Hortensins, Type 1 Ribosome-Inactivating Proteins from Seeds of Red Mountain Spinach: Isolation, Characterization, and Their Effect on Glioblastoma Cells
Abstract
1. Introduction
2. Results and Discussion
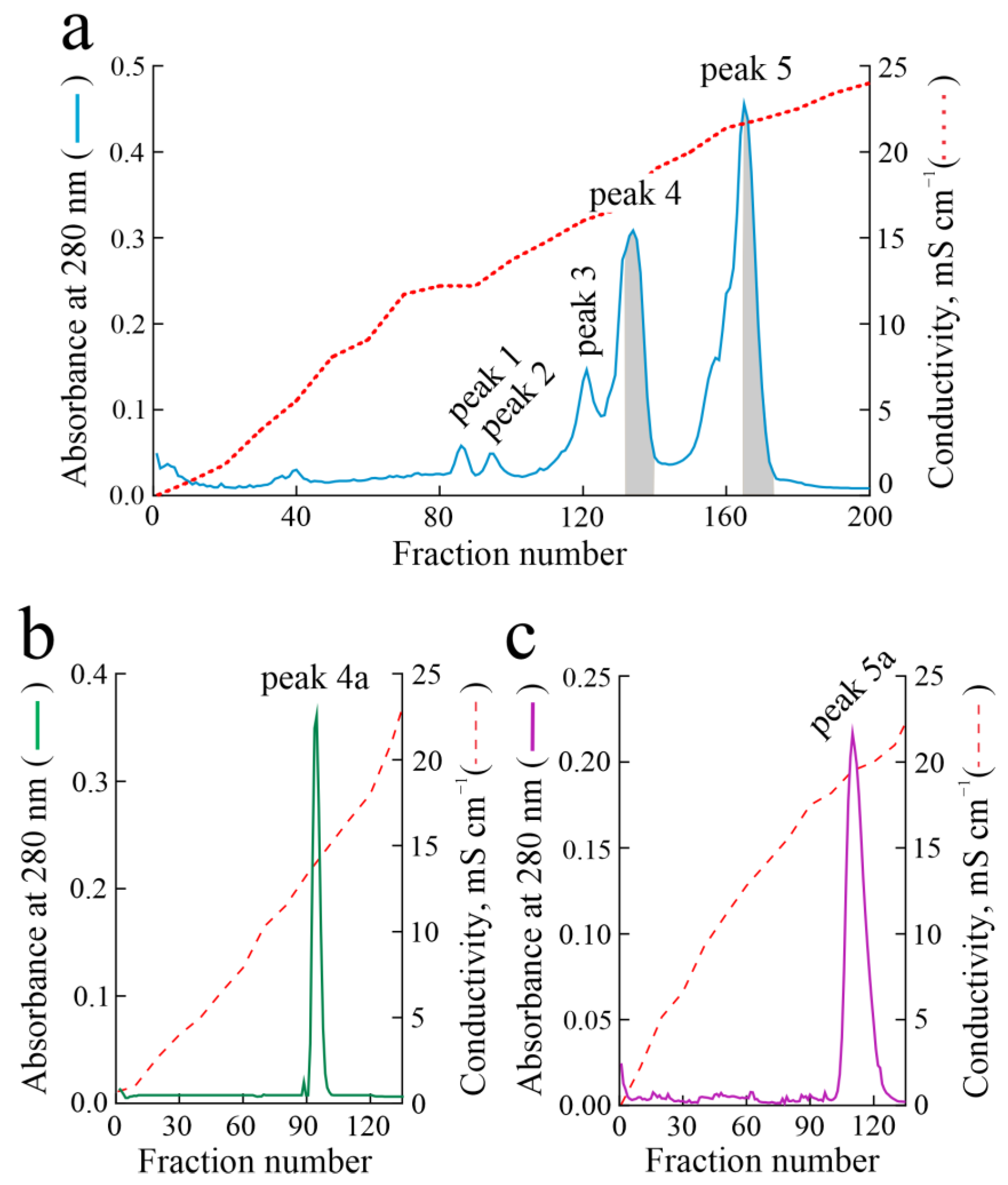
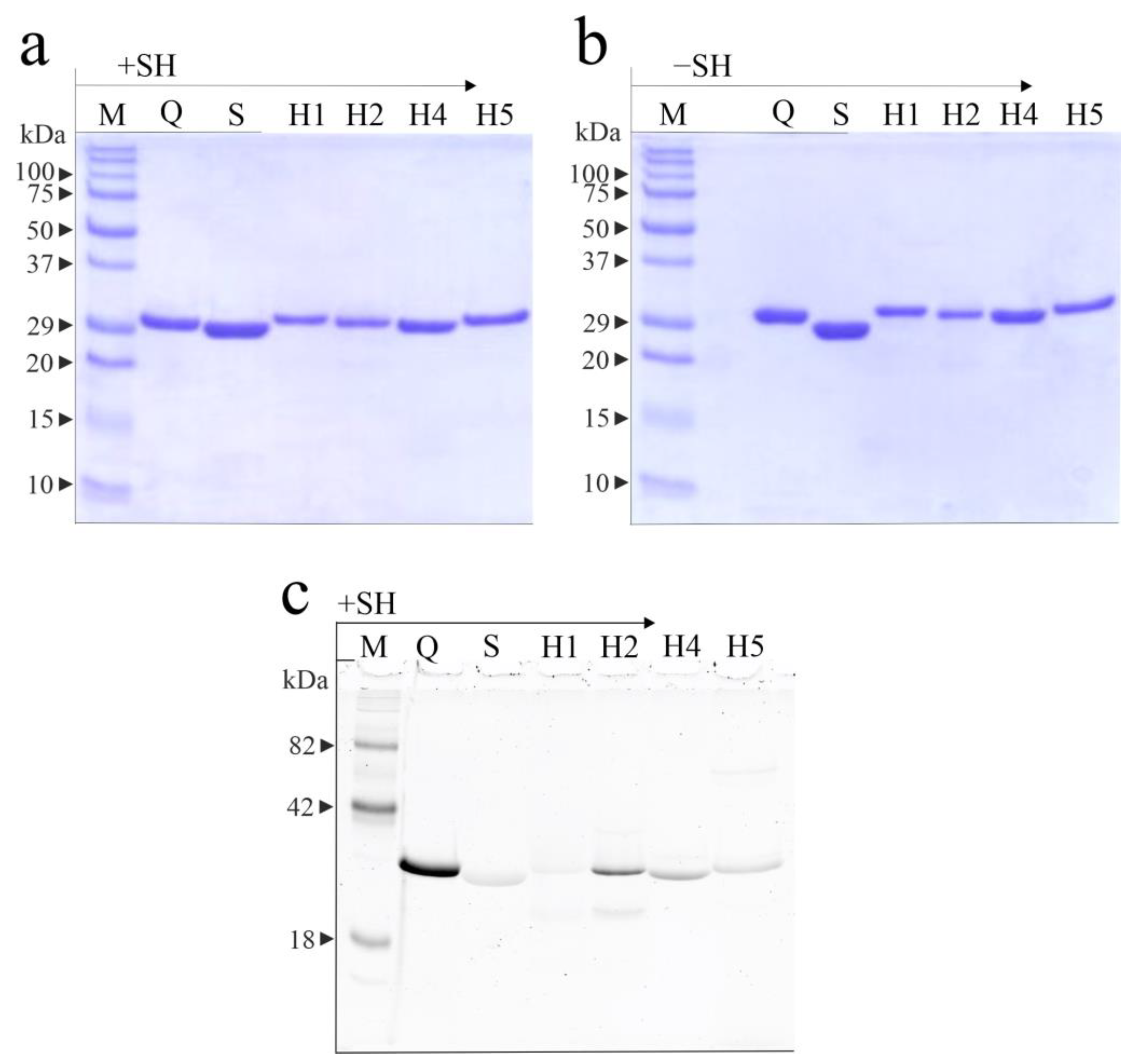
2.1. Isolation of Hortensins
2.2. Enzymatic Features of Hortensins 1, 2, 4, and 5
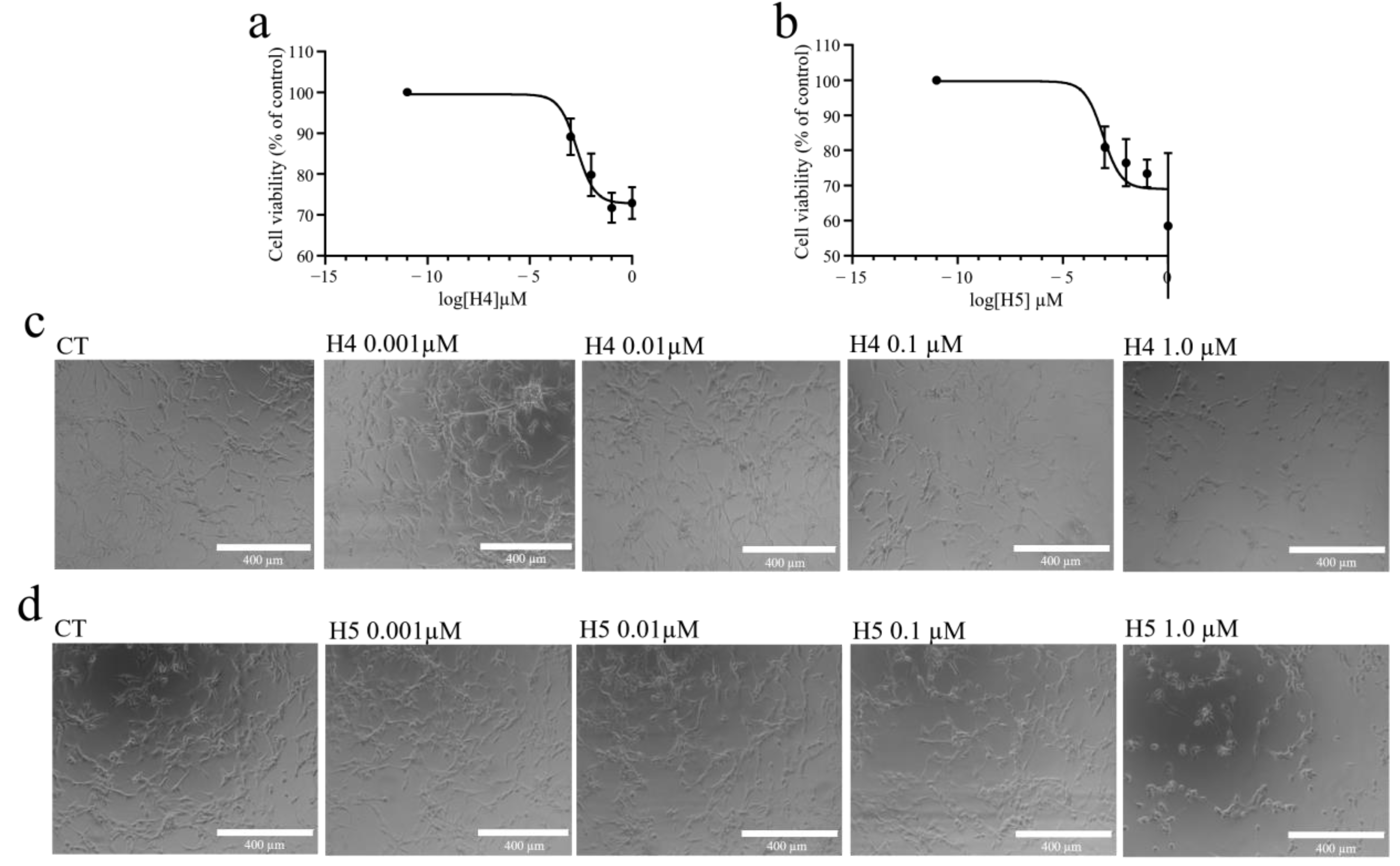
2.3. Inhibiting Effect of Hortensins 4 and 5 on Cell Growth and Morphology of Glioblastoma U87MG Cells
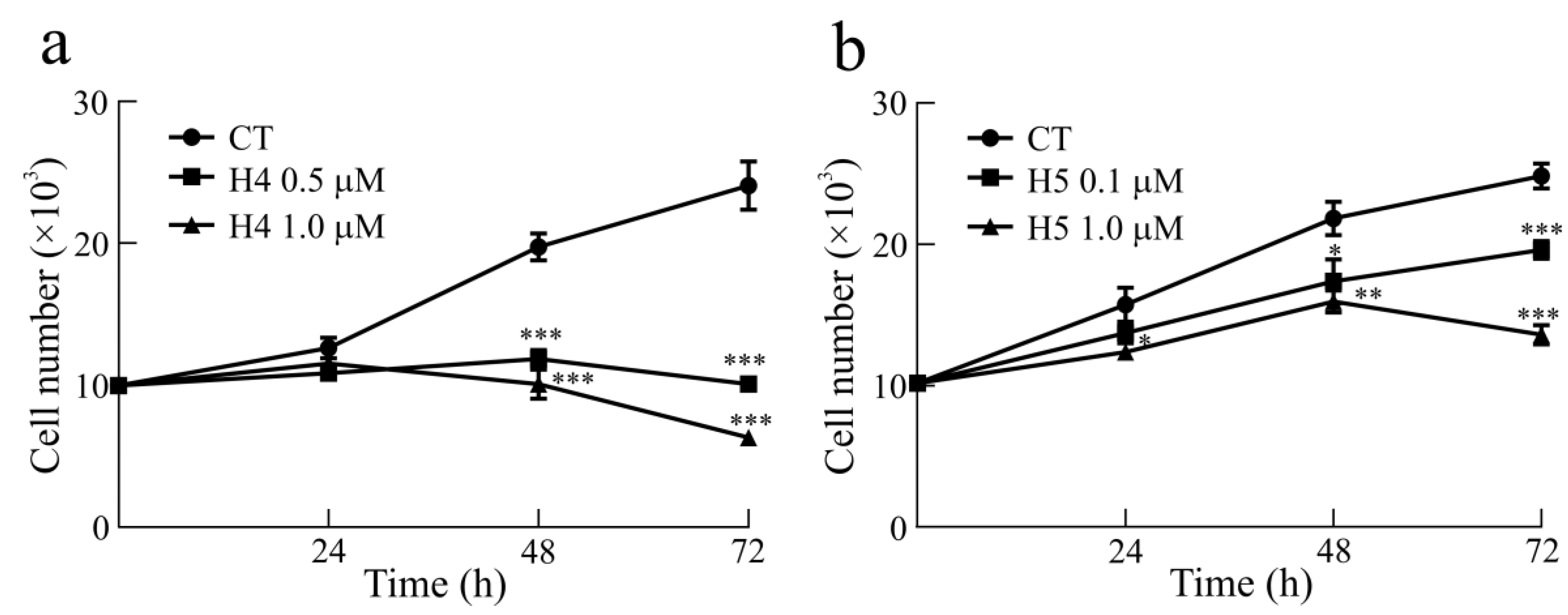
2.4. Cell Growth and Viability of Glioblastoma U87MG Cells after Treatment with Hortensins 4 and 5
2.5. TUNEL Assay of Glioblastoma U87MG Cells after Treatment with Hortensins 4 and 5
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Plant Seeds Samples
4.3. Protein Purification
4.4. Analytical Methods
4.5. Enzymatic Assays
4.6. Cell-Free Protein Synthesis Inhibition
4.7. Cell Culture
4.8. Dose–Response Effect of Hortensins 4 and 5 on U87MG Cells
4.9. Proliferation Assay
4.10. Apoptosis Evaluation in U87MG Cell Line using DNA Fragmentation Detection through TUNEL Assay
4.11. Microscopic Observation of Live Cells
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef]
- Endo, Y.; Tsurugi, K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 1988, 263, 8735–8739. [Google Scholar] [CrossRef]
- Grela, P.; Szajwaj, M.; Horbowicz-Drożdżal, P.; Tchórzewski, M. How Ricin Damages the Ribosome. Toxins 2019, 11, 241. [Google Scholar] [CrossRef]
- Barbieri, L.; Valbonesi, P.; Righi, F.; Zuccheri, G.; Monti, F.; Gorini, P.; Samorí, B.; Stirpe, F. Polynucleotide:Adenosine glycosidase is the sole activity of ribosome-inactivating proteins on DNA. J. Biochem. 2000, 128, 883–889. [Google Scholar] [CrossRef]
- Barbieri, L.; Valbonesi, P.; Bonora, E.; Gorini, P.; Bolognesi, A.; Stirpe, F. Polynucleotide: Adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucl. Acids Res. 1997, 25, 518–522. [Google Scholar] [CrossRef]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-Inactivating Proteins: An Overview. In Plant Toxins; Gopalakrishnakone, P., Carlini, C.R., Ligabue-Braun, R., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 1–29. [Google Scholar]
- Becker, W.; Apel, K. Isolation and characterization of a cDNA clone encoding a novel jasmonate-induced protein of barley (Hordeum vulgare L.). Plant Mol. Biol. 1992, 19, 1065–1067. [Google Scholar] [CrossRef]
- Walsh, T.A.; Morgan, A.E.; Hey, T.D. Characterization and molecular cloning of a proenzyme form of a ribosome-inactivating protein from maize. Novel mechanism of proenzyme activation by proteolytic removal of a 2.8-kilodalton internal peptide segment. J. Biol. Chem. 1991, 266, 23422–23427. [Google Scholar] [CrossRef]
- Stirpe, F.; Barbieri, L.; Gorini, P.; Valbonesi, P.; Bolognesi, A.; Polito, L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996, 382, 309–312. [Google Scholar] [CrossRef]
- Mishra, V.; Mishra, R.; Shamra, R.S. Ribosome inactivating proteins—An unfathomed biomolecule for developing multi-stress tolerant transgenic plants. Int. J. Biol. Macromol. 2022, 210, 107–122. [Google Scholar] [CrossRef]
- Zhu, F.; Zhou, Y.K.; Ji, Z.L.; Chen, X.R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef]
- Gilabert-Oriol, R.; Weng, A.; Mallinckrodt, B.; Melzig, M.F.; Fuchs, H.; Thakur, M. Immunotoxins constructed with ribosome-inactivating proteins and their enhancers: A lethal cocktail with tumor specific efficacy. Curr. Pharm. Des. 2014, 20, 6584–6643. [Google Scholar] [CrossRef]
- de Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins 2010, 2, 2699–2737. [Google Scholar] [CrossRef]
- Citores, L.; Iglesias, R.; Ferreras, J.M. Antiviral Activity of Ribosome-Inactivating Proteins. Toxins 2021, 13, 80. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, S.; Sharma, N.R.; Paul, K. Expanding role of ribosome-inactivating proteins: From toxins to therapeutics. IUBMB Life 2023, 75, 82–96. [Google Scholar] [CrossRef]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol. 2014, 85, 575–588. [Google Scholar] [CrossRef]
- Lapadula, W.J.; Ayub, M.J. Ribosome Inactivating Proteins from an evolutionary perspective. Toxicon 2017, 136, 6–14. [Google Scholar] [CrossRef]
- Dougherty, K.; Hudak, K.A. Phylogeny and domain architecture of plant ribosome inactivating proteins. Phytochemistry 2022, 202, 113337. [Google Scholar] [CrossRef]
- Landi, N.; Ruocco, M.R.; Ragucci, S.; Aliotta, F.; Nasso, R.; Pedone, P.V.; Di Maro, A. Quinoa as source of type 1 ribosome inactivating proteins: A novel knowledge for a revision of its consumption. Food Chem. 2021, 342, 128337. [Google Scholar] [CrossRef]
- Landi, N.; Ragucci, S.; Citores, L.; Clemente, A.; Hussain, H.Z.F.; Iglesias, R.; Ferreras, J.M.; Di Maro, A. Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L. Toxins 2022, 14, 566. [Google Scholar] [CrossRef]
- Schrot, J.; Weng, A.; Melzig, M.F. Ribosome-inactivating and related proteins. Toxins 2015, 7, 1556–1615. [Google Scholar] [CrossRef]
- Iglesias, R.; Pérez, Y.; de Torre, C.; Ferreras, J.M.; Antolín, P.; Jiménez, P.; Rojo, M.A.; Méndez, E.; Girbés, T. Molecular characterization and systemic induction of single-chain ribosome-inactivating proteins (RIPs) in sugar beet (Beta vulgaris) leaves. J. Exp. Bot. 2005, 56, 1675–1684. [Google Scholar] [CrossRef]
- Kwon, S.Y.; An, C.S.; Liu, J.R.; Kwak, S.S.; Lee, H.S.; Kim, J.K.; Paek, K.H. Molecular cloning of a cDNA encoding ribosome inactivating protein from Amaranthus viridis and its expression in E. coli. Mol. Cells 2000, 10, 8–12. [Google Scholar] [CrossRef]
- Kawade, K.; Ishizaki, T.; Masuda, K. Differential expression of ribosome-inactivating protein genes during somatic embryogenesis in spinach (Spinacia oleracea). Physiol. Plant 2008, 134, 270–281. [Google Scholar] [CrossRef]
- Jan, N.; Hussain, S.Z.; Naseer, B.; Bhat, T.A. Amaranth and quinoa as potential nutraceuticals: A review of anti-nutritional factors, health benefits and their applications in food, medicinal and cosmetic sectors. Food Chem. X 2023, 18, 100687. [Google Scholar] [CrossRef]
- Hunt, S.P.; Jarvis, D.E.; Larsen, D.J.; Mosyakin, S.L.; Kolano, B.A.; Jackson, E.W.; Martin, S.L.; Jellen, E.N.; Maughan, P.J. A Chromosome-Scale Assembly of the Garden Orach (Atriplex hortensis L.) Genome Using Oxford Nanopore Sequencing. Front. Plant Sci. 2020, 11, 624. [Google Scholar] [CrossRef]
- Wright, K.H.; Pike, O.A.; Fairbanks, D.J.; Huber, C.S. Composition of Atriplex hortensis, Sweet and Bitter Chenopodium quinoa Seeds. J. Food Sci. 2002, 67, 1383–1385. [Google Scholar] [CrossRef]
- Rinchen, T.; Singh, N.; Maurya, S.B.; Soni, V.; Phour, M.; Kumar, B. Morphological characterization of indigenous vegetable (Atriplex hortensis L.) from trans-Himalayan region of Ladakh. Australian J. Crop Sci. 2017, 11, 258–2631835. [Google Scholar] [CrossRef]
- Grieve, M. A Modern Herbal; Harcourt, Brace & Company: New York, NY, USA, 1931. [Google Scholar]
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282. [Google Scholar] [CrossRef]
- Stirpe, F.; Battelli, M.G. Ribosome-inactivating proteins: Progress and problems. Cell. Mol. Life Sci. 2006, 63, 1850–1866. [Google Scholar] [CrossRef]
- Rotondo, R.; Ragucci, S.; Castaldo, S.; Oliva, M.A.; Landi, N.; Pedone, P.V.; Arcella, A.; Di Maro, A. Cytotoxicity Effect of Quinoin, Type 1 Ribosome-Inactivating Protein from Quinoa Seeds, on Glioblastoma Cells. Toxins 2021, 13, 684. [Google Scholar] [CrossRef]
- Barbieri, L.; Stoppa, C.; Bolognesi, A. Large-scale chromatographic purification of ribosome-inactivating proteins. J. Chromatogr. A 1987, 408, 235–243. [Google Scholar] [CrossRef]
- Parente, A.; De Luca, P.; Bolognesi, A.; Barbieri, L.; Battelli, M.G.; Abbondanza, A.; Sande, M.J.; Gigliano, G.S.; Tazzari, P.L.; Stirpe, F. Purification and partial characterization of single-chain ribosome-inactivating proteins from the seeds of Phytolacca dioica L. Biochim. Biophys. Acta 1993, 1216, 43–49. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Di Maro, A.; Chambery, A.; Daniele, A.; Casoria, P.; Parente, A. Isolation and characterization of heterotepalins, type 1 ribosome-inactivating proteins from Phytolacca heterotepala leaves. Phytochemistry 2007, 68, 767–776. [Google Scholar] [CrossRef]
- Blank, A.; Sugiyama, R.H.; Dekker, C.A. Activity staining of nucleolytic enzymes after sodium dodecyl sulfate-polyacrylamide gel electrophoresis: Use of aqueous isopropanol to remove detergent from gels. Anal. Biochem. 1982, 120, 267–275. [Google Scholar] [CrossRef]
- Langer, M.; Rothe, M.; Eck, J.; Möckel, B.; Zinke, H. A nonradioactive assay for ribosome-inactivating proteins. Anal. Biochem. 1996, 243, 150–153. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragucci, S.; Russo, V.; Clemente, A.; Campanile, M.G.; Oliva, M.A.; Landi, N.; Pedone, P.V.; Arcella, A.; Di Maro, A. Hortensins, Type 1 Ribosome-Inactivating Proteins from Seeds of Red Mountain Spinach: Isolation, Characterization, and Their Effect on Glioblastoma Cells. Toxins 2024, 16, 135. https://doi.org/10.3390/toxins16030135
Ragucci S, Russo V, Clemente A, Campanile MG, Oliva MA, Landi N, Pedone PV, Arcella A, Di Maro A. Hortensins, Type 1 Ribosome-Inactivating Proteins from Seeds of Red Mountain Spinach: Isolation, Characterization, and Their Effect on Glioblastoma Cells. Toxins. 2024; 16(3):135. https://doi.org/10.3390/toxins16030135
Chicago/Turabian StyleRagucci, Sara, Veronica Russo, Angela Clemente, Maria Giuseppina Campanile, Maria Antonietta Oliva, Nicola Landi, Paolo Vincenzo Pedone, Antonietta Arcella, and Antimo Di Maro. 2024. "Hortensins, Type 1 Ribosome-Inactivating Proteins from Seeds of Red Mountain Spinach: Isolation, Characterization, and Their Effect on Glioblastoma Cells" Toxins 16, no. 3: 135. https://doi.org/10.3390/toxins16030135
APA StyleRagucci, S., Russo, V., Clemente, A., Campanile, M. G., Oliva, M. A., Landi, N., Pedone, P. V., Arcella, A., & Di Maro, A. (2024). Hortensins, Type 1 Ribosome-Inactivating Proteins from Seeds of Red Mountain Spinach: Isolation, Characterization, and Their Effect on Glioblastoma Cells. Toxins, 16(3), 135. https://doi.org/10.3390/toxins16030135