G.J.; Bulone, V.; Henrissat, B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 1997, 326, 929–942. [Google Scholar]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Tegl, G.; Nidetzky, B. Leloir glycosyltransferases of natural product C-glycosylation: Structure, mechanism and specificity. Biochem. Soc. Trans. 2020, 48, 1583–1598. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Hughes, M.A. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta crantz) cotyledons. DNA Seq. 1994, 5, 41–49. [Google Scholar] [CrossRef]
- Osmani, S.A.; Bak, S.; Imberty, A.; Olsen, C.E.; Møller, B.L. Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: Molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 2008, 148, 1295–1308. [Google Scholar] [PubMed]
- Brazier-Hicks, M.; Offen, W.A.; Gershater, M.C.; Revett, T.J.; Lim, E.-K.; Bowles, D.J.; Davies, G.J.; Edwards, R. Characterization and engineering of the bifunctional N-and O-glucosyltransferase involved in xenobiotic metabolism in plants. PNAS 2007, 104, 20238–20243. [Google Scholar]
- Teze, D.; Coines, J.; Fredslund, F.; Dubey, K.D.; Bidart, G.N.; Adams, P.D.; Dueber, J.E.; Svensson, B.; Rovira, C.; Welner, D.H. O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering. ACS Catal. 2021, 11, 1810–1815. [Google Scholar] [CrossRef]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium Mycotoxin Deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar]
- Schweiger, W.; Boddu, J.; Shin, S.; Poppenberger, B.; Berthiller, F.; Lemmens, M.; Muehlbauer, G.J.; Adam, G. Validation of a Candidate Deoxynivalenol-Inactivating UDP-Glucosyltransferase from Barley by Heterologous Expression in Yeast. Mol. Plant-Microbe Interact. 2010, 23, 977–986. [Google Scholar]
- He, Y.; Wu, L.; Liu, X.; Jiang, P.; Yu, L.; Qiu, J.; Wang, G.; Zhang, X.; Ma, H. TaUGT6, a Novel UDP-Glycosyltransferase Gene Enhances the Resistance to FHB and DON Accumulation in Wheat. Front. Plant Sci. 2020, 11, 574775. [Google Scholar] [CrossRef]
- Michlmayr, H.; Malachová, A.; Varga, E.; Kleinová, J.; Lemmens, M.; Newmister, S.; Rayment, I.; Berthiller, F.; Adam, G. Biochemical characterization of a recombinant UDP-glucosyltransferase from rice and enzymatic production of deoxynivalenol-3-O-β-D-glucoside. Toxins 2015, 7, 2685–2700. [Google Scholar] [CrossRef]
- Wetterhorn, K.M.; Gabardi, K.; Michlmayr, H.; Malachova, A.; Busman, M.; McCormick, S.P.; Berthiller, F.; Adam, G.; Rayment, I. Determinants and Expansion of Specificity in a Trichothecene UDP-Glucosyltransferase from Oryza sativa. Biochemistry 2017, 56, 6585–6596. [Google Scholar]
- Schweiger, W.; Pasquet, J.C.; Nussbaumer, T.; Kovalsky Paris, M.P.; Wiesenberger, G.; Macadré, C.; Amets, C.; Berthiller, F.; Lemmens, M.; Saindrenan, P.; et al. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Mol. Plant-Microbe Interact. 2013, 26, 781–792. [Google Scholar] [PubMed]
- Khairullina, A.; Renhuldt, N.T.; Wiesenberger, G.; Bentzer, J.; Collinge, D.B.; Adam, G.; Bülow, L. Identification and Functional Characterisation of Two Oat UDP-Glucosyltransferases Involved in Deoxynivalenol Detoxification. Toxins 2022, 14, 446. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Gao, L.; Chen, Q.; Pei, H.; Di, Z.; Xiao, J.; Wang, H.; Ma, L.; Chen, P.; Cao, A.; et al. Over-expressing a UDP-glucosyltransferase gene (Ta-UGT3) enhances Fusarium Head Blight resistance of wheat. Plant Growth Regul. 2018, 84, 561–571. [Google Scholar] [CrossRef]
- Michlmayr, H.; Varga, E.; Malachová, A.; Fruhmann, P.; Piatkowska, M.; Hametner, C.; Šofrová, J.; Jaunecker, G.; Häubl, G.; Lemmens, M.; et al. UDP-glucosyltransferases from rice, brachypodium, and barley: Substrate specificities and synthesis of type A and B Trichothecene-3-O-β-D-glucosides. Toxins 2018, 10, 111. [Google Scholar] [CrossRef] [PubMed]
- Wetterhorn, K.M.; Newmister, S.A.; Caniza, R.K.; Busman, M.; McCormick, S.P.; Berthiller, F.; Adam, G.; Rayment, I. Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol. Biochemistry 2016, 55, 6175–6186. [Google Scholar] [CrossRef]
- Mandala, G.; Tundo, S.; Francesconi, S.; Gevi, F.; Zolla, L.; Ceoloni, C.; D’Ovidio, R. Deoxynivalenol detoxification in transgenic wheat confers resistance to fusarium head blight and crown rot diseases. Mol. Plant-Microbe Interact. 2019, 32, 583–592. [Google Scholar]
- Li, X.; Michlmayr, H.; Schweiger, W.; Malachova, A.; Shin, S.; Huang, Y.; Dong, Y.; Wiesenberger, G.; McCormick, S.; Lemmens, M.; et al. A barley UDP-glucosyltransferase inactivates nivalenol and provides Fusarium Head Blight resistance in transgenic wheat. J. Exp. Bot. 2017, 68, 2187–2197. [Google Scholar] [CrossRef]
- Bethke, G.; Huang, Y.; Hensel, G.; Heinen, S.; Liu, C.; Wyant, S.R.; Li, X.; Quin, M.B.; McCormick, S.; Morrell, P.L.; et al. UDP-glucosyltransferase HvUGT13248 confers type II resistance to Fusarium graminearum in barley. Plant Physiol. 2023, 193, 2691–2710. [Google Scholar]
- Putkaradze, N.; Dato, L.; Kırtel, O.; Hansen, J.; Welner, D.H. Enzymatic glycosylation of aloesone performed by plant UDP-dependent glycosyltransferases. Glycobiology 2024, 34, cwae050. [Google Scholar] [CrossRef]
- Alemayehu, S.; Abera, F.A.; Ayimut, K.M.; Harvey, J.; Mahroof, R.; Subramanyam, B.; Ulmer, J.; Edema, R. Occurrence and Levels of Mycotoxins in On-Farm-Stored Sesame in Major Growing Districts of Ethiopia. Agriculture 2024, 14, 372. [Google Scholar]
- Anyogu, A.; Somorin, Y.M.; Oladipo, A.O.; Raheem, S. Food safety issues associated with sesame seed value chains: Current status and future perspectives. Heliyon 2024, 10, e36347. [Google Scholar]
- Harcourt, B. Multi mycotoxin profile of gamma-radiated sesame seeds from Abuja markets, Nigeria using LC-MS/MS. Nat. Sci. 2012, 10, 127–134. [Google Scholar]
- Villafana, R.T.; Rampersad, S.N. Signatures of TRI5, TRI8 and TRI11 protein sequences of Fusarium incarnatum-equiseti species complex (FIESC) indicate differential trichothecene analogue production. Toxins 2020, 12, 386. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.F.; Sun, J.; Zhao, S.F.; He, L. Black Spot Disease of Chinese Jujube (Ziziphus jujuba) Caused by Fusarium incarnatum in China. Plant Dis. 2016, 100, 529. [Google Scholar]
- Bosch, U.; Mirocha, C.J. Toxin Production by Fusarium Species from Sugar Beets and Natural Occurrence of Zearalenone in Beets and Beet Fiberst. Appl. Environ. Microbiol. 1992, 58, 3233–3239. [Google Scholar] [PubMed]
- Burlakoti, P.; Rivera, V.; Secor, G.A.; Qi, A.; Del rio-Mendoza, L.E.; Khan, M.F.R. Comparative pathogenicity and virulence of Fusarium species on sugar beet. Plant Dis. 2012, 96, 1291–1296. [Google Scholar]
- Bosch, U.; Mirocha, C.J.; Wen, Y. Production of Zearalenone, Moniliformin and Trichothecenes in Intact Sugar Beets Under Laboratory Conditions; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1992. [Google Scholar]
- Cao, S.; Yang, N.; Zhao, C.; Liu, J.; Han, C.; Wu, X. Diversity of Fusarium species associated with root rot of sugar beet in China. J. Gen. Plant Pathol. 2018, 84, 321–329. [Google Scholar]
- Raghavan, S.; Hansen, J.; Sonkar, S.; Kumar, S.; Panchapagesa, M.; Halkjaer Hansen, E.; Hansen, K.R. Methods and Materials for Recombinant Production of Saffron Compounds. U.S. Patent 20140248668A1, 4 September 2014. [Google Scholar]
- Berthiller, F.; Dall’Asta, C.; Schuhmacher, R.; Lemmens, M.; Adam, G.; Krska, A.R. Masked mycotoxins: Determination of a deoxynivalenol glucoside in artificially and naturally contaminated wheat by liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 2005, 53, 3421–3425. [Google Scholar] [CrossRef]
- Schmeitzl, C.; Warth, B.; Fruhmann, P.; Michlmayr, H.; Malachová, A.; Berthiller, F.; Schuhmacher, R.; Krska, R.; Adam, G. The metabolic fate of deoxynivalenol and its acetylated derivatives in a wheat suspension culture: Identification and detection of DON-15-O-glucoside, 15-acetyl-DON-3-O-glucoside and 15-acetyl-DON-3-sulfate. Toxins 2015, 7, 3112–3126. [Google Scholar] [CrossRef]
- Teze, D.; Bidart, G.N.; Welner, D.H. Family 1 glycosyltransferases (GT1, UGTs) are subject to dilution-induced inactivation and low chemo stability toward their own acceptor substrates. Front. Mol. Biosci. 2022, 9, 909659. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]
- Tan, Y.; Yang, J.; Jiang, Y.; Wang, J.; Liu, Y.; Zhao, Y.; Jin, B.; Wang, X.; Chen, T.; Kang, L.; et al. Functional Characterization of UDP-Glycosyltransferases Involved in Anti-viral Lignan Glycosides Biosynthesis in Isatis indigotica. Front. Plant Sci. 2022, 13, 921815. [Google Scholar] [CrossRef]
- McGraphery, K.; Schwab, W. Comparative analysis of high-throughput assays of family-1 plant glycosyltransferases. Int. J. Mol. Sci. 2020, 21, 2208. [Google Scholar] [CrossRef] [PubMed]
- Adiji, O.A.; Docampo-Palacios, M.L.; Alvarez-Hernandez, A.; Pasinetti, G.M.; Wang, X.; Dixon, R.A. UGT84F9 is the major flavonoid UDP-glucuronosyltransferase in Medicago truncatula. Plant Physiol. 2021, 185, 1617–1637. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Ewen, M.A. Toxic Effects of Deoxynivalenol on Ribosomes and Tissues of the Spring Wheat Cultivars Frontana and Casavant; Wiley-Liss, Inc.: Hoboken, NJ, USA, 1997. [Google Scholar]
- Meech, R.; Hu, D.G.; Mckinnon, R.A.; Mubarokah, N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, And Novel Paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef]
- Adam, G.; Mitterbauer, R.; Raditschnig, A.; Poppenberger, B.; Karl, T.; Goritschnig, S.; Weindorfer, H.; Glössl, J. Molecular Mechanisms of Deoxynivalenol Resistance in the Yeast Saccharomyces cerevisiae. Mycotoxin Res. 2001, 17 (Suppl. S1), 19–23. [Google Scholar] [CrossRef]
- Mitterbauer, R.; Heinrich, M.; Rauscher, R.; Lemmens, M.; Bijrstmayr, H.; Adam, G. Trichothecene Resistance in Wheat: Development of Molecular Markers for PDR-type ABC Transporter Genes. Mycotoxin Res. 2003, 19, 82–86. [Google Scholar] [CrossRef]
- Elena, C.; Ravasi, P.; Castelli, M.E.; Peirú, S.; Menzella, H.G. Expression of codon optimized genes in microbial systems: Current industrial applications and perspectives. Front. Microbiol. 2014, 5, 21. [Google Scholar] [CrossRef]
- Hohenblum, H.; Gasser, B.; Maurer, M.; Borth, N.; Mattanovich, D. Effects of Gene Dosage, Promoters, and Substrates on Unfolded Protein Stress of Recombinant Pichia pastoris. Biotechnol. Bioeng. 2004, 85, 367–375. [Google Scholar] [CrossRef]
- Sander, P.; Grunewald, S.; Bach, M.; Haase, W.; Reilander, H.; Michel, H. Heterologous expression of the human D, dopamine receptor in protease-deficient Saccharomyces cerevisiae strains. Eur. J. Biochem. 1994, 226, 697–705. [Google Scholar] [PubMed]
- Zhang, N.; An, Z. Heterologous Protein Expression in Yeasts and Filamentous Fungi. In Manual of Industrial Microbiology and Biotechnology, 3rd ed.; Wiley: Hoboken, NJ, USA, 2010; pp. 145–156. [Google Scholar]
- Ge, F.; Zhu, L.; Aang, A.; Song, P.; Li, W.; Tao, Y.; Du, G. Recent advances in enhanced enzyme activity, thermostability and secretion by N-glycosylation regulation in yeast. Biotechnol. Lett. 2018, 40, 847–854. [Google Scholar] [CrossRef]
- Pisithkul, T.; Patel, N.M.; Amador-Noguez, D. Post-translational modifications as key regulators of bacterial metabolic fluxes. Curr. Opin. Microbiol. 2015, 24, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Ringe, D.; Petsko, G.A. How enzymes work. Science 2008, 320, 1428–1429. [Google Scholar]
- Van Eunen, K.; Bouwman, J.; Daran-Lapujade, P.; Postmus, J.; Canelas, A.B.; Mensonides, F.I.C.; Orij, R.; Tuzun, I.; van den Brink, J.; Smits, G.J.; et al. Measuring enzyme activities under standardized in vivo-like conditions for systems biology. FEBS J. 2010, 277, 749–760. [Google Scholar]
- Goodey, N.M.; Benkovic, S.J. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008, 4, 474–482. [Google Scholar] [PubMed]
- Markina, N.M.; Kotlobay, A.A.; Tsarkova, A.S. Heterologous Metabolic Pathways: Strategies for Optimal Expression in Eukaryotic Hosts. Acta Naturae 2020, 12, 28–39. [Google Scholar]
- Crowe, S.A.; Zhao, X.; Gan, F.; Chen, X.; Hudson, G.A.; Astolfi, M.C.T.; Scheller, H.V.; Liu, Y.; Keasling, J.D. Engineered Saccharomyces cerevisiae as a Biosynthetic Platform of Nucleotide Sugars. ACS Synth. Biol. 2024, 13, 1215–1224. [Google Scholar]
- Lim, E.K.; Bowles, D.J. A class of plant glycosyltransferases involved in cellular homeostatis. EMBO J. 2004, 23, 2915–2922. [Google Scholar]
- Kirana, R.P.; Gaurav, K.; Arora, S.; Wiesenberger, G.; Doppler, M.; Michel, S.; Zimmerl, S.; Matic, M.; Eze, C.E.; Kumar, M.; et al. Identification of a UDP-glucosyltransferase conferring deoxynivalenol resistance in Aegilops tauschii and wheat. Plant Biotechnol. J. 2022, 21, 109–121. [Google Scholar]
- Shin, S.; Torres-Acosta, J.A.; Heinen, S.J.; McCormick, S.; Lemmens, M.; Paris, M.P.K.; Berthiller, F.; Adam, G.; Muehlbauer, G.J. Transgenic Arabidopsis thaliana expressing a barley UDP-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J. Exp. Bot. 2012, 63, 4731–4740. [Google Scholar] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio Team: Boston, MA, USA, 2020. [Google Scholar]

| Enzyme | Organism | Subfamily | Group | Common Name | Accession (NCBI) |
|---|---|---|---|---|---|
| SrUGT | Stevia rebaudiana | 73 | D | Stevia | SEQ ID NO 12 [54] |
| SiUGT | Sesamum indicum | 73 | E | Sesame | XP_011077288 |
| SpUGT | Solanum pennellii | 73 | C | Wild tomato | XP_015088999 |
| EgUGT | Eucalyptus grandis | 76 | H | Red grandis | XP_010033065 |
| PtUGT | Populus trichocarpa | 73 | D | Black cottonwood | KAI5604194 |
| BvUGT | Beta vulgaris subsp. vulgaris | 73 | D | Beet | KMT08362 |
| ZjUGT | Ziziphus jujuba var. spinosa | 71 | E | Red date | WFR85808 |
| AcUGT | Ananas comosus | 73 | C | Pineapple | XP_020089421 |
| Enzyme | Optimal pH | Optimal Temperature (°C) | TM (°C) | Km (mM) | kcat (s−1) | kcat/Km (M−1 s−1) | Reference |
|---|---|---|---|---|---|---|---|
| SrUGT | 8 | 45 | 59.4 ± 0.6 | 0.89 ± 0.18 | 0.80 ± 0.20 | 8.83 × 102 | This study |
| SpUGT | 8 | 32 | 54.3 ± 0.9 | 0.66 ± 0.14 | 0.40 ± 0.02 | 6.28 × 102 | This study |
| EgUGT | 8.5 | 30 | 46.9 ± 0.4 | 0.94 ± 0.19 | 0.04 ± 0.02 | 4.22 × 101 | This study |
| ZjUGT | 8 | 39 | 48.1 ± 0.2 | 0.42 ± 0.14 | 0.93 ± 0.06 | 2.45 × 103 | This study |
| AcUGT | 8 | 45 | 63.0 ± 0.5 | 0.68 ± 0.12 | 0.02 ± 0.01 | 3.05 × 101 | This study |
| OsUGT79 | n.a. | n.a. | n.a. | 0.23 ± 0.06 | 0.57 | 2.48 × 103 | [34] |
| OsUGT79 | n.a. | n.a. | n.a. | 0.061 | 1.07 ± 0.04 | 1.75 × 104 | [40] |
| HvUGT13248 | n.a. | n.a. | n.a. | 3.0 ± 0.6 | 0.78 | 2.60 × 102 | [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Della Gala, V.; Dato, L.; Wiesenberger, G.; Jæger, D.; Adam, G.; Hansen, J.; Welner, D.H. Plant-Derived UDP-Glycosyltransferases for Glycosylation-Mediated Detoxification of Deoxynivalenol: Enzyme Discovery, Characterization, and In Vivo Resistance Assessment. Toxins 2025, 17, 153. https://doi.org/10.3390/toxins17040153
Della Gala V, Dato L, Wiesenberger G, Jæger D, Adam G, Hansen J, Welner DH. Plant-Derived UDP-Glycosyltransferases for Glycosylation-Mediated Detoxification of Deoxynivalenol: Enzyme Discovery, Characterization, and In Vivo Resistance Assessment. Toxins. 2025; 17(4):153. https://doi.org/10.3390/toxins17040153
Chicago/Turabian StyleDella Gala, Valeria, Laura Dato, Gerlinde Wiesenberger, Diana Jæger, Gerhard Adam, Jørgen Hansen, and Ditte Hededam Welner. 2025. "Plant-Derived UDP-Glycosyltransferases for Glycosylation-Mediated Detoxification of Deoxynivalenol: Enzyme Discovery, Characterization, and In Vivo Resistance Assessment" Toxins 17, no. 4: 153. https://doi.org/10.3390/toxins17040153
APA StyleDella Gala, V., Dato, L., Wiesenberger, G., Jæger, D., Adam, G., Hansen, J., & Welner, D. H. (2025). Plant-Derived UDP-Glycosyltransferases for Glycosylation-Mediated Detoxification of Deoxynivalenol: Enzyme Discovery, Characterization, and In Vivo Resistance Assessment. Toxins, 17(4), 153. https://doi.org/10.3390/toxins17040153







