Immunological Cross-Reactivity and Preclinical Assessment of a Colombian Anticoral Antivenom against the Venoms of Three Micrurus Species
Abstract
1. Introduction
2. Results and Discussion
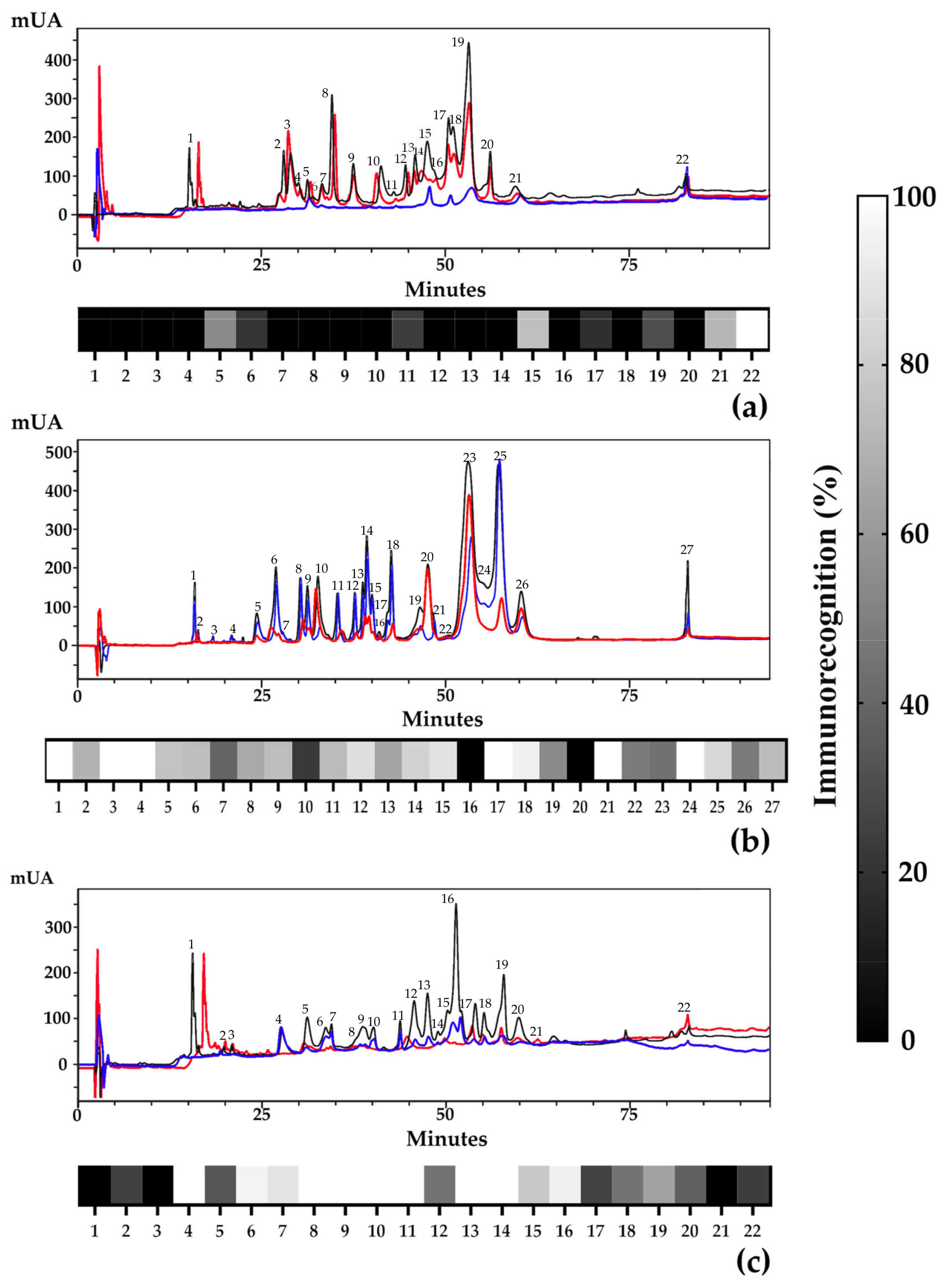
2.1. Preferential Recognition towards Complete Venoms of the Monad Group
2.2. High-Molecular-Weight Components Are Better Immunogens
2.3. Hydrophobicity and Large Molecular Size as Determinants of Recognition by the INS Antivenom
2.4. Observed Immunoreactivity against M. sangilensis and M. medemi Is Confirmed by the In Vivo Neutralization Assay
3. Conclusions
4. Materials and Methods
4.1. Venoms and Antivenom
4.2. Protein Quantification
4.3. Polyacrylamide Gel Electrophoresis (SDS-PAGE)
4.4. Reverse-Phase Liquid Chromatography (RP-HPLC)
4.5. Antivenom Assessment
4.5.1. Affinity Chromatography
4.5.2. Western Blot
4.5.3. Enzyme-Linked Immunosorbent Assay—ELISA and EC50 Determination
4.5.4. In Vivo Neutralization
4.6. Statistical Analysis
4.7. Ethical Statement
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gutiérrez, J.M.; Calvete, J.; Habib, A.; Harrison, R.; Williams, D.; Warrell, D. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017, 3, 17063. [Google Scholar] [CrossRef] [PubMed]
- Kasturiratne, A.; Wickremasinghe, A.R.; De Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; De Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, 1591–1604. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J. Accidente Ofídico, Período Epidemiológico XIII de 2022, Colombia; Instituto Nacional de Salud: Bogotá, Colombia, 2022.
- Lynch, J.; Angarita-Sierra, T.; Ruiz Gomez, F. Programa Nacional para la Conservación de las Serpientes Presentes en Colombia; Ministerio de Ambiente y Desarrollo Sostenible, Universidad Nacional de Colombia, Instituto Nacional de Salud: Bogotá, Colombia, 2014; ISBN 978-958-8901-18-3.
- Campbell, J.A.; Lamar, W. The Venomous Reptiles of the Western Hemisphere, 1st ed.; Comstock Press: Ithaca, NY, USA, 2004; ISBN 0-8014-4141-2. [Google Scholar]
- Savage, J.A.Y.M.; Slowinski, J.B. The colouration of the venomous coral snakes (family Elapidae) and their mimics (families Aniliidae and Colubridae). Biol. J. Linn. Soc. 1992, 45, 235–254. [Google Scholar] [CrossRef]
- Roze, J.A. Coral Snakes of the Americas: Biology, Identification, and Venoms; Krieger Publishing Company: Malabar, FL, USA, 1996; ISBN 0-89464-847-0. [Google Scholar]
- Gómez, J.P.H.; Ramírez, M.V.; Gómez, F.J.R.; Fouquet, A.; Fritz, U. Multilocus phylogeny clarifies relationships and diversity within the Micrurus lemniscatus complex (Serpentes: Elapidae). Salamandra 2021, 57, 229–239. [Google Scholar]
- Morales-Betancourt, M.; Lasso, C.; Páez, V.; Bock, B. Libro Rojo de Reptiles de Colombia; Instituto de Investigación de Recursos Biológicos Alexander von Humboldt (IAvH), Universidad de Antioquia: Bogotá, Colombia, 2015; ISBN 9789588889795. [Google Scholar]
- Uetz, P.; Freed, P.; Aguilar, R.; Reyes, F.; Hošek, J. A Quarter Century of Reptile and Amphibian Databases. Herpetol. Rev. 2021, 52, 246–255. [Google Scholar]
- Gómez, J. Informe del Evento: Accidente Ofídico, Colombia. 2021. Available online: https://www.ins.gov.co/buscador-eventos/Informesdeevento/ACCIDENTEOFIDICOINFORME2021.pdf (accessed on 12 May 2023).
- Da Silva, N.J., Jr.; Bucaretchi, F. Mecanismo de açao do veneno elapídico e aspectos clínicos dos acidentes. In Animais Peçonhentos no Brasil; Cardoso, J., de Siqueira, F., Wen, F., Sant´Ana, C., Haddad, V., Eds.; Sarvier: São Paulo, Brazil, 2009; pp. 116–124. [Google Scholar]
- Rey-Suárez, P.; Núñez, V.; Gutiérrez, J.M.; Lomonte, B. Proteomic and biological characterization of the venom of the redtail coral snake, Micrurus mipartitus (Elapidae), from Colombia and Costa Rica. J. Proteom. 2011, 75, 655–667. [Google Scholar] [CrossRef]
- Rey-Suárez, P.; Núñez, V.; Fernández, J.; Lomonte, B.; Nuñez, V.; Fernández, J.; Lomonte, B. Integrative characterization of the venom of the coral snake Micrurus dumerilii (Elapidae) from Colombia: Proteome, toxicity, and cross-neutralization by antivenom. J. Proteom. 2016, 136, 262–273. [Google Scholar] [CrossRef]
- Caicedo-Portilla, J.R.; Lynch, J.D. Micrurus medemi. In Libro Rojo de Reptiles de Colombia (2015); Orales-Betancourt, M.A., Lasso, C.A., Páez, V.P., Bock, B.C., Eds.; Instituto de Investigación de Recursos Biológicos Alexander von Humboldt (IAvH), Universidad de Antioquia: Bogotá, Colombia, 2015; pp. 98–100. ISBN 978-958-888-979-5. [Google Scholar]
- Montoya, A.; Díaz Flórez, R.A.; Mancera, D. A new departmental and elevational record for the Villavicencio Coralsnake, Micrurus medemi Roze 1967 (Squamata: Elapidae), in Colombia. Reptil. Amphib. 2022, 29, 189–190. [Google Scholar] [CrossRef]
- Angarita-Sierra, T.; Montaño-Londoño, L.F.; Bravo-Vega, C.A. ID please: Evaluating the utility of Facebook as a source of data for snake research and conservation. An. Acad. Bras. Cienc. 2022, 94, e20211043. [Google Scholar] [CrossRef]
- Niceforo-Maria, H. Los ofidios de Colombia. Rev. Acad. Colomb. Ciencias Exactas Físicas Nat. 1942, 5, 89–101. [Google Scholar]
- Lalloo, D.G.; Theakston, R.D.G. Snake Antivenoms. J. Toxicol. Clin. Toxicol. 2003, 41, 277–290. [Google Scholar] [CrossRef]
- Bolaños, R.; Cerdas, L.; Abalos, J.W. Venoms of coral snakes (Micrurus spp.): Report on a multivalent antivenin for the Americas. Bull. Pan Am. Health Organ. 1978, 12, 23–27. [Google Scholar] [PubMed]
- Kini, R.M.; Doley, R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef]
- Lomonte, B.; Sasa, M.; Rey-Suárez, P.; Bryan, W.; Gutiérrez, J.M.J.M. Venom of the coral snake Micrurus clarki: Proteomic profile, toxicity, immunological cross-neutralization, and characterization of a three-finger Toxin. Toxins 2016, 8, 138. [Google Scholar] [CrossRef]
- Rodríguez-Vargas, A.; Franco-Vásquez, A.M.; Bolívar-Barbosa, J.A.; Vega, N.; Reyes-Montaño, E.; Arreguín-Espinosa, R.; Carbajal-Saucedo, A.; Angarita-Sierra, T.; Ruiz-Gómez, F. Unveiling the Venom Composition of the Colombian Coral Snakes Micrurus helleri, M. medemi, and M. sangilensis. Toxins 2023, 15, 622. [Google Scholar] [CrossRef]
- Guerrero-Garzón, J.F.; Bénard-Valle, M.; Restano-Cassulini, R.; Zamudio, F.; Corzo, G.; Alagón, A.; Olvera-Rodríguez, A. Cloning and sequencing of three-finger toxins from the venom glands of four Micrurus species from Mexico and heterologous expression of an alpha-neurotoxin from Micrurus diastema. Biochimie 2018, 147, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.L.; Arbuckle, K. Coevolution of snake venom toxic activities and diet: Evidence that ecological generalism favours toxicological diversity. Toxins 2019, 11, 711. [Google Scholar] [CrossRef]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Dashevsky, D.; Fry, B.G. Ancient Diversification of Three-Finger Toxins in Micrurus Coral Snakes. J. Mol. Evol. 2018, 86, 58–67. [Google Scholar] [CrossRef]
- Sanz, L.; Quesada-Bernat, S.; Ramos, T.; Casais-e-Silva, L.L.; Corrêa-Netto, C.; Silva-Haad, J.J.; Sasa, M.; Lomonte, B.; Calvete, J.J. New insights into the phylogeographic distribution of the 3FTx/PLA 2 venom dichotomy across genus Micrurus in South America. J. Proteom. 2019, 200, 90–101. [Google Scholar] [CrossRef]
- Fernández, J.; Alape-Girón, A.; Angulo, Y.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.; Lomonte, B.; Fernández, J.; Alape-Girón, A.; Angulo, Y.; et al. Venomic and Antivenomic Analyses of the Central American Coral Snake, Micrurus nigrocinctus (Elapidae). J. Proteome Res. 2011, 10, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Lomonte, B.; Lohse, B.; Fernández, J.; Gutiérrez, J.M. Unveiling the nature of black mamba (Dendroaspis polylepis) venom through venomics and antivenom immunoprofiling: Identification of key toxin targets for antivenom development. J. Proteom. 2015, 119, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Sells, P.G.; Jones, R.G.; Laing, G.D.; Smith, D.C.; Theakston, R.D. Experimental evaluation of ovine antisera to Thai cobra (Naja kaouthia) venom and its alpha-neurotoxin. Toxicon 1994, 32, 1657–1665. [Google Scholar] [CrossRef]
- de la Rosa, G.; Olvera, F.; Archundia, I.G.; Lomonte, B.; Alagón, A.; Corzo, G. Horse immunization with short-chain consensus α-neurotoxin generates antibodies against broad spectrum of elapid venomous species. Nat. Commun. 2019, 10, 3642. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, G.D.; Furtado, M.D.F.D.; Portaro, F.C.V.; Sant’Anna, O.A.; Tambourgi, D.V. Diversity of Micrurus snake species related to their venom toxic effects and the prospective of antivenom neutralization. PLoS Negl. Trop. Dis. 2010, 4, e622. [Google Scholar] [CrossRef] [PubMed]
- Isbister, G.K. Antivenom efficacy or effectiveness: The Australian experience. Toxicology 2010, 268, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, G.D.; Sant’anna, O.A.; Marcelino, J.R.; Lustoza Da Luz, A.C.; Teixeira Da Rocha, M.M.; Tambourgi, D.V. Micrurus snake species: Venom immunogenicity, antiserum cross-reactivity and neutralization potential. Toxicon 2016, 117, 59–68. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Solano, G.; Pla, D.; Herrera, M.; Segura, Á.; Villalta, M.; Vargas, M.; Sanz, L.; Lomonte, B.; Calvete, J.; et al. Assessing the preclinical efficacy of antivenoms: From the lethality neutralization assay to antivenomics. Toxicon 2013, 69, 168–179. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Lomonte, B.; Sanz, L.; Calvete, J.; Pla, D. Immunological profile of antivenoms: Preclinical analysis of the efficacy of a polyspecific antivenom through antivenomics and neutralization assays. J. Proteom. 2014, 105, 340–350. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Solano, G.; Pla, D.; Herrera, M.; Segura, Á.; Vargas, M.; Villalta, M.; Sánchez, A.; Sanz, L.; Lomonte, B.; et al. Preclinical evaluation of the efficacy of antivenoms for snakebite envenoming: State-of-the-art and challenges ahead. Toxins 2017, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; María Gutiérrez, J.; Calvete, J. Second generation snake antivenomics: Comparing immunoaffinity and immunodepletion protocols. Toxicon 2012, 60, 688–699. [Google Scholar] [CrossRef]
- Calvete, J.; Sanz, L.; Pla, D.; Lomonte, B.; Gutiérrez, J.M. Omics meets biology: Application to the design and preclinical assessment of antivenoms. Toxins 2014, 6, 3388–3405. [Google Scholar] [CrossRef]
- Instituto Nacional de Salud. Antiveneno Anticoral Polivalente INS, Colombia, Medication Package Insert. 2016; pp. 1–3. Available online: https://www.ins.gov.co/lineas-de-accion/Produccion/SiteAssets/Paginas/suero-antiofidico-polivalente/Inserto%20Antiveneno%20Anticoral%20Polivalente.pdf (accessed on 21 December 2023).
- Castillo-Beltrán, M.C.; Hurtado-Gómez, J.P.; Corredor-Espinel, V.; Ruiz-Gómez, F.J. A polyvalent coral snake antivenom with broad neutralization capacity. PLoS Negl. Trop. Dis. 2018, 13, e0007250. [Google Scholar] [CrossRef]
- Gómez-Robles, J.; Rey-Suárez, P.; Pereañez, J.A.; Lomonte, B.; Núñez, V. Antibodies against a single fraction of Micrurus dumerilii venom neutralize the lethal effect of whole venom. Toxicol. Lett. 2023, 374, 77–84. [Google Scholar] [CrossRef]
- Pulido-Méndez, M.M.; Azuaje, E.; Rodríguez-Acosta, A. Immunotoxicological effects triggered by the rattlesnake Crotalus durissus cumanensis, mapanare (Bothrops colombiensis) venoms and its purified fractions on spleen and lymph nodes cells. Immunopharmacol. Immunotoxicol. 2020, 42, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Laboratorios Probiol. Suero Antiofídico Anticoral Liofilizado. 2023. Available online: https://www.probiol.com/linea-humana.html (accessed on 21 December 2023).
- de Roodt, R.A.; Lanari, L.C.; Ramírez, J.E.; Gómez, C.; Barragán, J.; Litwin, S.; Henriët van Grootheest, J.; Desio, M.; Dokmetjian, J.C.; Dolab, J.A.; et al. Cross-reactivity of some Micrurus venoms against experimental and therapeutic anti-Micrurus antivenoms. Toxicon 2021, 200, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Ramos, H.R.; Vassão, R.C.; de Roodt, A.R.; Santos E Silva, E.C.; Mirtschin, P.; Ho, P.L.; Spencer, P.J. Cross neutralization of coral snake venoms by commercial Australian snake antivenoms. Clin. Toxicol. 2017, 55, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Lim, S.E.; Tan, N.H. Venomics of the beaked sea snake, Hydrophis schistosus: A minimalist toxin arsenal and its cross-neutralization by heterologous antivenoms. J. Proteom. 2015, 126, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Manson, E.Z.; Kyama, M.C.; Kimani, J.; Bocian, A.; Hus, K.K.; Leg, J.; Kimotho, J.H. Development and Characterization of Anti-Naja ashei Three-Finger Toxins (3FTxs)-Specific Monoclonal Antibodies and Evaluation of Their In Vitro Inhibition Activity. Toxins 2022, 14, 285. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Calvete, J. Strategies in “snake venomics” aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Rey-Suárez, P.; Mónica, S.; Torres, U.; Marin-villa, M.; Lomonte, B.; Núñez, V. Novel three-finger toxins from Micrurus dumerilii and Micrurus mipartitus coral snake venoms: Phylogenetic relationships and characterization of Clarkitoxin-I-Mdum. Toxicon 2019, 170, 85–93. [Google Scholar] [CrossRef]
- Rey-Suárez, P.; Floriano, R.S.; Rostelato-Ferreira, S.; Saldarriaga-Córdoba, M.; Núñez, V.; Rodrigues-Simioni, L.; Lomonte, B. Mipartoxin-I, a novel three-finger toxin, is the major neurotoxic component in the venom of the redtail coral snake Micrurus mipartitus (Elapidae). Toxicon 2012, 60, 851–863. [Google Scholar] [CrossRef]
- Instituto Clodomiro Picado. Suero Antiofídico Anti-Coral. Available online: https://www.kerwa.ucr.ac.cr/bitstream/handle/10669/866/INSERTO_ACLQ.pdf?sequence=1&isAllowed=y (accessed on 21 December 2023).
- Sánchez, E.; Lopez-Johnston, J.C.; Rodríguez-Acosta, A.; Pérez, J.C. Neutralization of two North American coral snake venoms with United States and Mexican antivenoms. Toxicon 2008, 51, 297–303. [Google Scholar] [CrossRef]
- de Roodt, A.R.; Paniagua-Solis, J.F.; Dolab, J.A.; Estévez-Ramiréz, J.; Ramos-Cerrillo, B.; Litwin, S.; Dokmetjian, J.C.; Alagón, A. Effectiveness of two common antivenoms for North, Central, and South American Micrurus envenomations. J. Toxicol. Clin. Toxicol. 2004, 42, 171–178. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gutiérrez, J.M.; Fan, H.W.; Silvera, C.L.M.; Angulo, Y. Stability, distribution and use of antivenoms for snakebite envenomation in Latin America: Report of a workshop. Toxicon 2009, 53, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Hermanson, G.T.; Mallia, A.K.; Smith, P.K. Immobilized Affinity Ligand Techniques; Academic Press: San Diego, Brazil, 1992; ISBN 0123423309. [Google Scholar]
- Burnette, W.N. “Western Blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 1981, 112, 195–203. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Guidelines for the Production, Control and Regulation of Snake Antivenom Inmunoglubolins; WHO: Geneva, Switzerland, 2010. [Google Scholar]
- World Health Organization. Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins Annex 5; Replacement of Annex 2 of WHO Technical Report Series, No. 964. WHO Technical Report Series; World Health Organization: Geneva, Switzerland, 2017; pp. 197–388. [Google Scholar]
- ICP. Determinación de Actividades Tóxicas de Venenos de Serpientes y Neutralización por Antivenenos. Manual de Métodos de Laboratorio; Universidad de Costa Rica: San Diego, Brazil, 2007. [Google Scholar]
- Spearman, C. The method of ‘right and wrong cases’ (‘constant stimuli’) without Gauss’s formulae. Br. J. Psychol. 1908, 2, 227–242. [Google Scholar] [CrossRef]
- Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn. Schmiedebergs. Arch. Exp. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar] [CrossRef]


| Venom | Median Effective Dose (ED50) * |
|---|---|
| Micrurus helleri | 0.58 (0.4–0.84) ** |
| Micrurus medemi | 0.68 (0.44–1.06) ** |
| Micrurus sangilensis | 0.75 (0.53–1.07) ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Vargas, A.; Franco-Vásquez, A.M.; Triana-Cerón, M.; Alam-Rojas, S.N.; Escobar-Wilches, D.C.; Corzo, G.; Lazcano-Pérez, F.; Arreguín-Espinosa, R.; Ruiz-Gómez, F. Immunological Cross-Reactivity and Preclinical Assessment of a Colombian Anticoral Antivenom against the Venoms of Three Micrurus Species. Toxins 2024, 16, 104. https://doi.org/10.3390/toxins16020104
Rodríguez-Vargas A, Franco-Vásquez AM, Triana-Cerón M, Alam-Rojas SN, Escobar-Wilches DC, Corzo G, Lazcano-Pérez F, Arreguín-Espinosa R, Ruiz-Gómez F. Immunological Cross-Reactivity and Preclinical Assessment of a Colombian Anticoral Antivenom against the Venoms of Three Micrurus Species. Toxins. 2024; 16(2):104. https://doi.org/10.3390/toxins16020104
Chicago/Turabian StyleRodríguez-Vargas, Ariadna, Adrián Marcelo Franco-Vásquez, Miguel Triana-Cerón, Shaha Noor Alam-Rojas, Derly C. Escobar-Wilches, Gerardo Corzo, Fernando Lazcano-Pérez, Roberto Arreguín-Espinosa, and Francisco Ruiz-Gómez. 2024. "Immunological Cross-Reactivity and Preclinical Assessment of a Colombian Anticoral Antivenom against the Venoms of Three Micrurus Species" Toxins 16, no. 2: 104. https://doi.org/10.3390/toxins16020104
APA StyleRodríguez-Vargas, A., Franco-Vásquez, A. M., Triana-Cerón, M., Alam-Rojas, S. N., Escobar-Wilches, D. C., Corzo, G., Lazcano-Pérez, F., Arreguín-Espinosa, R., & Ruiz-Gómez, F. (2024). Immunological Cross-Reactivity and Preclinical Assessment of a Colombian Anticoral Antivenom against the Venoms of Three Micrurus Species. Toxins, 16(2), 104. https://doi.org/10.3390/toxins16020104






