Abstract
Xenorhabdus and Photorhabdus, bacterial symbionts of entomopathogenic nematodes Steinernema and Heterorhabditis, respectively, have several biological activities including insecticidal and antimicrobial activities. Thus, XnChi, XhChi, and PtChi, chitinases of X. nematophila, X. hominickii, and P. temperata isolated from Korean indigenous EPNs S. carpocapsae GJ1-2, S. monticolum GJ11-1, and H. megidis GJ1-2 were cloned and expressed in Escherichia coli BL21 to compare their biological activities. Chitinase proteins of these bacterial symbionts purified using the Ni-NTA system showed different chitobiosidase and endochitinase activities, but N-acetylglucosamidinase activities were not shown in the measuring of chitinolytic activity through N-acetyl-D-glucosarmine oligomers. In addition, the proteins showed different insecticidal and antifungal activities. XnChi showed the highest insecticidal activity against Galleria mellonella, followed by PtChi and XhChi. In antifungal activity, XhChi showed the highest half-maximal inhibitory concentration (IC50) against Fusarium oxysporum with 0.031 mg/mL, followed by PtChi with 0.046 mg/mL, and XnChi with 0.072 mg/mL. XhChi also showed the highest IC50 against F. graminearum with 0.040 mg/mL, but XnChi was more toxic than PtChi with 0.055 mg/mL and 0.133 mg/mL, respectively. This study provides an innovative approach to the biological control of insect pests and fungal diseases of plants with the biological activity of symbiotic bacterial chitinases of entomopathogenic nematodes.
Keywords:
chitinase; Entomopathogenic nematode; Xenorhabdus nematophilia; Xenorhabdus hominickii; Photorhabdus temperata Key Contribution:
XnChi; XhChi; and PtChi isolated from Korean indigenous entomopathogenic nematodes S. carpocapsae GJ1-2; S. monticolum GJ11-1; and H. megidis GJ1-2 were cloned and expressed using an E. coli recombinant expression system. Recombinant chitinases had different biological activities; namely, they showed not only different chitobiosidase and endochitinase activities, but also different insecticidal and antifungal activities against G. mellonella, F. oxysporum, and F. graminearum, respectively.
1. Introduction
Xenorhabdus and Photorhabdus are symbiotically and pathologically associated with the entomopathogenic nematodes (EPNs), Steinernema and Heterorhabditis, respectively [1]. The symbiotic relationship between EPN and bacterium begins with infection of an insect host with free-living, third-stage infective juvenile EPNs which house the mutualistic symbiotic bacterium in its intestine.
Once the infective juvenile enters the insect host through natural openings (mouth, anus, or spiracles) and penetrates into the hemocoel, the infective juvenile releases the bacterial cells from its intestine in the insect’s hemocoel, and symbiotic bacterium produces a range of secondary metabolites killing the host within 48 h by septicemia [2,3,4]. Recently, insecticidal proteins, as well as secondary metabolites of Xenorhabdus and Photorhabdus, have been targeted for their potential use in agricultural-pest-management investigation of the virulence mechanism [5,6,7]. The representative insecticidal proteins of Xenorhabdus include Xpt [8], Txp40 toxin [9], XaxAB [10], XnGroE [11], and PirAB [12]. Photorhabdus include a wide range of insecticidal proteins including multiunit toxin complexes (Tc), Photorhabdus insect-related (PirAB) toxins, XaxAB, and Photox binary toxins, Makes caterpillar floppy (Mcf), Photorhabdus virulence cassettes (PVC), Photorhabdus insecticidal toxin (Pit), and a ubiquitous Txp40 toxin [13,14,15,16,17]. In addition to the insecticidal proteins without catalytic activity of symbiotic bacteria, digestive enzymes that degrade an insect body are produced by bacteria to provide food for both bacteria and nematodes [18]. However, bacterial enzymes of EPNs associated with insecticidal activities have rarely been investigated.
Chitin, the second most abundant biopolymer in nature after cellulose, is a polymer composed of repeating units of β-1,4-N-acetylglucosamine (GlcNAc). Chitin serves as the main structural component of the extracellular matrix and is found in organisms, including insect exoskeletons, fungal cell walls, crustacean shells, and nematode eggshells [19,20]. Thus, chitinase’s key enzyme has received increasing attention as biopesticide for the control of insect pests and fungal diseases because chitin synthesis is performed using a range of organisms, including fungi and insects, and key enzyme in biosynthesis [21,22]. Chitinase is a Glycoside hydrolase (GH) that acts to degrade chitin using hydrolyzing glycosidic bonds [23], and it is classified into two main groups, chitinase (EC3.2.1.14) and β-acetylhexosaminidase (EC 3.2.1.52), officially called endochitinase and exochitinase, respectively, depending on the products produced during the hydrolysis process [24,25]. Endochitinase cleaves into the chitin chain at an internal site. Exochitinase contains two subcatogories, called chitobiosidases (EC 3.2.1.29) and β-N-acetylglucosaminidases (EC 3.2.1.30) that are now included with (EC 3.2.1.52), β-N-acetylhexosaminidase [24]. While chitobiosidases catalyze progressive release of di-acetylchitobiose from a terminal non-reducing end, β-N-acetylglucosaminidases cleave oligomeric products, such as (GlcNAc)2, (GlcNAc)3, (GlcNAc)4, obtained transforming endochitinase into monomers of N-actyl glucosamine (GlcNAc) [24,26]. These enzymes are grouped into different Glycoside Hydrolase (GH) Families namely GH18, 19 and 20, based on the amino acid similarity of chitinase from various organisms [27]. In particular, as chitinases belong to the GH18 family, they are widely distributed in almost all organisms and were found to be involved in many physiological processes including tissue degradation, developmental regulation, pathogenicity, and immune defense [28]. These chitinases from a variety of microorganisms which facilitate the invasion of pathogens by causing structural changes in the peritrophic membrane of insects, which is primarily composed of chitin, promoting the accessibility of the substrate for the pathogen into the haemocoel, and leading to the interception of nutrient absorption in the midgut [23,29]. Furthermore, chitinases can facilitate the binding process of toxins to specific receptors in the midgut epithelium of insects and enhance the insecticidal activity of entomopathogenic bacteria [30]. Bacillus thuringiensis chitinases (BtChi) have been previously reported to have a contribution to pathogenicity through synergistic effects in combination with other components including Cry proteins [24]. In bacterium Yersinia entomophaga MH96 isolated from diseased grass grub larva of Costelytra zealandica (Coleoptera: Scarabaeidae), the 3D structure of Tc toxins includes two putative chitinases (Chi1 and Chi2) which are essential for complex formation [31]. In addition, two chitinase (chi60 and chi70) genes were found in the toxin complex locus of X. nematophila [31] and the corresponding proteins were shown to be vital for the insecticidal activity against Helicoverpa armigera in the toxicity of Tc toxins [23]. Moreover, chitinases are able to inhibit the elongation and growth of mycelia and spore germination of fungi [32], that is, chitinase Chi2A and chitin binding protein (CBP) in the secondary cells of P. luminescens are necessary to inhibit the growth of Fusarium graminearum [33]. Thus, chitinases could be potential and effective virulence factors for the biological control of insect pests and plant pathogenic fungi.
Xenorhabdus and Photorhabdus show high similarity between clades, as 16S rRNA (16S rDNA) sequences indicate a close phylogenetic relationship between these two genera [34,35,36], but differ in the life cycle and in the pathogenic mechanisms including evasion of the insect immune system and expression of virulence factors [36]. To date, only comparative analyses of insecticidal activity has been performed for Xenorhabdus and Photorhabdus between strains of the same species or different genera [37,38]. However, factors accounting for the different pathogenicity among them have not been clearly investigated. In the recent comparative analysis of genes related to pathogenicity at different genus levels, insecticidal activities were different even in the same pathogenic protein [39]. This suggests the possibility that even the same pathogenic protein may have different biological activities, e.g., due to amino acid variability. Accordingly, chitinases are important toxic and antifungal factors against insect larvae and plant pathogenic fungi [32,33,40]; nevertheless, symbiotic bacterial chitnases of EPNs have not been studied except X. nematophila and P. luminescens. Therefore, chitinases of X. nematophila (GeneBank access number: OR724704), X. hominickii (GeneBank access number: OR724706), and P. temperata (GeneBank access number: OR724705), isolated from Korean indigenous EPNs S. carpocapsae GJ1-2, S. monticolum GJ11-1, and H. megidis GJ1-2 were cloned and expressed to compare biological activity. In addition, Phylogenetic analysis and multiple sequence alignment were performed to characterize the chitinase genes. Chitinolytic activity of their chitinases was measured to compare insecticidal and antifungal activity against Galleria mellonella, F. oxysporum, and F. graminearum.
2. Results
2.1. Sequencing and Phylogenetic Analysis of Chitinases of Xenorhabdus and Photorhabdus Strains Isolated from Korean Indigenous EPNs
Sequence analysis of the chitinase genes confirmed that chitinases of X. nematophila (XnChi), X. hominickii (XhChi), and P. temperata (PhChi) were of the GH18 family, but their length of polypeptides were different, i.e., 648, 498, and 637 aa, respectively (Table S3).
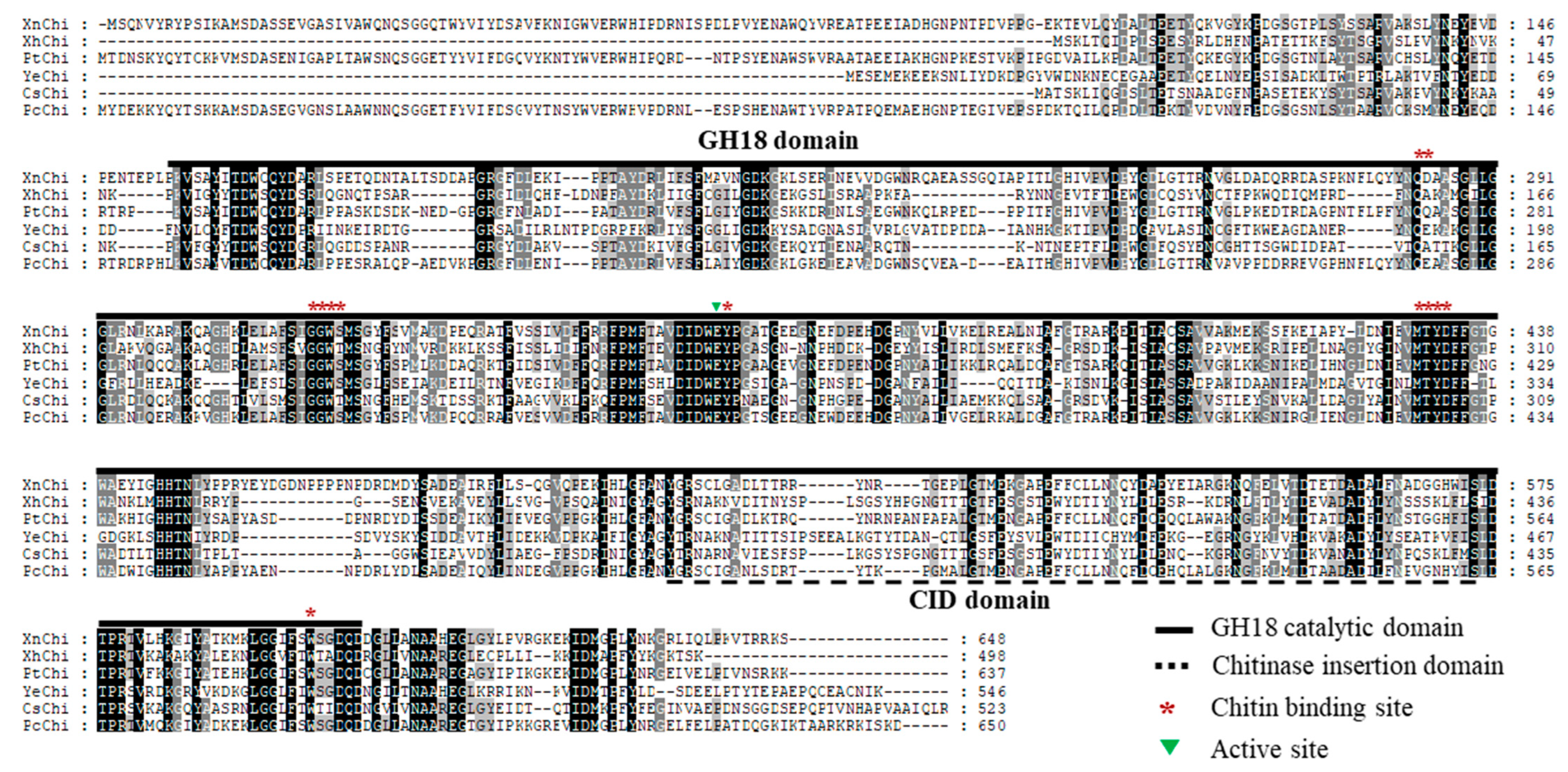
In the multiple sequence alignment of chitinases of X. nematophila, X. hominickii, and P. temperata, including previously reported bacteria Pseudomonas chloriraphis B25, Cronobacter sakazakii wls2261, and Y. entomophaga MH96, chitinases of these six bacteria contained a GH18 catalytic domain, chitinase insertion domain (CID), chitin binding site (CBDs), and active site. Although sequences of the GH18 catalytic domain and the CID were significantly different among them, the CBDs and active site of chitinase were highly conserved. However, XhChi showed a relatively large difference due to its short length compared to the other five bacteria sequences (Figure 1).
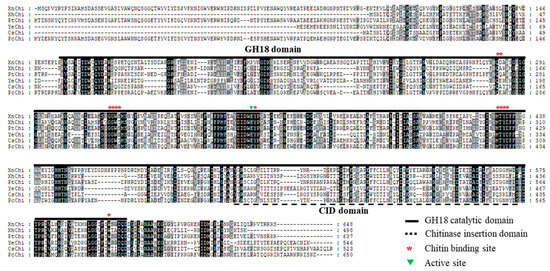
Figure 1.
Multiple sequence alignment for the amino acid sequences of chitinase. The multiple sequence alignment comparison analysis of chitinase amino acid sequences including XnChi, XhChi, PtChi, and three other GH18 chitinases, was performed through the GeneDoc program. Identical and similar residues are shaded black and shaded gray, respectively. The GH18 domain is represented by a bold black line, CID domain is represented by a black dotted line, chitin binding site is represented by a red asterisk, and active site is represented by a green triangle. YeChi, CsChi, and PcChi represent their respective chitinase for Yersinia entomophaga MH96 (GeneBank accession number: ANI28952.1), Cronobacter sakazakii wls2261 (GeneBank accession number: WP241700376.1), and Pseudomonas chloriraphis B25 (GeneBank accession number: WP124321822.1).
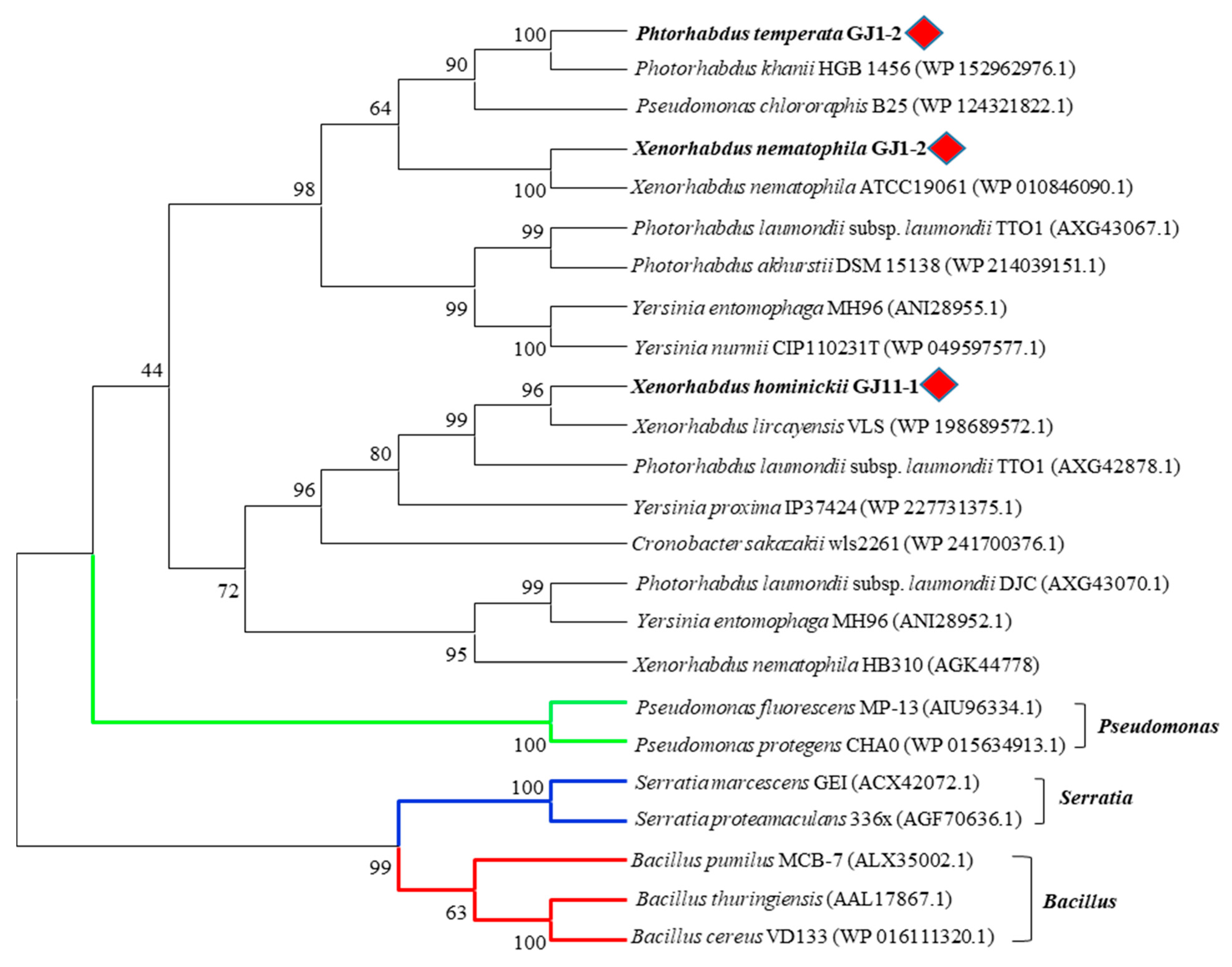
In addition, chitinases of XnChi, XhChi, and PtChi did not belong to the same clade as those of previously studied Pseudomonas, Bacillus, and Serratia in the phylogenetic analysis. The same genus of EPN symbiotic bacteria were generally grouped into relatively close clades, but XnChi, XhChi, and PtChi distinctly formed their respective clades (Figure 2). Such tree topology indicates lineage-specific gene duplication and losses.
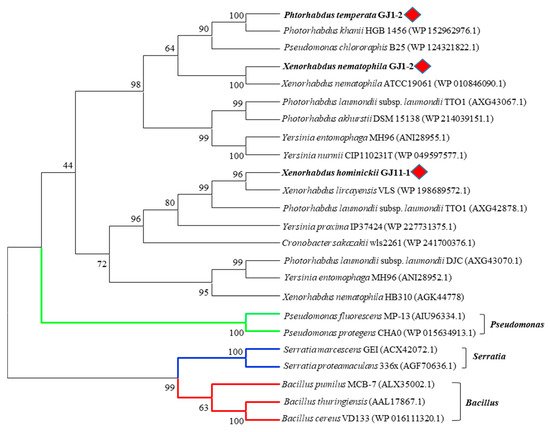
Figure 2.
Phylogenetic analysis of various bacterial chitinases. The phylogenetic tree was constructed using a Maximum Likelihood method based on the amino acid sequence alignment in MEGA X program. The aligned GH18 family chitinase amino acid sequence was obtained from the NCBI database. The red rhombus represents XnChi, XhChi, and PtChi used in this study and green, blue, and red lines indicated Pseudomonas spp., Serratia spp. and Bacillus spp., respectively. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicate) are shown next to the branches. All positions containing gaps and missing data were eliminated (complete deletion option).
2.2. Cloning, Expression and Purification of Recombinant Chitnases
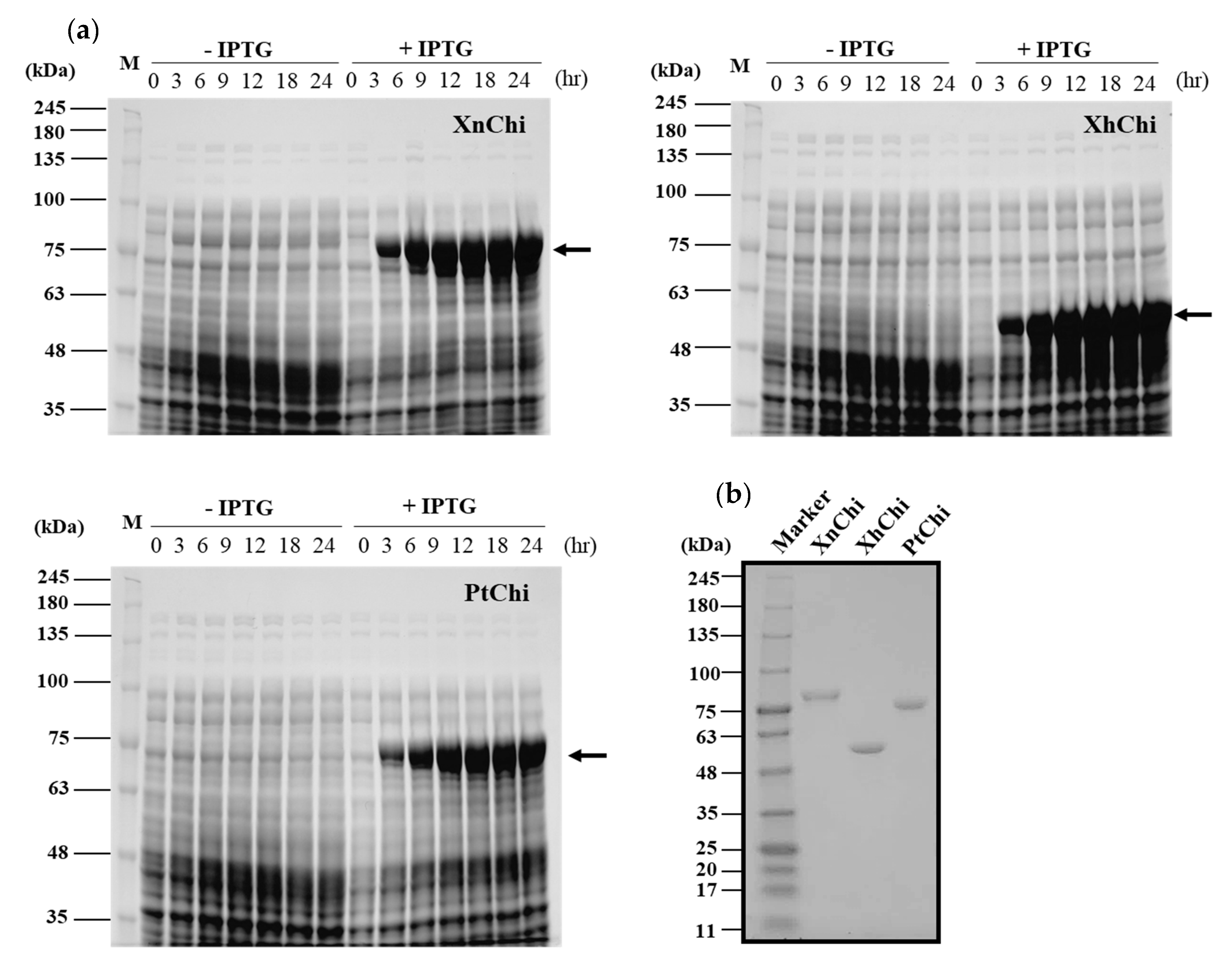
XnChi, XhChi, and PtChi were cloned and expressed using Escherichia coli expression system to compare their biological functions. The prominent bands of the recombinant proteins for XnChi, XhChi, and PtChi on the SDS–PAGE were detected at ca. 76 kDa, 59 kDa, and 75 kDa, respectively. The highest protein expression level was observed at 24 h after IPTG induction (Figure 3a). Because high-levels of protein expression could lead to inclusion body formation [41], samples were used at 18 h after IPTG induction, which showed similar expression levels (Figure 3a). Recombinant proteins were purified using Ni-NTA superflow resin under native conditions (Figure S1). In order to fully characterize the XnChi, XhChi and phChi, the eluted fractions were concentrated and desalted using an Amicon Ultra centrifugal filter (30 kDa cut-off). The purified recombinant chitinases showed the single individual bands with 76 kDa, 59 kDa, and 75 kDa on SDS–PAGE (Figure 3b).
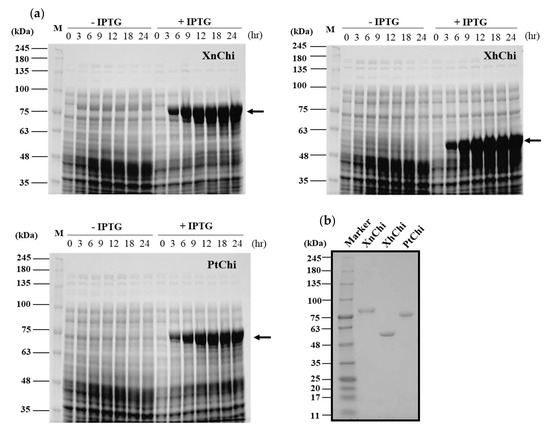
Figure 3.
Expression and purification of the recombinant XnChi, XhChi, and PtChi. (a) A total of 10% SDS–PAGE analysis of chitinase protein induced by 1 mM IPTG through sampling for 3 h, 6 h, 9 h, 12 h, 18 h, and 24 h, respectively. From the left, XnChi, XhChi, and PtChi are shown. (b) Chitinase protein purified using Ni–NTA resin was concentrated and desalted using an Amicon tube (30 kDa-cut off). Concentrated chitinase protein (2 µg) was analyzed using 4–15% SDS–PAGE. Lane M: Protein marker. The protein bands corresponding to chitinase protein are indicated with an black arrow.
2.3. Chitinolytic Activity Assay with Recombinant Chitinases
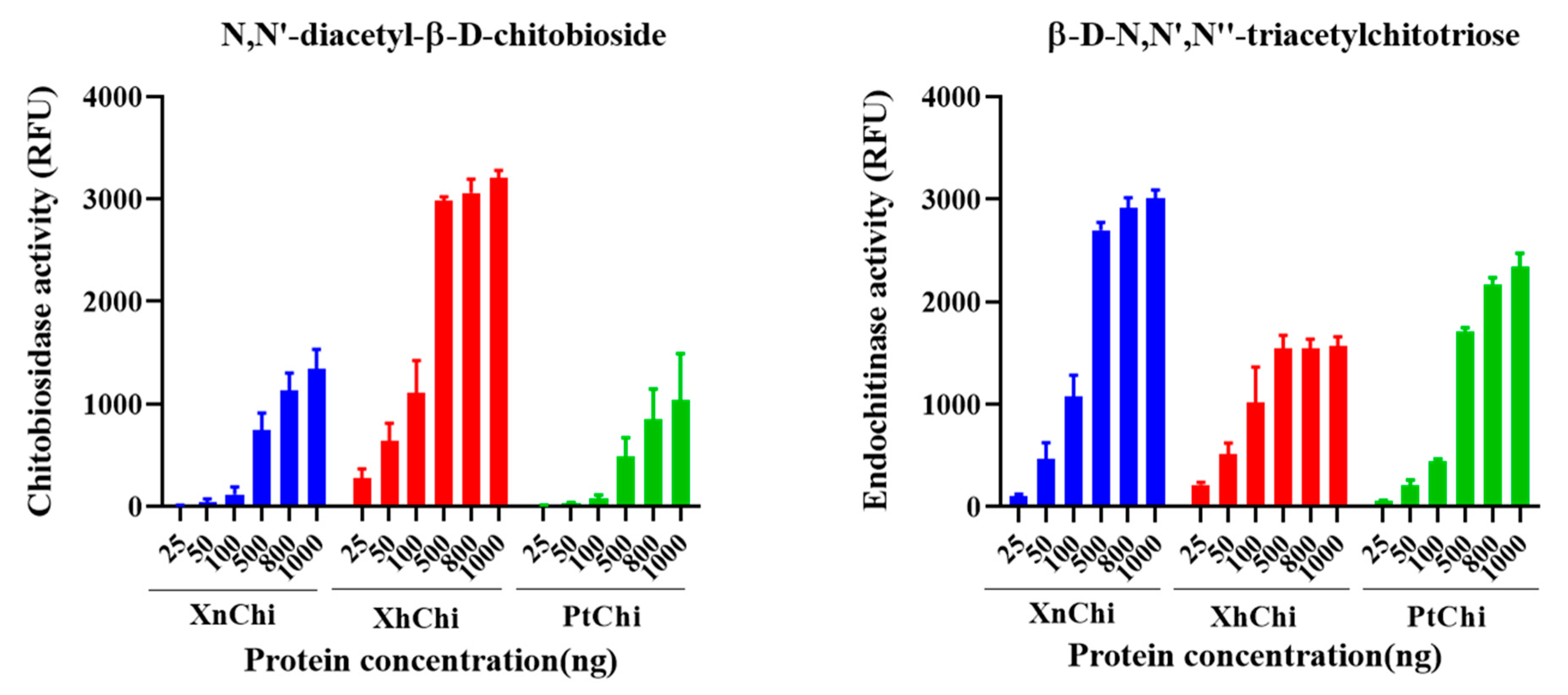
Chitinase is classified into exochitinase and endochitinase according to enzymatic action on chitin substrate. XnChi, XhChi, and PtChi showed chitiobiosidase, which is an exochitinase, and endochitinase activities in a concentration-dependent manner on the substrate, respectively. However, β-1,4-N-acetylglucosaminidases activity was not observed. At a concentration of 1000 ng/µL, which exhibited the highest activity of chitinase, chitobiosidase activity of XhChi was 2.4-fold and 2.1-fold higher than that of XnChi and PtChi, respectively. However, endochitinase activities of XnChi and PtChi were higher than those of XhChi by 0.5-fold and 0.7-fold, respectively (Figure 4).
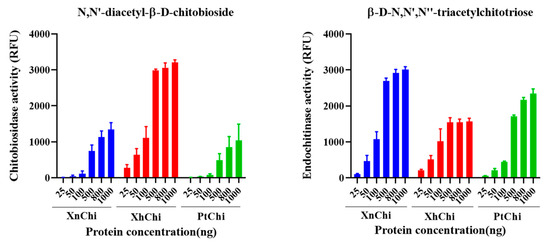
Figure 4.
Chitinolytic activity of the recombinant XnChi, XhChi, and PtChi. Enzyme activity of chitinase using two substrates known as chitoligosaccharide analogs: 4-methylumbeliferyl N,N′-diacetyl-β-D-chitobioside [4-MU-(GlcNAc)2] and 4-Methylumbelliferyl β-D-N,N′,N″-triacetylchitotriose [4-MU-(GlcNAc)3]. The kinetic curve for the degree of hydrolysis of the substrate (0.5 mg/mL at the final volume of 100 μL) was measured according to the standard curve prepared using fluorescence readings of five standard solutions. The different concentrations of the samples used were 25, 50, 100, 500, 800, and 1000 ng/μL.
2.4. Evaluation of Insecticidal Activity of Chitinase against Galleria mellonella
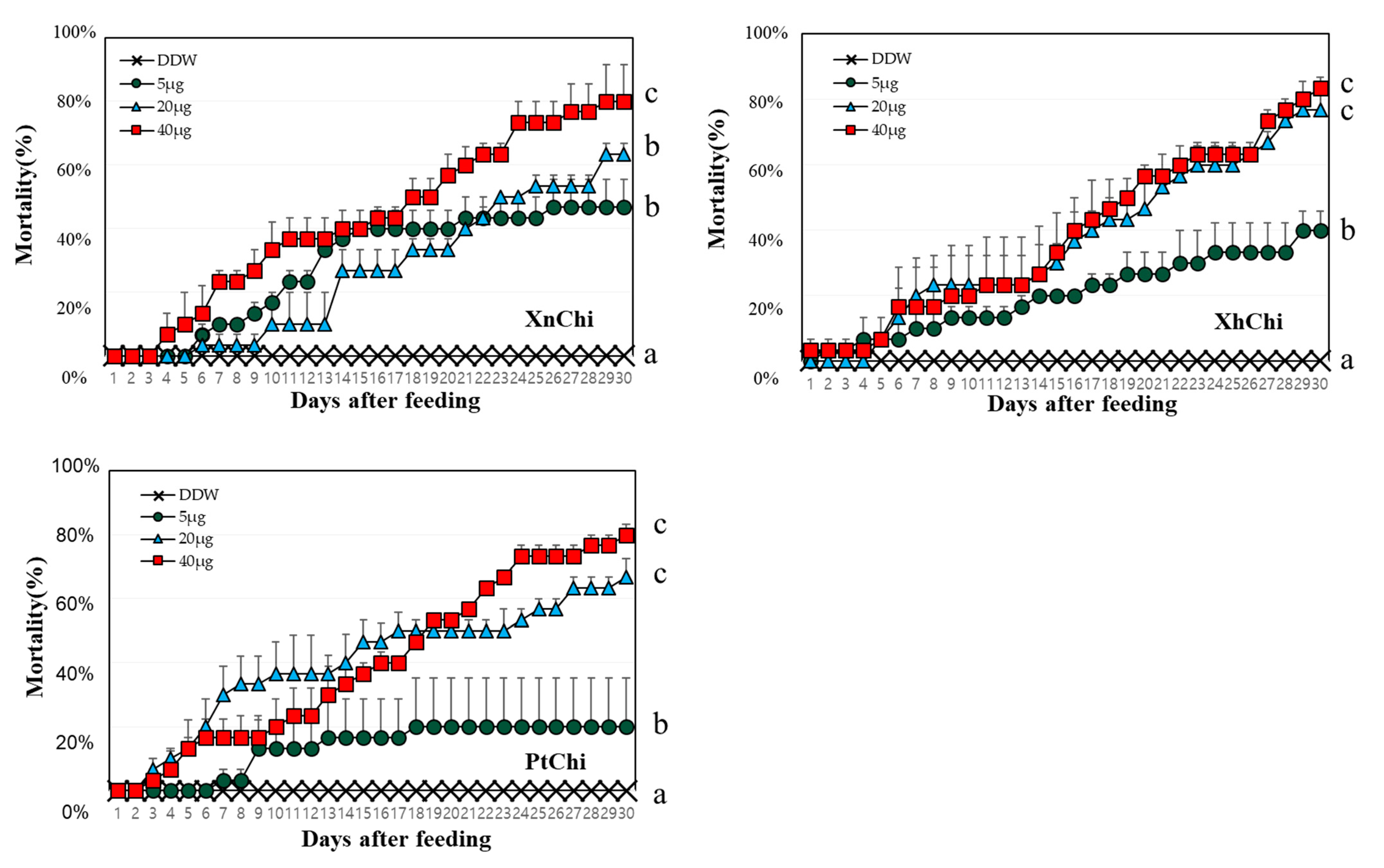
XnChi, XhChi, and PtChi were highly toxic against G. mellonella. Although mortality was different depending on the chitinase concentration, larval mortality was significantly higher at the rate of 40 µg (Figure 5). The median lethal time (LT50) value was calculated to compare insect mortality. LT50 values of XnChi, XhChi, and PtChi were ca. 16, 18, and 17 at the 40 μg, respectively (Table 1). In addition, chitinase affected the development of G. mellonella, and was not limited to a specific stage (Figure S2). Galleria larvae developed into adults in the control, whereas chitinase-fed larvae did not developed to pupae or adults. Most of them were dead in the larval or pupal stage (Figure S2).
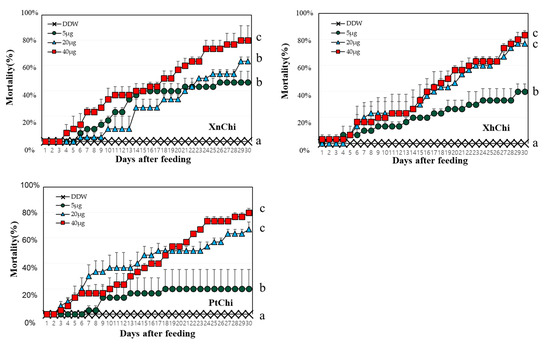
Figure 5.
Mortality according to concentration of chitinases against G. mellonella. 10 of second larval instar reared on different concentration of protein (5, 20 and 40 µg/larva) for 30 days. Mortality was calculated as the number of dead larvae/total number of larvae. Deionized purified water (DDW) was used as control. Turkey’s test was used as a post hoc method to identify the concentration of high insecticidal activity that showed significant differences. Values within a figure with different letters are significantly different according to the Tukey’s test (p < 0.05) in the IBM SPSS Statistics 27 software program.

Table 1.
LT50 and LT70 values of orally administered XnChi, XhChi, and PtChi against larvae of G. mellonella.
2.5. Assessment of Antifungal Activity of Chitinase Proteins against Fusarium
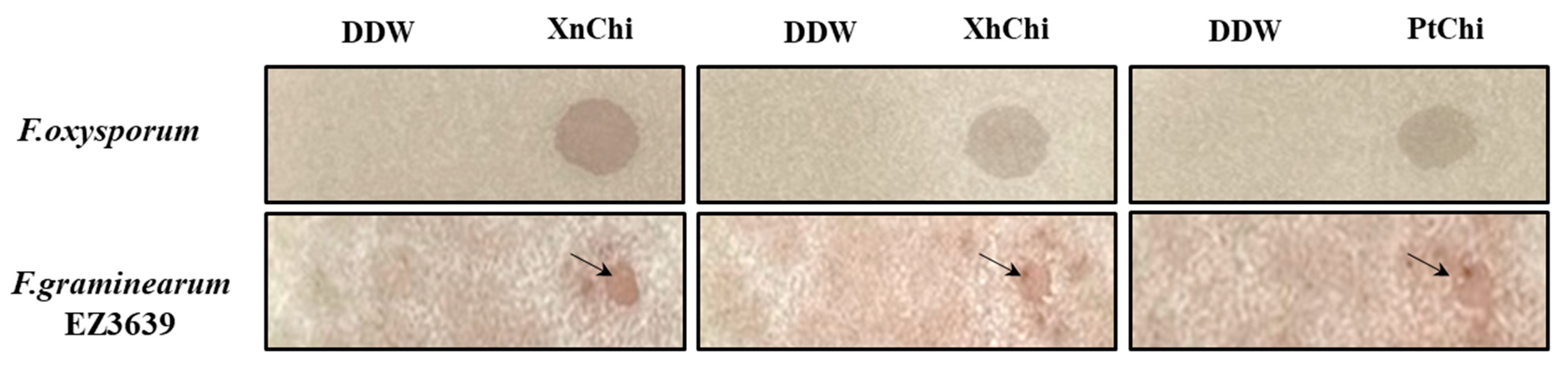
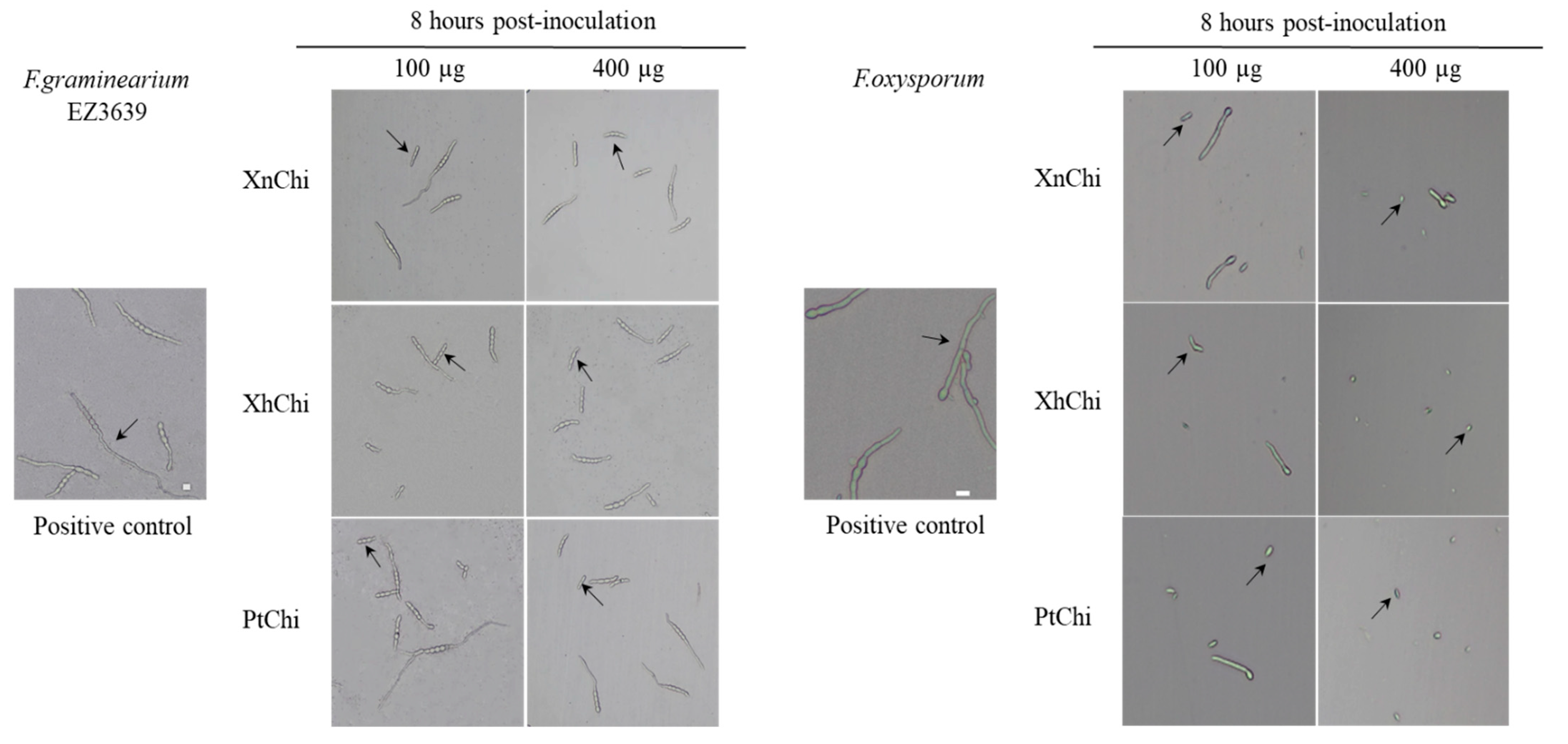
In the disc-diffusion susceptibility assay of the chitinases against F. oxysporum and F. graminearum, plant pathogenic fungi, at the concentration of 20 or 40 µg of XnChi, XhChi, and PtChi, most inhibition of mycelium formation showed at a concentration of 40 µg of the chitinases (Figure 6). In particular, XnChi led to the formation of a clear inhibition zone against F. oxysporum even at 20 µg (Figure S3). The conidial germination rates of F. graminearum and F. oxysporum were measured for 4 h or 8 h with 50, 100, 200, and 400 µg of XnChi, XhChi, and PtChi. The highest inhibition of conidial germination of both fungi was observed at the concentration of 400 µg for 8 h (Figure S4), and the growth inhibition of conidial germ tubes is shown in Figure 7. However, the antifungal activity of XnChi, XhChi, and PtChi was different depending on fungus even at the same concentration. The IC50 value of XnChi, XhChi, and PtChi against F. graminearum were 0.055 mg/mL, 0.040 mg/mL, and 0.133 mg/mL, respectively, compared with 0.072 mg/mL, 0.031 mg/mL and 0.046 mg/mL against F. oxysporum, respectively (Table 2).
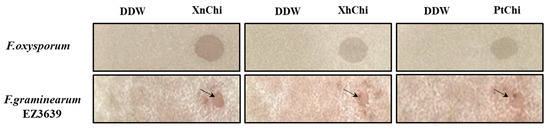
Figure 6.
Antifungal activity of XnChi, XhChi, and PtChi (40 µg) against F. oxysporum and F. graminearum. Inhibition of mycelium growth by chitinase was shown as the inhibition zone through disc-diffusion susceptibility assay. No growth inhibition was observed in the control. The arrow indicates the zone of inhibition for mycelium growth of the fungi.
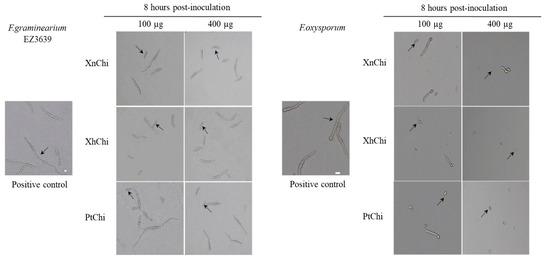
Figure 7.
Effect of XnChi, XhChi, and PtChi on conidial germination of F. graminearum and F.oxysporum. Chitinases were added to Potato dextrose broth (PDB) at 100 µg and 400 µg concentrations, and conidial germination was measured at 8 h after inoculation with 2 × 105 conidia/mL. Fungal inoculum was used as positive control. The germinated conidia were observed through light microscopy. The germinated conidia were judged to have grown more than 1/2 of the conidial length. The scale bar was measured through the image J program. Scale bar = 10 μm. The arrow indicates the difference of conidia between the treatments and control.

Table 2.
Comparison of antifungal activity of XnChi, XhChi, and PtChi against F. graminearum and F. oxysporum.
3. Discussion
Chitinases identified from various bacteria have insecticidal and antifungal activities. These pesticidal chitinases have been also identified from EPN symbiotic bacteria, Xenorhabdus and Photorhabdus [32,33,40,42,43]. Xenorhabdus and Photorhabdus, having insecticidal activity, exhibit different pathogenicity depending on the species or strain [37,39]. Symbiotic bacterial chitinases may also be involved in different insecticidal activities against target pests, although they have the same functions in biological activities. Thus, chitinases of X. nematophila, X. hominickii, and P. temperata isolated from Korean indigenous EPNs, S. carpocapsae GJ1-2, S. monticolum GJ11-1, and H. megidis GJ1-2, respectively, were identified and compared to determine gene characterization using multiple sequence alignment and phylogenetic analysis. These bacterial chitinases, XnChi, XhChi, and PtChi, showed significant differences in the GH18 catalytic domain and chitinase insertion domain (CID). However, active site and chitin binding site (CBDs) were conserved (Figure 1). CBDs play an important role in interacting with insoluble chitin and promoting microbial adhesion to chitin for subsequent degradation, while CID promotes orientation and binding of longer carbohydrate substrates [24].
In the phylogenetic analysis, as shown in Figure 2, XnChi, XhChi, and PtChi were divided into distinct clades from previously studied Pseudomonas, Bacillus, and Serratia [44,45,46,47,48,49]. Out of three bacterial chitinases, XnChi was closer to PtChi rather than XhChi. This may be resulted from the amino acid sequence similarity (XnChi vs. PtChi = 77.47%; XnChi vs. XhChi = 52.82%). In particular, XnChi is relatively close to the X. nematophila ATCC19061 chitinase (Genbank accession number: WP 010846090.1), which shows 99.69% homology to Chi70 (Genbank accession number: GK44779.1) of X. nematophila. Chi70 was reported to have insecticidal and antifungal activities against certain pests and fungi, and to improve insecticidal activity of BtCryAc and Tc toxins [23,31,32]. Thus, symbiotic bacterial chitinases of EPNs could be promising control agents of insect pests and plant pathogenic fungi by playing an important role in pesticidal activity. However, even though Xenorhadus and Photorhadus have multiple insect-model host-specific pathogenicity, different pathogenicity factors between Xenorhadus and Photorhadus have not yet been investigated [37,39]. Thus, to compare the insecticidal activity of their chitinases from different strains within EPN species, the chitinases of X. nematophila, X. hominickii, and P. temperata were expressed and purified. The molecular weights of three recombinant chitinases were higher (Figure 3) than the predicted weight, and this might be caused by O-glycosylation, one of the post-translational modifications of the chitinase protein that occurs in some bacterial enzymes [50,51].
In the results of measuring chitinolytic activity of purified recombinant chitinases through two chitoligosaccharide analogues, endochitinase activity degrading the trimer substrate [MU-(GlcNAc)3 ] and chitibiosidase activity degrading the dimer substrate [MU-(GlcNAc)2] were observed (Figure 4). The fact that chitinases of bacterial origin exhibit both endochitinase activity and exochitinase activity, respectively, has been well-established in previous studies [52,53]. However, for Chen et al., after the study of [54], in the results of recent studies [55,56] show both endochitinase activity and exochitinase activity, similar to our chitinolytic assay results, and which are consistent with our results. Furthermore, Mahmood et al. [40] also demonstrated that chitinase from X. nematophila exhibits high β-N-acetylglucosaminidase activity and endochitinase activity, compared to chitiobiosidase activity. In general, previous studies have reported that exochitinase is not very efficient in chitin degradation because access to substrates is limited [57]. Therefore, the combination of exo- and endo-chitinase activity is typically several times more potent than single activity [58], implying that these chitinases may be more effective in pest control compared to chitinases from other sources. However, activity of β-1,4,N-acetylglucosaminidase, which degrades the monomer substrate [MU-GlcNAc], was not observed in XnChi, XhChi, and PtChi. This has also been observed in the measurement of the activity of metagenome-sourced chitinase Chi18H8 [55,59]. In particular, XhChi showed higher chitobiosidase activity, whereas endochitinase activity was higher in XnChi and PtChi. This might be due to different biological activities due to a difference of chitinolytic activity. Furthermore, the different amino acid sequences of the three chitinases suggest that they may have affected the structure of the substrate binding pocket/gap, which will require protein structure analysis [60].
In a previous study, a novel chitinase from X. nematophila was found to exhibit oral insecticidal activity against Helicoverpa armigera [40]. In this study, the oral insecticidal activity of XnChi, XhChi, and PtChi against G.mellonela was compared. We found that the insecticidal activity of three chitinases was observed in all of the developmental stages of G. mellonella (Figure S2), especially showing that XnChi has the highest efficacy against G. mellonella (Table 1). This suggests that external chitinases disrupted the controlled degradation of chitin, playing an essential role in the molting and pupal processes, which are the growth and developmental processes of insects [61,62]. The difference in virulence activity is probably as a result of their chitinase activity depending on the chitin component of the target insect as a substrate. This is consistent with the chitinolytic activity results showing that the endochitinase activity (Figure 4), which possesses the ability to cleave all parts of the chitin polymer it comes in contact with, is most prominent in XnChi. The insecticidal activity of endochitinases against plant pests has been well reported previously [30,63].
Chitin is also a major component of most fungal cell walls, so enzymes in the chitinolytic system play an important role in controlling fungi. The enzymatic lysis of the fungal cell wall via extracellular chitinase is implicated as a biological control mechanism by bacterial agents [64]. Thus, antifungal activity of three chitinases against F. oxysporum and F. graminearum was compared through disc-diffusion susceptibility assay and a conidial germination test. Inhibition of mycelial growth was observed for both fungi (Figure 6), with the size of the inhibition zone being larger for F.oxysporum compared to F.graminearum. Previous studies have reported that, even within the Fusarium genus, differences exist in the cell wall components and contents. F. graminearum contains a higher quantity of N-acetylglucosamine, phosphorus, and minerals (ash), which positively contribute to the safety and strength of the cell wall compared to F. oxysporum [65,66]. These results suggest that F. oxysporum may have been more sensitive to chitinase than F. graminearum. Previous studies have also shown that even within the same Fusarium genus, the degree of mycelial growth inhibition is different [67,68]. Inhibition of conidial germ tube elongation (Figure 7) of both fungi were observed. Out of three chitinases, XhChi showed the highest antifungal activity against F. oxysporum and F. graminearum, followed by XnChi and PtChi (Table 2). This suggests that chitinases with distinct amino acid sequences may influence specific activities based on the chitin composition of the target fungi. These results could also be inferred from Figure 4, which shows that although XhChi showed slightly lower endochitinase activity than PtChi and XhChi, chitobiosidase activity was significantly higher in XhChi than XnChi and PtChi. The result is assumed to be consistent with previous research, which reported that the combination of endochitinase and chitobiosidase showed higher antifungal activity [58]. In fact, antifungal activity of chitinases from symbiotic bacteria of EPN has been reported in several studies [32,33]. For example, the partially purified chitinase enzyme from X. bovienii has exhibited strong antifungal activity against Botrytis cinerea by inhibiting conidial germination and germ tube elongation or lysing the germ tube [54]. Additionally, the rapid increase in the chitinase activity during the first 24–48 h of bacterial culture suggests that these chitinases may play an important role in early protection against fungal invasion of dying insects [54].
The entomopathogenicity-related genes with large molecular weights (i.e., Mcf, Tc, Xpt, PVC) have been well studied in Xenorhabdus and Photorhabdus [69,70,71], but enzymatic toxins, including chitinases have been rarely studied. Furthermore, the antifungal activity of chitinases have been little studied, except for X. nematophila and P. luminescence [32,33]. In addition, studies comparing the activity of pathogenicity factors among EPN symbiotic bacteria have not yet been investigated. Therefore, this study suggest that even if they have similar biological activity, these chitinases, depending on different strains, may exhibit different pathogenicity activity against target insect and plant pathogenic fungi. Different biological activities of symbiotic bacterial chitinases indicate that these chitinases can be promising candidates for the development of pest-resistant crops [72]. Thus, further studies on the production of insect-resistant and fungus-resistant transgenic crops with EPN symbiotic bacterial chitinases and mass-production of chitinase proteins as bio-control agent are recommended.
4. Materials and Methods
4.1. Selection of Type Strain to Identify Chitinase Gene of EPN Symbiotic Bacteria
Chitinase genes of type strains were selected to identify those of symbiotic bacteria X. nematophila, X. hominickii, and P. temperata isolated from Korean indigenous EPNs S. carpocapsae GJ1-2, S. monticolum GJ11-1, and H. megidis GJ1-2, respectively.
Chitinase genes of X. nematophila ATCC19061 and P. temperata J3 were selected because not only does chitinase of X. nematophila ATCC19061 (GeneBank accession number: WP010846090.1) have insecticidal properties [40] but also that of P. temperata J3 (GeneBank accession number: WP023045972.1) has the highest similarity with X. nematophila ATCC19061 in P. temperata strains by the NCBI blastp program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch & LINK_LOC=blasthome, accessed on 20 November 2023).
These chitinase of two bacteria belong to the Glycoside Hydrolase Family 18 (GH18). Thus, chitinase of X. hominickii ANU1 belonging to GH 18 (GeneBank access number WP: 069316843.1) which is the only known strain of X. hominickii group in the NCBI database was used as the type strain of studied X. hominickii (Table S2).
4.2. Chitinase Sequence Multi-Alignment and Phylogenetic Analysis
GH 18 catalytic domain, chitinase insertion domain, active site, and chitin binding site of chitinase of X. nematophila, X. hominickii, and P. temperata were identified through the Expasy program website (https://prosite.expasy.org/, accessed on 20 November 2023) and InterPro website (https://www.ebi.ac.uk/interpro/, accessed on on 20 November 2023) to compare amino acid sequence. The aligned chitinase amino acid sequences obtained through the MultAlign website (http://multalin.toulouse.inra.fr/multalin/, accessed on 20 November 2023) were visualized using a GeneDoc program for multiple sequence alignment analysis (Table S4). A phylogenetic tree was constructed using the Maximum Likelihood method (bootstrap test 1000 replicate) in MEGA X bioinformatics tool. Amino acid sequences of the GH 18 family of chitinases used in the analysis were acquired from the PROTEIN category of the NCBI database (Table S5).
4.3. Construction of Recombinant Chitinase-Encoding Genes
Genomic DNA of X. nematophila, X. hominickii, and P. temperata was extracted using DNeasy Ultra Clean Microbial Kit (Qiagen, Hilden, Germany). The primers were designed with CDS sequences of three type strains selected previously to identify a chitinase gene from genomic DNA of the above three bacteria. In addition, specific restriction enzymes were linked to the front and rear sequences of the primer for cloning (Table S1). TOPO™ XL-2 Complete PCR Cloning Kit (Invitrogen, Carlsbad, CA, USA) was used to clone chitinase gene by performing PCR with Platinum™ SuperFi™ Green PCR Master Mix [2X] (Invitrogen, Carlsbad, CA, USA) under the following conditions: 98 °C for 30 s, 30 cycles of 98 °C for 10 s, 61 °C for 10 s, 72 °C for 1 min, and 72 °C for 10 min. The PCR products were purified using the QIAquick PCR Purification kit (Qiagen, Hilden, Germany) and cloned into pCR-XL-2-TOPO™ Vector (Invitrogen, Carlsbad, CA, USA). Then, One Shot™ OmniMAX™ 2 T1R Chemically Competent E. coli (invitrogen, Carlsbad, CA, USA) was transformed. Plasmids were extracted using QIAprep spin mini prep kit (Qiagen, Hilden, Germany) and sequencing was performed using Genetic analyzer 3730 xl (Applied Biosystems, Waltham, MA, USA). For the comparison of protein expression level of chitinase, a pCold II vector protein expression (TaKaRa Bio, Shiga, Japan) system was used. Topo vector-cloned plasmids were digested by SacⅠ (TaKaRa Bio, Shiga, Japan) and purified using QIAquick PCR Purification Kit. After that, the products were digested with each of XbaⅠ, SalⅠ, and PstⅠ (TaKaRa Bio, Shiga, Japan) (Table S1) restriction enzymes and gel electrophoresis was performed on 0.8% agarose gel and purified through a QIAquick Gel extraction kit (Qiagen, Hilden, Germany). Similarly, pColdⅡ vector was also double digested and purified to prevent re-ligation. A DNA ligation kit (TaKaRa Bio, Shiga, Japan) was used to subclone chitinase pcr fragments into pColdⅡ vector (TaKaRa Bio, Shiga, Japan). The final volume was 10 μL (DNA solution:Ligation Mixture = 1:1) at a ratio of 3:1 (insert: vector molar ratio). To increase the ligation efficiency after heating at 65 °C and cooling on ice for 2–3 min, overnight incubation was performed at 16 °C. The ligation product was transformed into 100 μL of DH 5α Chemically Competent E. coli (Enzynomics, Daejeon, Republic of Korea), and the product was screened with a selective DifoTM LB Broth Miler (Bectone, Dickinson and Company, Sparks, MD, USA) medium containing 100 μg/mL ampicillin (MB cell, Seoul, Republic of Korea). Finally, the plasmids were extracted from the selected colony and transformed into a protein expression E. coli, BL21 (TaKaRa Bio, Shiga, Japan).
4.4. Expression and Purification of Recombinant Chitinase Protein
A single colony of BL21-transformant was picked up to express pCold-chitinase construct and cultured in LB medium including 100 μg/mL ampicillin for 16 to 20 h at 37 °C with a shaking incubator at 225 rpm. Then, approximately 15% of the culture was added to fresh LB medium containing 100 μg/mL ampicillin and cultured for 2 to 3 h at 37 °C in a shaking incubator until OD600 = 0.4 to 0.6. After being cold-shocked on ice for 30 min, cultures added with 1 mM isopropyl-β-D-thiogalactopyranoside (Thermo Scientific, Lithuania, Italy) were incubated at 15 °C in a shaking incubator to induce protein expression. The cultures were collected to confirm the level of protein expression over time at 3, 6, 9, 12, 18, and 24 h after IPTG addition using M 15R centrifuges (Hanil, Daejeon, Republic of Korea) at 13,000 rpm at 4 °C. In the control, cultures were collected at the same time without adding IPTG. The collected bacterial cells were washed twice in 1 × PBS (Bioneer, Daejeon, Republic of Korea) and mixed with 1 × FastBreak Cell Lysis Reagent (Promega, Madison, WI, USA), Lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), and 0.1 mg/mL Lysozyme Solution (Thermo Scientific, Rockford, IL, USA). These were incubated in a 37 °C shaking incubator for 30 min. After that, cell mixture was subjected to cell lysis using a 21 G syringe (Koreavaccine, Seoul, Republic of Korea), and the cell lysate was centrifuged (Hanil, Daejeon, Republic of Korea) at 13,000 rpm for 30 min at 4 °C. The supernatants of cell lysates were incubated with Ni-NTA Superflow (Qiagen, Hilden, Germany) at 4 °C for 1 h at 225 rpm in a shaking incubator. The soluble fraction was dispensed onto a disposable polypropylene column (Thermo Scientific, Rockford, IL, USA) and subjected to Ni2+-NTA affinity chromatography through an imidazole-dependent gradient method (Qiagen manual) as follows; the loaded suspension was washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM or 20 mM imidazole, pH 8.0), and proteins were eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl; 100 mM, 150 mM or 200 mM or 300 mM imidazole; pH 8.0). Then, 2 × sample buffer (Bio-Rad, Hercules, CA, USA) supplemented with 2- Mercaptoethanol (Bio-Rad, Hanghai, China) and the purified protein were mixed at a ratio of 1:1. The mixture was boiled using a Microprocessor block heater (Barnstead/Lab-LINE, Melrose park, IL, USA) for 5 min, followed by cooling on ice for 2 min and centrifuging at 13,000 rpm at 4 °C. Finally, 20 μL of the supernatant was loaded onto a 4–15% SDS–PAGE (Bio-Red, Hercules, CA, USA). After electrophoresis, staining and destaining were performed through Coomassie Brilliant Blue R—250 Solution (Ezynomics, Daejeon, Republic of Korea) and Coomassie Brilliant Blue R—250 destaining Solution (Ezynomics, Daejeon, Republic of Korea), respectively. Some 30 kDa Ultra-15 centrifuge Filter Devices (Amicon, Darmstadt, Germany) were also used for the concentration and desalting of protein. After pre-rinsing filter devices with Ambion DEPC-treated Water (Invitogen, Waltham, MA, USA), purified protein was added and centrifuged 3500× g for 20 min at 4 °C in Combi R 515 Swinging bucket rotor (Hanil, Daejeon, Republic of Korea). Subsequently, the filtrate was discarded, and we added DEPC-treated water. After that, centrifugation was performed under the same conditions as above. After washing twice, the protein concentrated in the filter was recovered and quantified manually using the Pierce BCA Protein Assay kit (Thermo Scientific, Rockford, IL, USA).
4.5. Chitinolytic Activity Assay with Recombinant Chitinase Protein
The activity of purified chitinase was measured using a chitinase assay kit, fluorimetric (Sigma-Aldrich, St. Louis, MO, USA). The enzyme activity of chitinase was measured by detecting the fluorescence of 4-methylumbelliferone (MU), a fluorescent dye generated during the hydrolysis of chitooligosaccharide analogs, which are substrates of chitinase: 4-Methylumbelliferyl β-D-N,N′,N″-triacetylchitotriose [4-MU-(GlcNAc)3], 4-Methylumbelliferyl N,N′-diacetyl-β-D-chitobioside [4-MU-(GlcNAc)2], and 4-methylumbelliferyl N-acetyl-β-D-glucosaminide [4-MU-GlcNAc]. The emission of 4-methylumbelliferone (MU) was measured by fluorescence detection with a PerkinElmer 2030 multilabel plate reader (PerkinElmer, Waltham, MA, USA) under the conditions of pH 5.0 and 360 nm excitation wavelength and 450 nm emission wavelength. 4-Methylumbelliferyl N,N′-diacetyl-β-D-chitobioside [4-MU-(GlcNAc)2] and 4-methylumbelliferyl N-acetyl-β-D-glucosaminide [4-MU-GlcNAc] were used to measure exochitinase activity, while 4-Methylumbelliferyl β-D-N,N′,N″-triacetylchitotriose [4-MU-(GlcNAc)3] was used to measure endochitinase activity. Simultaneously, 0, 25, 100, 500, 800, and 1000 ng/µl of purified chitinase protein were added to MicroWell™ 96-well black plates (Sigma Aldrich, Roskilde, Denmark) to assess concentration-dependent substrate degradation with the above three substrates. After 30 min incubation of the above mixture at 37 °C, Stop Solution (Sodium Carbonate) was added to each well to measure substrate resolution.
4.6. Insecticidal Activity Bioassay of Recombinant Chitinase Protein against Galleria mellonella
Insecticidal activity of purified chitinase proteins of three symbiotic bacteria was evaluated against G. mellonella using oral feeding assay on 1 g artificial diet (Rice bran 33.1%, Wheat bran 33.1%, Yeast Extract 0.2%, Calcium propionate 0.6%, Honey 13.2%, Glycerin 13.2%, Vitamin 0.05% and 6.65%) mixed with various concentrations (0.8, 0.4, and 0.1 mg/mL) of purified chitinase protein dissolved in 0.5 mL of deionized purified water (DDW) in 60 mm Petri dishes for 2nd to 3rd instars and 100 mm Petri dishes for 4th instar. Larvae in the control were fed on only an artificial diet treated with the same volume of DDW. The artificial diet treated with DDW and chitinase proteins was dried on a clean bench for one day before use.
Ten 24 h-fasted 2nd instar larvae were placed in each Petri dish and reared in environmentally controlled conditions at 26 ± 2 °C and 69% relative humidity. Larval mortality was checked every day, and food was replaced as needed. The experiment was performed in triplicate using ten larvae per replicate. Larval mortality depending on the concentration of each bacterium was compared by the by as a post hoc Turkey’s test, and LT50 values were calculated using probit analysis through IBM SPSS Statistics 27 software program.
4.7. Antifungal Activity Assay of Recombinant Chitinase Protein against Plant Pathogenic Fungi
The antifungal activity of purified chitinase proteins of three symbiotic bacteria was evaluated against plant pathogenic fungi F. graminearum EZ 3639 and F. oxysporum by conidial germination test and disc-diffusion susceptibility assay. The fungi used in the test were provided from the Fungal Plant Pathology Laboratory, Dong-A University, Busan, Republic of Korea. Mycelial blocks of fungi cultivated on potato dextrose agar (PDA) for 3 days were inoculated into carboxyl methyl cellulose (CMC) medium [73] to induce conidia and were cultured at 200 rpm at 25 °C for 5 days [74]. Cultivated conidia were collected using centrifugation at 13,000 rpm at 4 °C for 30 min, and then washed twice using DDW. Chitinase proteins diluted serially with concentrations of 0.05, 0.1, 0.2, and 0.4 mg/mL (final volume of 1 mL) were inoculated into 25 mL of potato dextrose broth (PDB) adjusted to a final concentration of 2 × 105 conidia/mL. DDW of the same volume was used as the control. The ratio of germinated conidia was counted using microscopy (Nikon ECLIPSE Ci, India) at 4 and 8 h after inoculation. The presence or absence of conidial germination was judged to indicate germination when fungi reached more than 1/2 of the spore length. The IC50 values of conidia inhibition rate were calculated using probit analysis and germination rate (germinated conidia/total conidia) for the chitinase of each bacterium was compared with a post hoc Turkey’s test using the IBM SPSS Statistics 27 software program [74]. In the disc-diffusion susceptibility assay, fungal conidia were collected in the same way as above for measuring the conidial germination rate. The collected conidia were inoculated into a sterilized PDA medium at a final concentration of 2 × 104 conidia/mL, and then were cultured at 25 °C for one day to solidify the medium. Finally, 40 µg (final volume 40 µL) of purified chitinase protein was dropped into a spore-exposed medium, dried for 5 min, cultured at 25 °C, and observed on the next day. An equal volume of DDW was used as a negative control.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/toxins16010026/s1. Figure S1. Cell lysate, a soluble fraction containing 6xHis-tagged chitinase protein, was purified using an imidazole concentration-dependent method using Ni-NTA superflow resin; Figure S2. Oral toxicity of XnChi, XhChi, and PtChi against G. mellonella was observed on 25 days after treatment; Figure S3. Antifungal activity of XnChi, XhChi, and PtChi (20 µg) against F. oxysporum and F. graminearum; Figure S4. Effect of chitinase on conidial germination of F. graminearum (a) and F. oxysporum (b); Table S1. Primer used in the study; Table S2. Amino acid sequences of chitinases of type strain used in the study; Table S3. Amino acid sequences of chitinases used in the study; Table S4. Amino acid sequences of chitinases used in multiple sequence alignment analysis; Table S5. Amino acid sequences of chitinases used in phylogenetic analysis.
Author Contributions
Conceptualization, D.-J.S. and Y.-M.C.; methodology, D.-J.S., G.-G.K. and Y.-M.C.; investigation, D.-J.S., G.-G.K. and N.-J.C.; resources, N.-J.C. and G.-G.K.; data curation, D.-J.S. and Y.-M.C.; writing—original draft preparation, D.-J.S. and Y.-M.C.; writing—review and editing, D.-J.S., H.-Y.C. and Y.-M.C.; visualization, D.-J.S.; supervision, H.-Y.C., N.-J.C. and Y.-M.C.; project administration, D.-J.S., N.-J.C. and Y.-M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Ministry of Agriculture, Food and Rural Affairs, Grant number 120081-05”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or supplementary material.
Acknowledgments
The authors want to thank everyone who helped with the experiment. Then, we want to thank the J. G. Lee of Dong-A University for providing plant pathogenic fungi. Finally, we want to thank to the reviewers for their valuable comments and suggestions for improving the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Boemare, N. Interactions between the Partners of the Entomopathogenic Bacterium Nematode Complexes, Steinernema-Xenorhabdus and Heterorhabditis-Photorhabdus. Nematology 2002, 4, 601–603. [Google Scholar] [CrossRef]
- Choo, H.Y.; Lee, S.M.; Lee, D.W.; Kim, H.H. Ecology of entomopathogenic nematodes. In Microbial Insecticides: Principles and Applications; Borgio, J.F., Sahayaraj, K., Susurluk, I.A., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2011; ISBN 978-1-61209-223-2. [Google Scholar]
- Park, Y.; Kim, Y. Eicosanoids Rescue Spodoptera exigua Infected with Xenorhabdus nematophilus, the Symbiotic Bacteria to the Entomopathogenic Nematode Steinernema carpocapsae. J. Insect Physiol. 2000, 46, 1469–1476. [Google Scholar] [CrossRef]
- Labaude, S.; Griffin, C. Transmission Success of Entomopathogenic Nematodes Used in Pest Control. Insects 2018, 9, 72. [Google Scholar] [CrossRef]
- Kumari, P.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. Ectopic Expression of GroEL from Xenorhabdus nematophila in Tomato Enhances Resistance against Helicoverpa armigera and Salt and Thermal Stress. Transgenic Res. 2015, 24, 859–873. [Google Scholar] [CrossRef]
- Stock, S.P.; Kusakabe, A.; Orozco, R.A. Secondary Metabolites Produced by Heterorhabditis Symbionts and Their Application in Agriculture: What We Know and What to Do Next. J. Nematol. 2017, 49, 373–383. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, J.; Liu, F.; Zeng, F. Expression of a Nematode Symbiotic Bacterium-Derived Protease Inhibitor Protein in Tobacco Enhanced Tolerance against Myzus persicae. Plant Cell Rep. 2012, 31, 1981–1989. [Google Scholar] [CrossRef]
- Sergeant, M.; Jarrett, P.; Ousley, M.; Morgan, J.A.W. Interactions of Insecticidal Toxin Gene Products from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 2003, 69, 3344–3349. [Google Scholar] [CrossRef]
- Brown, S.E.; Cao, A.T.; Dobson, P.; Hines, E.R.; Akhurst, R.J.; East, P.D. Txp40, a Ubiquitous Insecticidal Toxin Protein from Xenorhabdus and Photorhabdus Bacteria. Appl. Environ. Microbiol. 2006, 72, 1653–1662. [Google Scholar] [CrossRef]
- Vigneux, F.; Zumbihl, R.; Jubelin, G.; Ribeiro, C.; Poncet, J.; Baghdiguian, S.; Givaudan, A.; Brehélin, M. The xaxAB Genes Encoding a New Apoptotic Toxin from the Insect Pathogen Xenorhabdus nematophila Are Present in Plant and Human Pathogens. J. Biol. Chem. 2007, 282, 9571–9580. [Google Scholar] [CrossRef]
- Kumari, P.; Kant, S.; Zaman, S.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. A Novel Insecticidal GroEL Protein from Xenorhabdus nematophila Confers Insect Resistance in Tobacco. Transgenic Res. 2014, 23, 99–107. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, J.; Li, T.; Liu, S.; Song, P.; Nangong, Z.; Wang, Q. PirAB Protein from Xenorhabdus nematophila HB310 Exhibits a Binary Toxin with Insecticidal Activity and Cytotoxicity in Galleria mellonella. J. Invertebr. Pathol. 2017, 148, 43–50. [Google Scholar] [CrossRef]
- Santhoshkumar, K.; Mathur, C.; Mandal, A.; Dutta, T.K. A Toxin Complex Protein from Photorhabdus akhurstii Conferred Oral Insecticidal Activity against Galleria mellonella by Targeting the Midgut Epithelium. Microbiol. Res. 2021, 242, 126642. [Google Scholar] [CrossRef]
- Jang, E.-K.; Kwon Jung, B.; Park, G.-S.; Rahim Khan, A.; Hong, S.-J.; Park, Y.-J.; Kim, W.-C.; Shin, J.-H.; Al-Ghamdi, K.M.S.; Oudh Al-Johny, B.; et al. Cloning and Expression of the Insecticidal Toxin Gene “tccB” from Photorhabdus temperata M1021 in Escherichia coli Expression System. J. Asia-Pac. Entomol. 2020, 23, 172–176. [Google Scholar] [CrossRef]
- Mathur, C.; Phani, V.; Kushwah, J.; Somvanshi, V.S.; Dutta, T.K. TcaB, an Insecticidal Protein from Photorhabdus akhurstii Causes Cytotoxicity in the Greater Wax Moth, Galleria mellonella. Pestic. Biochem. Physiol. 2019, 157, 219–229. [Google Scholar] [CrossRef]
- Mathur, C.; Kushwah, J.; Somvanshi, V.S.; Dutta, T.K. A 37 kDa Txp40 Protein Characterized from Photorhabdus luminescens Sub Sp. Akhurstii Conferred Injectable and Oral Toxicity to Greater Wax Moth, Galleria mellonella. Toxicon 2018, 154, 69–73. [Google Scholar] [CrossRef]
- Ullah, I.; Jang, E.-K.; Kim, M.-S.; Shin, J.-H.; Park, G.-S.; Khan, A.; Hong, S.-J.; Jung, B.-K.; Choi, J.; Park, Y.; et al. Identification and Characterization of the Insecticidal Toxin “Makes Caterpillars Floppy” in Photorhabdus temperata M1021 Using a Cosmid Library. Toxins 2014, 6, 2024–2040. [Google Scholar] [CrossRef]
- Forst, S.; Nealson, K. Molecular Biology of the Symbiotic-Pathogenic Bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 1996, 60, 21–43. [Google Scholar] [CrossRef]
- Elieh-Ali-Komi, D.; Hamblin, M.R. Chitin and Chitosan: Production and Application of Versatile Biomedical Nanomaterials. Int. J. Adv. Res. 2017, 4, 411. [Google Scholar]
- Moussian, B. Chitin: Structure, Chemistry and Biology. Adv. Exp. Med. Biol. 2019, 1142, 5–18. [Google Scholar]
- Kramer, K.J.; Muthukrishnan, S. Insect Chitinases: Molecular Biology and Potential Use as Biopesticides. Insect Biochem. Mol. Biol. 1997, 27, 887–900. [Google Scholar] [CrossRef]
- Merzendorfer, H. The Cellular Basis of Chitin Synthesis in Fungi and Insects: Common Principles and Differences. Eur. J. Cell Biol. 2011, 90, 759–769. [Google Scholar] [CrossRef]
- Liu, J.; Bai, H.; Song, P.; Nangong, Z.; Dong, Z.; Li, Z.; Wang, Q. Insecticidal Activity of Chitinases from Xenorhabdus nematophila HB310 and Its Relationship with the Toxin Complex. Toxins 2022, 14, 646. [Google Scholar] [CrossRef]
- Martínez-Zavala, S.A.; Barboza-Pérez, U.E.; Hernández-Guzmán, G.; Bideshi, D.K.; Barboza-Corona, J.E. Chitinases of Bacillus Thuringiensis: Phylogeny, Modular Structure, and Applied Potentials. Front. Microbiol. 2020, 10, 3032. [Google Scholar] [CrossRef]
- He, B.; Yang, L.; Yang, D.; Jiang, M.; Ling, C.; Chen, H.; Ji, F.; Pan, L. Biochemical Purification and Characterization of a Truncated Acidic, Thermostable Chitinase from Marine Fungus for N-Acetylglucosamine Production. Front. Bioeng. Biotechnol. 2022, 10, 1013313. [Google Scholar] [CrossRef]
- Rathore, A.S.; Gupta, R.D. Chitinases from Bacteria to Human: Properties, Applications, and Future Perspectives. Enzym. Res. 2015, 2015, 1–8. [Google Scholar] [CrossRef]
- Henrissat, B.; Bairoch, A. New Families in the Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1993, 293, 781–788. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, X.; Yang, Q. Glycoside Hydrolase Family 18 Chitinases: The Known and the Unknown. Biotechnol. Adv. 2020, 43, 107553. [Google Scholar] [CrossRef]
- Bahar, A.A.; Sezen, K.; Demirbağ, Z.; Nalçacioğlu, R. The Relationship between Insecticidal Effects and Chitinase Activities of Coleopteran-Originated Entomopathogens and Their Chitinolytic Profile. Ann. Microbiol. 2012, 62, 647–653. [Google Scholar] [CrossRef]
- Regev, A.; Keller, M.; Strizhov, N.; Sneh, B.; Prudovsky, E.; Chet, I.; Ginzberg, I.; Koncz-Kalman, Z.; Koncz, C.; Schell, J.; et al. Synergistic Activity of a Bacillus thuringiensis δ-Endotoxin and a Bacterial Endochitinase against Spodoptera littoralis Larvae. Appl. Environ. Microbiol. 1996, 62. [Google Scholar] [CrossRef]
- Busby, J.N.; Landsberg, M.J.; Simpson, R.M.; Jones, S.A.; Hankamer, B.; Hurst, M.R.H.; Lott, J.S. Structural Analysis of Chi1 Chitinase from Yen-Tc: The Multisubunit Insecticidal ABC Toxin Complex of Yersinia entomophaga. J. Mol. Biol. 2012, 415, 359–371. [Google Scholar] [CrossRef]
- Liu, J.; NanGong, Z.; Zhang, J.; Song, P.; Tang, Y.; Gao, Y.; Wang, Q. Expression and Characterization of Two Chitinases with Synergistic Effect and Antifungal Activity from Xenorhabdus nematophila. World J. Microbiol. Biotechnol. 2019, 35, 106. [Google Scholar] [CrossRef]
- Dominelli, N.; Platz, F.; Heermann, R. The Insect Pathogen Photorhabdus luminescens Protects Plants from Phytopathogenic Fusarium Graminearum via Chitin Degradation. Appl. Environ. Microbiol. 2022, 88, e00645-22. [Google Scholar] [CrossRef]
- Sajnaga, E.; Kazimierczak, W. Evolution and Taxonomy of Nematode-Associated Entomopathogenic Bacteria of the Genera Xenorhabdus and Photorhabdus: An Overview. Symbiosis 2020, 80, 1–13. [Google Scholar] [CrossRef]
- Boemare, N.; Akhurst, R. The Genera Photorhabdus and Xenorhabdus. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 451–494. ISBN 978-0-387-25496-8. [Google Scholar]
- Hinchliffe, S.J. Insecticidal Toxins from the Photorhabdus and Xenorhabdus Bacteria. Open Toxinol. J. 2013, 3, 101–118. [Google Scholar] [CrossRef]
- Fukruksa, C.; Yimthin, T.; Suwannaroj, M.; Muangpat, P.; Tandhavanant, S.; Thanwisai, A.; Vitta, A. Isolation and Identification of Xenorhabdus and Photorhabdus Bacteria Associated with Entomopathogenic Nematodes and Their Larvicidal Activity against Aedes aegypti. Parasit. Vectors 2017, 10, 440. [Google Scholar] [CrossRef]
- Mollah, M.M.I.; Roy, M.C.; Choi, D.-Y.; Hasan, M.A.; Al Baki, M.A.; Yeom, H.-S.; Kim, Y. Variations of Indole Metabolites and NRPS-PKS Loci in Two Different Virulent Strains of Xenorhabdus hominickii. Front. Microbiol. 2020, 11, 583594. [Google Scholar] [CrossRef]
- Rivera-Ramírez, A.; Salgado-Morales, R.; Jiménez-Pérez, A.; Pérez-Martínez, R.; García-Gómez, B.I.; Dantán-González, E. Comparative Genomics and Pathogenicity Analysis of Two Bacterial Symbionts of Entomopathogenic Nematodes: The Role of The GroEL Protein in Virulence. Microorganisms 2022, 10, 486. [Google Scholar] [CrossRef]
- Mahmood, S.; Kumar, M.; Kumari, P.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. Novel Insecticidal Chitinase from the Insect Pathogen Xenorhabdus nematophila. Int. J. Biol. Macromol. 2020, 159, 394–401. [Google Scholar] [CrossRef]
- Palmer, I.; Wingfield, P.T. Preparation and Extraction of Insoluble (Inclusion-Body) Proteins from Escherichia coli. Curr. Protoc. Protein Sci. 2012, 70, 6.3.1–6.3.20. [Google Scholar] [CrossRef]
- Singh, G.; Arya, S.K. Antifungal and Insecticidal Potential of Chitinases: A Credible Choice for the Eco-Friendly Farming. Biocatal. Agric. Biotechnol. 2019, 20, 101289. [Google Scholar] [CrossRef]
- Prasanna, L.; Eijsink, V.G.H.; Meadow, R.; Gåseidnes, S. A Novel Strain of Brevibacillus laterosporus Produces Chitinases That Contribute to Its Biocontrol Potential. Appl. Microbiol. Biotechnol. 2013, 97, 1601–1611. [Google Scholar] [CrossRef]
- Suganthi, M.; Senthilkumar, P.; Arvinth, S.; Chandrashekara, K.N. Chitinase from Pseudomonas Fluorescens and Its Insecticidal Activity against Helopeltis theivora. J. Gen. Appl. Microbiol. 2017, 63, 222–227. [Google Scholar] [CrossRef]
- Stutz, E.W.; Défago, G.; Kern, H. Naturally Occurring Fluorescent Pseudomonads Involved in Suppression. Phytopathology 1986, 76, 181–185. [Google Scholar] [CrossRef]
- Tu, S.; Qiu, X.; Cao, L.; Han, R.; Zhang, Y.; Liu, X. Expression and Characterization of the Chitinases from Serratia Marcescens GEI Strain for the Control of Varroa Destructor, a Honey Bee Parasite. J. Invertebr. Pathol. 2010, 104, 75–82. [Google Scholar] [CrossRef]
- Wang, M.; Xing, Y.; Wang, J.; Xu, Y.; Wang, G. The Role of the Chi1 Gene from the Endophytic Bacteria Serratia proteamaculans 336x in the Biological Control of Wheat Take-All. Can. J. Microbiol. 2014, 60, 533–540. [Google Scholar] [CrossRef]
- Rishad, K.S.; Varghese, S.; Jisha, M.S. Sequence Analysis and Docking Performance of Extracellular Chitinase from Bacillus Pumilus MCB-7, a Novel Mangrove Isolate. Enzym. Microb. Technol. 2020, 140, 109624. [Google Scholar] [CrossRef]
- Barboza-Corona, J.E.; Nieto-Mazzocco, E.; Velázquez-Robledo, R.; Salcedo-Hernandez, R.; Bautista, M.; Jiménez, B.; Ibarra, J.E. Cloning, Sequencing, and Expression of the Chitinase Gene chiA74 from Bacillus thuringiensis. Appl. Environ. Microbiol. 2003, 69, 1023–1029. [Google Scholar] [CrossRef]
- Russell, J.; Kim, S.-K.; Duma, J.; Nothaft, H.; Himmel, M.E.; Bomble, Y.J.; Szymanski, C.M.; Westpheling, J. Deletion of a Single Glycosyltransferase in Caldicellulosiruptor bescii Eliminates Protein Glycosylation and Growth on Crystalline Cellulose. Biotechnol. Biofuels 2018, 11, 259. [Google Scholar] [CrossRef]
- Boysen, A.; Palmisano, G.; Krogh, T.J.; Duggin, I.G.; Larsen, M.R.; Møller-Jensen, J. A Novel Mass Spectrometric Strategy “BEMAP” Reveals Extensive O-Linked Protein Glycosylation in Enterotoxigenic Escherichia coli. Sci. Rep. 2016, 6, 32016. [Google Scholar] [CrossRef]
- Beier, S.; Bertilsson, S. Bacterial Chitin Degradation—Mechanisms and Ecophysiological Strategies. Front. Microbiol. 2013, 4, 149. [Google Scholar] [CrossRef]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, Limitations, and Trends in Engineering for Suitable Applications. Biosci. Rep. 2018, 38, BSR2018032300. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, Y.; Li, J.; Dunphy, G.B.; Punja, Z.K.; Webster, J.M. Chitinase Activity of Xenorhabdus and Photorhabdus Species, Bacterial Associates of Entomopathogenic Nematodes. J. Invertebr. Pathol. 1996, 68, 101–108. [Google Scholar] [CrossRef]
- Berini, F.; Casartelli, M.; Montali, A.; Reguzzoni, M.; Tettamanti, G.; Marinelli, F. Metagenome-Sourced Microbial Chitinases as Potential Insecticide Proteins. Front. Microbiol. 2019, 10, 1358. [Google Scholar] [CrossRef]
- Churklam, W.; Aunpad, R. Enzymatic Characterization and Structure-Function Relationship of Two Chitinases, LmChiA and LmChiB, from Listeria monocytogenes. Heliyon 2020, 6, e04252. [Google Scholar] [CrossRef]
- Roberts, W.K.; Selitrennikoff, C.P. Plant and Bacterial Chitinases Differ in Antifungal Activity. Microbiology 1988, 134, 169–176. [Google Scholar] [CrossRef]
- Lorito, M.; Harman, G.E.; Hayes, C.K.; Broadway, R.M.; Tronsmo, A.; Woo, S.L.; Di Pietro, A. Chitinolytic Enzymes Produced by Trichoderma harzianum: Antifungal Activity of Purified Endochitinase and Chitobiosidase. Phytopathology 1993, 83, 302–307. [Google Scholar] [CrossRef]
- Hjort, K.; Presti, I.; Elväng, A.; Marinelli, F.; Sjöling, S. Bacterial Chitinase with Phytopathogen Control Capacity from Suppressive Soil Revealed by Functional Metagenomics. Appl. Microbiol. Biotechnol. 2014, 98, 2819–2828. [Google Scholar] [CrossRef]
- Jiménez-Ortega, E.; Kidibule, P.E.; Fernández-Lobato, M.; Sanz-Aparicio, J. Structure–Function Insights into the Fungal Endo-Chitinase Chit33 Depict Its Mechanism on Chitinous Material. Int. J. Mol. Sci. 2022, 23, 7599. [Google Scholar] [CrossRef]
- Koga, D.; Funakoshi, T.; Mizuki, K.; Ide, A.; Kramer, K.J.; Zen, K.-C.; Choi, H.; Muthukrishnan, S. Immunoblot Analysis of Chitinolytic Enzymes in Integument and Molting Fluid of the Silkworm, Bombyx Mori, and the Tobacco Hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 1992, 22, 305–311. [Google Scholar] [CrossRef]
- Koga, D.; Jilka, J.; Kramer, K.J. Insect Endochitinases: Glycoproteins from Moulting Fluid, Integument and Pupal Haemolymph of Manduca sexta L. Insect Biochem. 1983, 13, 295–305. [Google Scholar] [CrossRef]
- Navarro-González, S.S.; Ramírez-Trujillo, J.A.; Peña-Chora, G.; Gaytán, P.; Roldán-Salgado, A.; Corzo, G.; Lina-García, L.P.; Hernández-Velázquez, V.M.; Suárez-Rodríguez, R. Enhanced Tolerance against a Fungal Pathogen and Insect Resistance in Transgenic Tobacco Plants Overexpressing an Endochitinase Gene from Serratia marcescens. Int. J. Mol. Sci. 2019, 20, 3482. [Google Scholar] [CrossRef]
- Zarei, M.; Aminzadeh, S.; Zolgharnein, H.; Safahieh, A.; Daliri, M.; Noghabi, K.A.; Ghoroghi, A.; Motallebi, A. Characterization of a Chitinase with Antifungal Activity from a Native Serratia marcescens B4A. Braz. J. Microbiol. 2011, 42, 1017–1029. [Google Scholar] [CrossRef]
- Schoffelmeer, E.A.M.; Klis, F.M.; Sietsma, J.H.; Cornelissen, B.J.C. The Cell Wall of Fusarium oxysporum. Fungal Genet. Biol. 1999, 27, 275–282. [Google Scholar] [CrossRef]
- Barbosa, I.P.; Kemmelmeier, C. Chemical Composition of the Hyphal Wall from Fusarium graminearum. Exp. Mycol. 1993, 17, 274–283. [Google Scholar] [CrossRef]
- Liu, M.; Gong, Y.; Sun, H.; Zhang, J.; Zhang, L.; Sun, J.; Han, Y.; Huang, J.; Wu, Q.; Zhang, C.; et al. Characterization of a Novel Chitinase from Sweet Potato and Its Fungicidal Effect against Ceratocystis fimbriata. J. Agric. Food Chem. 2020, 68, 7591–7600. [Google Scholar] [CrossRef]
- Kotb, E.; Alabdalall, A.H.; Alghamdi, A.I.; Ababutain, I.M.; Aldakeel, S.A.; Al-Zuwaid, S.K.; Algarudi, B.M.; Algarudi, S.M.; Ahmed, A.A.; Albarrag, A.M. Screening for Chitin Degrading Bacteria in the Environment of Saudi Arabia and Characterization of the Most Potent Chitinase from Streptomyces Variabilis Am1. Sci. Rep. 2023, 13, 11723. [Google Scholar] [CrossRef]
- Rodou, A.; Ankrah, D.O.; Stathopoulos, C. Toxins and Secretion Systems of Photorhabdus luminescens. Toxins 2010, 2, 1250–1264. [Google Scholar] [CrossRef]
- Sheets, J.J.; Hey, T.D.; Fencil, K.J.; Burton, S.L.; Ni, W.; Lang, A.E.; Benz, R.; Aktories, K. Insecticidal Toxin Complex Proteins from Xenorhabdus nematophilus. J. Biol. Chem. 2011, 286, 22742–22749. [Google Scholar] [CrossRef]
- Ffrench-Constant, R.H.; Dowling, A.; Waterfield, N.R. Insecticidal Toxins from Photorhabdus Bacteria and Their Potential Use in Agriculture. Toxicon 2007, 49, 436–451. [Google Scholar] [CrossRef]
- Mahmood, S.; Kumari, P.; Kisku, A.V.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. Ectopic Expression of Xenorhabdus nematophila Chitinase in Tobacco Confers Resistance against Helicoverpa armigera. Plant Cell Tissue Organ Cult. (PCTOC) 2022, 151, 593–604. [Google Scholar] [CrossRef]
- Cappellini, R.A.; Peterson, J.L. Macroconidium Formation in Submerged Cultures by a Nonsporulating Strain of Gibberella zeae. Mycologia 1965, 57, 962–966. [Google Scholar] [CrossRef]
- Li, T.; Jung, B.; Park, S.-Y.; Lee, J. Survival Factor Gene FgSvf1 Is Required for Normal Growth and Stress Resistance in Fusarium graminearum. Plant Pathol. J. 2019, 35, 393–405. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).