Okadaic Acid Detection through a Rapid and Sensitive Amplified Luminescent Proximity Homogeneous Assay
Abstract
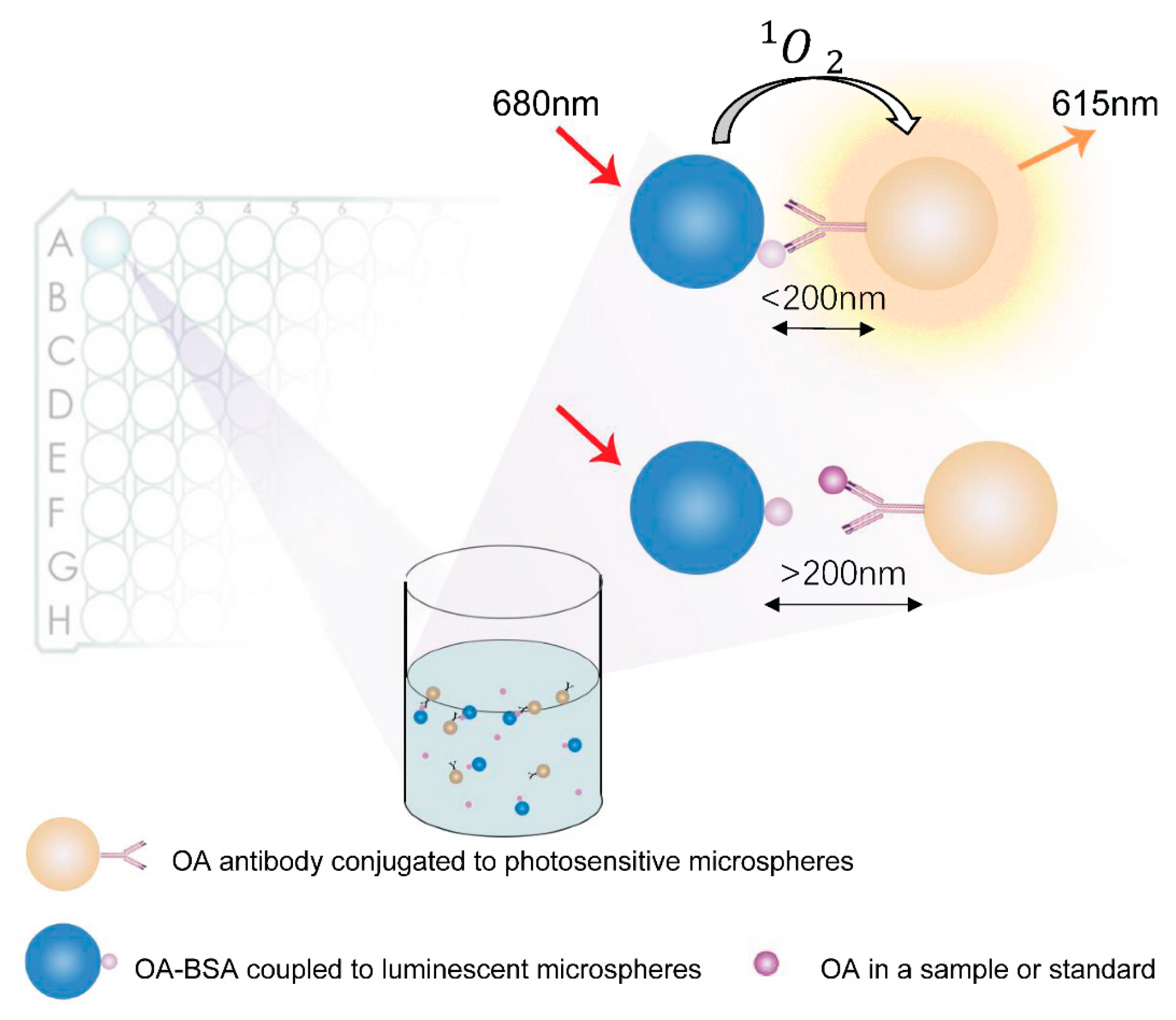
1. Introduction
2. Results
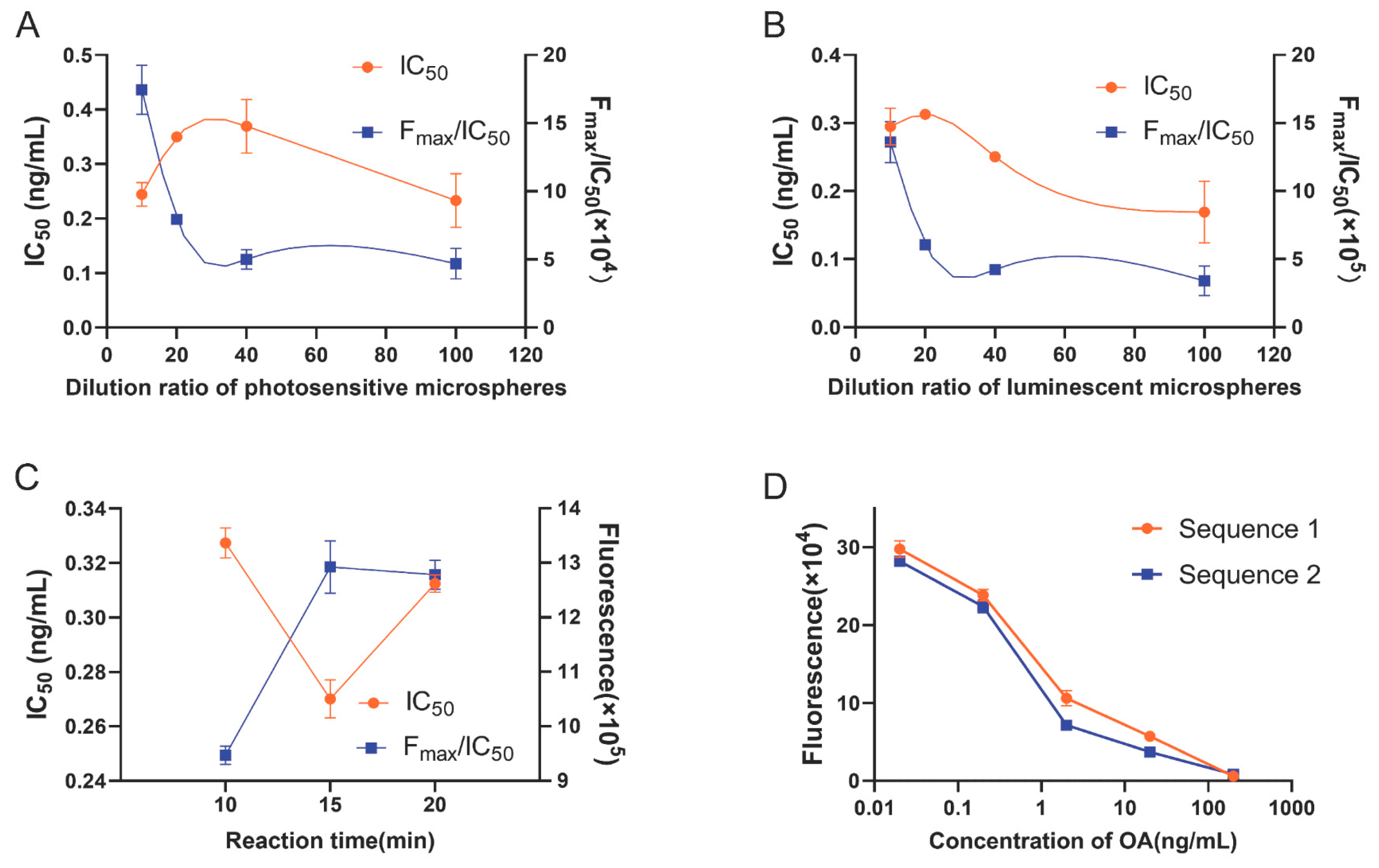
2.1. Optimization of OA-AlphaLISA
2.2. Identification of OA-AlphaLISA
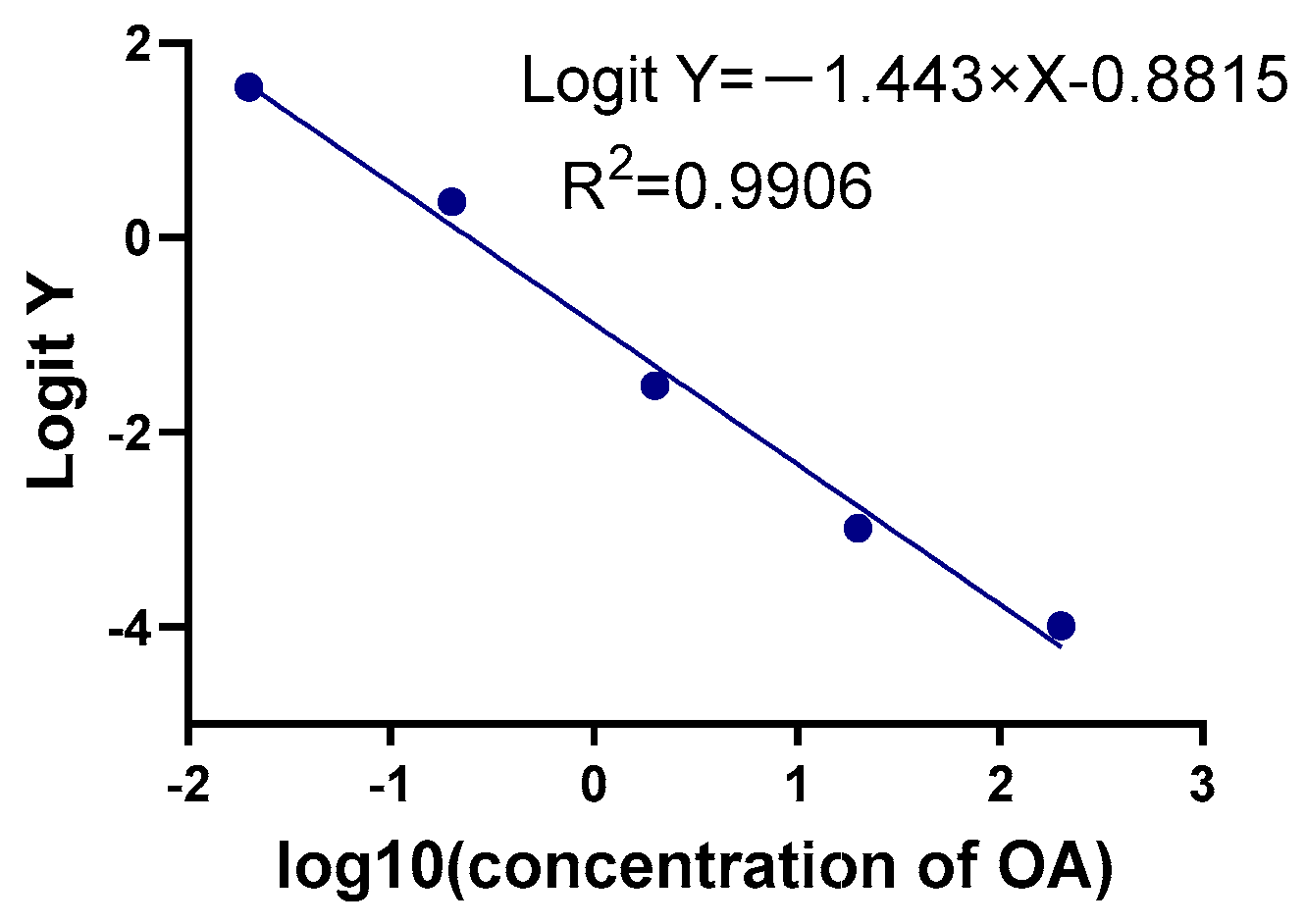
2.2.1. Sensitivity, Precision, Specificity, and Recovery Rate
2.2.2. Stability
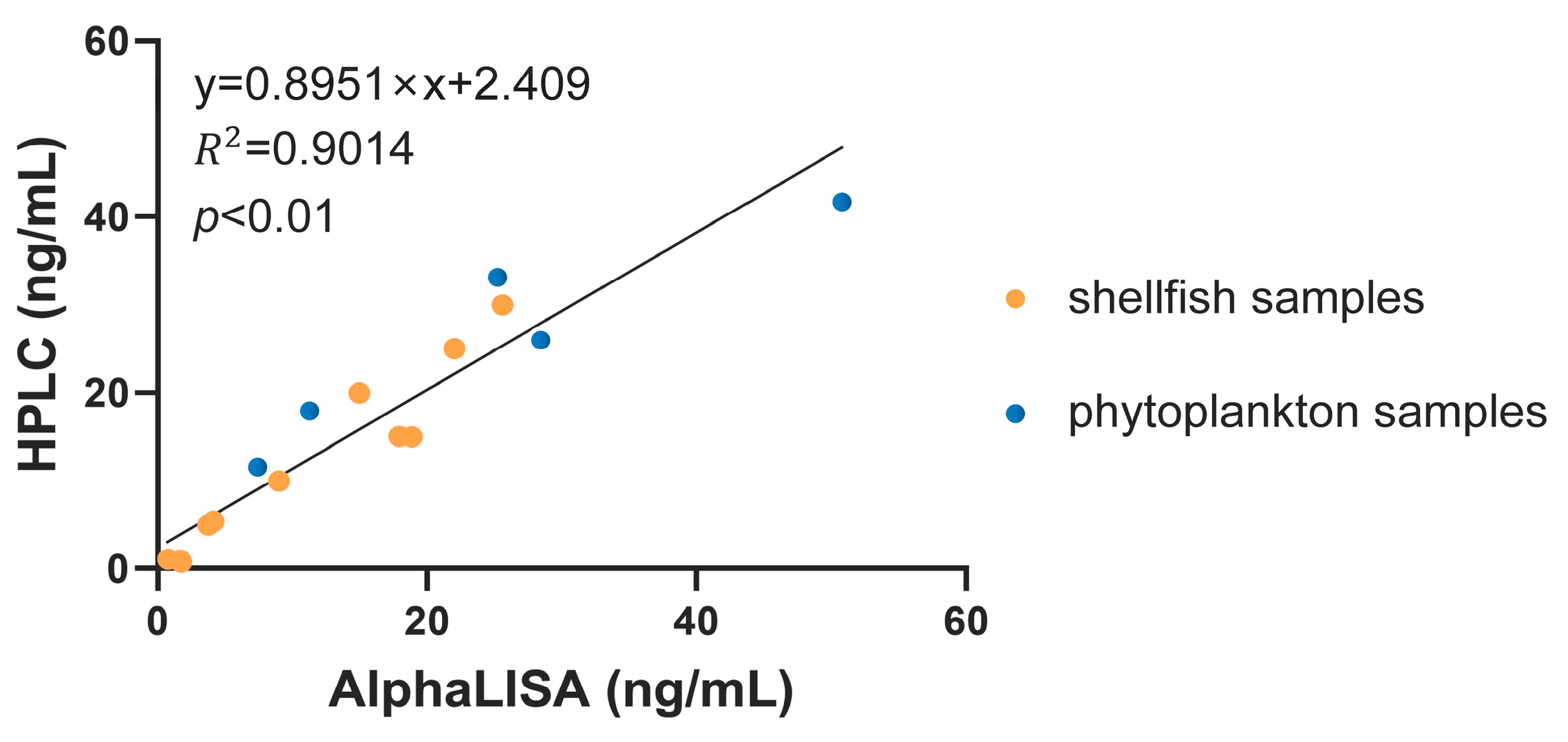
2.3. Application of OA-AlphaLISA
3. Discussion
4. Materials and Methods
4.1. Reagents and Instruments
4.2. Conjugated Microspheres
4.3. AlphaLISA Detection Protocol for OA
4.4. Optimization
4.5. Method Evaluation
4.5.1. Precision
4.5.2. Sensitivity
4.5.3. Specificity
4.5.4. Stability
4.5.5. Recovery Rate
4.6. Processing and Testing of Samples
4.7. HPLC Detection Protocol for OA
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferreiro, S.F.; Carrera, C.; Vilariño, N.; Louzao, M.C.; Santamarina, G.; Cantalapiedra, A.G.; Botana, L.M. Acute cardiotoxicity evaluation of the marine biotoxins OA, DTX-1 and YTX. Toxins 2015, 7, 1030–1047. [Google Scholar] [CrossRef] [PubMed]
- Salonen, L.M.; Pinela, S.R.; Fernandes, S.P.S.; Louçano, J.; Carbó-Argibay, E.; Sarriá, M.P.; Rodríguez-Abreu, C.; Peixoto, J.; Espiña, B. Adsorption of marine phycotoxin okadaic acid on a covalent organic framework. J. Chromatogr. A 2017, 1525, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, B.; Kim, H.; Abassi, S.; Ki, J.S. Toxic Effects and Tumor Promotion Activity of Marine Phytoplankton Toxins: A Review. Toxins 2022, 14, 397. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Muñoz, D.; Lawton, L.A.; Edwards, C. Degradation of okadaic acid in seawater by UV/TiO(2) photocatalysis-Proof of concept. Sci. Total Environ. 2020, 733, 139346. [Google Scholar] [CrossRef]
- Torgersen, T.; Lindegarth, S.; Ungfors, A.; Sandvik, M. Profiles and levels of fatty acid esters of okadaic acid group toxins and pectenotoxins during toxin depuration. Part I: Brown crab (Cancer pagurus). Toxicon 2008, 52, 407–417. [Google Scholar] [CrossRef]
- McCarthy, M.; O’Halloran, J.; O’Brien, N.M.; van Pelt, F. Does the marine biotoxin okadaic acid cause DNA fragmentation in the blue mussel and the pacific oyster? Mar. Environ. Res. 2014, 101, 153–160. [Google Scholar] [CrossRef]
- Wu, H.; Chen, J.; Peng, J.; Zhong, Y.; Zheng, G.; Guo, M.; Tan, Z.; Zhai, Y.; Lu, S. Nontarget Screening and Toxicity Evaluation of Diol Esters of Okadaic Acid and Dinophysistoxins Reveal Intraspecies Difference of Prorocentrum lima. Environ. Sci. Technol. 2020, 54, 12366–12375. [Google Scholar] [CrossRef]
- Suganuma, M.; Fujiki, H.; Suguri, H.; Yoshizawa, S.; Hirota, M.; Nakayasu, M.; Ojika, M.; Wakamatsu, K.; Yamada, K.; Sugimura, T. Okadaic acid: An additional non-phorbol-12-tetradecanoate-13-acetate-type tumor promoter. Proc. Natl. Acad. Sci. USA 1988, 85, 1768–1771. [Google Scholar] [CrossRef]
- García, C.; Pereira, P.; Valle, L.; Lagos, N. Quantitation of diarrhetic shellfish poisoning toxins in Chilean mussel using pyrenyldiazomethane as fluorescent labeling reagent. Biol. Res. 2003, 36, 171–183. [Google Scholar] [CrossRef][Green Version]
- Dietrich, J.; Sommersdorf, C.; Gohlke, S.; Poetz, O.; Traenkle, B.; Rothbauer, U.; Hessel-Pras, S.; Lampen, A.; Braeuning, A. Okadaic acid activates Wnt/β-catenin-signaling in human HepaRG cells. Arch. Toxicol. 2019, 93, 1927–1939. [Google Scholar] [CrossRef]
- Vilariño, N.; Louzao, M.C.; Abal, P.; Cagide, E.; Carrera, C.; Vieytes, M.R.; Botana, L.M. Human Poisoning from Marine Toxins: Unknowns for Optimal Consumer Protection. Toxins 2018, 10, 324. [Google Scholar] [CrossRef] [PubMed]
- González, J.C.; Fontal, O.I.; Vieytes, M.R.; Vieites, J.M.; Botana, L.M. Basis for a new procedure to eliminate diarrheic shellfish toxins from a contaminated matrix. J. Agric. Food Chem. 2002, 50, 400–405. [Google Scholar] [CrossRef]
- Reguera, B.; Riobó, P.; Rodríguez, F.; Díaz, P.A.; Pizarro, G.; Paz, B.; Franco, J.M.; Blanco, J. Dinophysis toxins: Causative organisms, distribution and fate in shellfish. Mar. Drugs 2014, 12, 394–461. [Google Scholar] [CrossRef]
- Ramalingam, S.; Chand, R.; Singh, C.B.; Singh, A. Phosphorene-gold nanocomposite based microfluidic aptasensor for the detection of okadaic acid. Biosens. Bioelectron. 2019, 135, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Ferron, P.J.; Hogeveen, K.; Fessard, V.; Le Hégarat, L. Comparative analysis of the cytotoxic effects of okadaic acid-group toxins on human intestinal cell lines. Mar. Drugs 2014, 12, 4616–4634. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Freitas, M.; de Almeida, A.M.; Martins, J.C.; Domínguez-Pérez, D.; Osório, H.; Vasconcelos, V.; Reis Costa, P. OMICs Approaches in Diarrhetic Shellfish Toxins Research. Toxins 2020, 12, 493. [Google Scholar] [CrossRef]
- Farrell, H.; Ajani, P.; Murray, S.; Baker, P.; Webster, G.; Brett, S.; Zammit, A. Diarrhetic Shellfish Toxin Monitoring in Commercial Wild Harvest Bivalve Shellfish in New South Wales, Australia. Toxins 2018, 10, 446. [Google Scholar] [CrossRef]
- Mouratidou, T.; Kaniou-Grigoriadou, I.; Samara, C.; Kouimtzis, T. Detection of the marine toxin okadaic acid in mussels during a diarrhetic shellfish poisoning (DSP) episode in Thermaikos Gulf, Greece, using biological, chemical and immunological methods. Sci. Total Environ. 2006, 366, 894–904. [Google Scholar] [CrossRef]
- Prego-Faraldo, M.V.; Valdiglesias, V.; Méndez, J.; Eirín-López, J.M. Okadaic acid meet and greet: An insight into detection methods, response strategies and genotoxic effects in marine invertebrates. Mar. Drugs 2013, 11, 2829–2845. [Google Scholar] [CrossRef]
- Suzuki, H. Drastic hypothermia after intraperitoneal injection of okadaic acid, a diarrhetic shellfish poisoning toxin, in mice. Exp. Anim. 2021, 70, 412–418. [Google Scholar] [CrossRef]
- Otero, P.; Alfonso, A.; Alfonso, C.; Rodríguez, P.; Vieytes, M.R.; Botana, L.M. Effect of uncontrolled factors in a validated liquid chromatography-tandem mass spectrometry method question its use as a reference method for marine toxins: Major causes for concern. Anal. Chem. 2011, 83, 5903–5911. [Google Scholar] [CrossRef]
- Dhanji-Rapkova, M.; O’Neill, A.; Maskrey, B.H.; Coates, L.; Teixeira Alves, M.; Kelly, R.J.; Hatfield, R.G.; Rowland-Pilgrim, S.J.; Lewis, A.M.; Algoet, M.; et al. Variability and profiles of lipophilic toxins in bivalves from Great Britain during five and a half years of monitoring: Okadaic acid, dinophysis toxins and pectenotoxins. Harmful Algae 2018, 77, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Vdovenko, M.M.; Hung, C.T.; Sakharov, I.Y.; Yu, F.Y. Determination of okadaic acid in shellfish by using a novel chemiluminescent enzyme-linked immunosorbent assay method. Talanta 2013, 116, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zeng, L.; Yang, H.; Zhong, Y.; Wang, J.; Ling, S.; Saeed, A.F.; Yuan, J.; Wang, S. Detection of okadaic acid (OA) using ELISA and colloidal gold immunoassay based on monoclonal antibody. J. Hazard. Mater. 2017, 339, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lv, Q.; Liu, P.; Guo, L.; Zhang, L.; Zheng, Y.; Ming, L.; Kong, D.; Jiang, H.; Jiang, Y. AlphaLISA for detection of staphylococcal enterotoxin B free from interference by protein A. Toxicon 2019, 165, 62–68. [Google Scholar] [CrossRef]
- Armstrong, C.M.; Ruth, L.E.; Capobianco, J.A.; Strobaugh, T.P., Jr.; Rubio, F.M.; Gehring, A.G. Detection of Shiga Toxin 2 Produced by Escherichia coli in Foods Using a Novel AlphaLISA. Toxins 2018, 10, 422. [Google Scholar] [CrossRef]
- Xiang, Z.; Chen, X.; Zhou, X.; Qin, Y.; Zhao, X.; Wang, Y.; Li, Q.; Huang, B. Development and application of a novel aldehyde nanoparticle-based amplified luminescent proximity homogeneous assay for rapid quantitation of pancreatic stone protein. Clin. Chim. Acta 2022, 535, 120–130. [Google Scholar] [CrossRef]
- Zong, H.; Zhang, S.; Shang, X.; Jiang, H.; Zhao, Z.; Chen, S.; Wang, X.; Wang, Y.; Jiang, Y.; Li, X.; et al. Development of an AlphaLISA assay for sensitive and accurate detection of influenza B virus. Front. Med. 2023, 10, 1155551. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, B.; Zhang, J.; Wang, K.; Jin, J. Development of a homogeneous immunoassay based on the AlphaLISA method for the detection of chloramphenicol in milk, honey and eggs. J. Sci. Food Agric. 2012, 92, 1944–1947. [Google Scholar] [CrossRef]
- Chen, J.; Fu, B.; Xiang, Z.; Chen, X.; Wang, L.; Qin, Y.; Zhao, X.; Zhou, X.; Liu, P.; Huang, B. Sensitive amplified luminescent proximity homogeneous assay for the quantitative detection of CA242. J. Immunol. Methods 2023, 517, 113487. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, J.; Zou, L.; Zou, Y.; Lang, L.; Gao, F.; Hu, N.; Wang, P. A novel sensitive cell-based Love Wave biosensor for marine toxin detection. Biosens. Bioelectron. 2016, 77, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Morton, S.L.; Vershinin, A.; Smith, L.L.; Leighfield, T.A.; Pankov, S.; Quilliam, M.A. Seasonality of Dinophysis spp. and Prorocentrum lima in Black Sea phytoplankton and associated shellfish toxicity. Harmful Algae 2009, 8, 629–636. [Google Scholar] [CrossRef]
- Fabro, E.; Almandoz, G.O.; Ferrario, M.; Tillmann, U.; Cembella, A.; Krock, B. Distribution of Dinophysis species and their association with lipophilic phycotoxins in plankton from the Argentine Sea. Harmful Algae 2016, 59, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Ji, Y.; Fang, Y.; Zhao, M.; Wang, S.; Ai, Q.; Li, A. Response of fatty acids and lipid metabolism enzymes during accumulation, depuration and esterification of diarrhetic shellfish toxins in mussels (Mytilus galloprovincialis). Ecotoxicol. Environ. Saf. 2020, 206, 111223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lv, Q.; Zheng, Y.; Chen, X.; Kong, D.; Huang, W.; Liu, P.; Jiang, H.; Jiang, Y. A rapid and accurate method for screening T-2 toxin in food and feed using competitive AlphaLISA. FEMS Microbiol. Lett. 2021, 368, fnab029. [Google Scholar] [CrossRef]
- Qin, Y.; Li, J.Y.; Kuang, J.N.; Shen, S.C.; Jiang, J.W.; Zhang, Z.; Zhao, C.H.; Zhou, X.M.; Huang, B.; Han, B.N. Sensitive time-resolved fluoroimmunoassay for the quantitative detection of okadaic acid. Front. Mar. Sci. 2022, 9, 961751. [Google Scholar] [CrossRef]
- Wang, R.R.; Qi, H.L.; Liang, M.; Liao, G.X.; Yang, F. Rapid enzyme-linked immunosorbent assay and colloidal gold immunoassay for assessing okadaic acid and its derivatives in shellfish. Food Agric. Immunol. 2022, 33, 677–691. [Google Scholar] [CrossRef]
- Wang, X.M.; Yang, C.; Jiang, W.; Zhang, M.Y.; Li, R.F.; Lin, Y.J.; Wang, Q. Rapid quantitative detection of okadaic acid in shellfish using lanthanide-labelled fluorescent-nanoparticle immunochromatographic test strips. Food Control 2023, 148, 109635. [Google Scholar] [CrossRef]
- Blanco, J.; Alvarez, G.; Rengel, J.; Diaz, R.; Marino, C.; Martin, H.; Uribe, E. Accumulation and Biotransformation of Dinophysis Toxins by the Surf Clam Mesodesma donacium. Toxins 2018, 10, 314. [Google Scholar] [CrossRef]
- Furumochi, S.; Onoda, T.; Cho, Y.; Fuwa, H.; Sasaki, M.; Yotsu-Yamashita, M.; Konoki, K. Effect of carbon chain length in acyl coenzyme A on the efficiency of enzymatic transformation of okadaic acid to 7-O-acyl okadaic acid. Bioorg. Med. Chem. Lett. 2016, 26, 2992–2996. [Google Scholar] [CrossRef]
- Pelin, M.; Sosa, S.; Brovedani, V.; Fusco, L.; Poli, M.; Tubaro, A. A Novel Sensitive Cell-Based Immunoenzymatic Assay for Palytoxin Quantitation in Mussels. Toxins 2018, 10, 329. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Chen, J.; Wang, J.; He, X.; Xin, M.; Wang, B. Monitoring and warning of lipophilic marine algal toxins in mariculture zone based on toxin profiles of phytoplankton. Ecotoxicol. Environ. Saf. 2020, 197, 110647. [Google Scholar] [CrossRef] [PubMed]
- Fux, E.; McMillan, D.; Bire, R.; Hess, P. Development of an ultra-performance liquid chromatography-mass spectrometry method for the detection of lipophilic marine toxins. J. Chromatogr. A 2007, 1157, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Bouaïcha, N.; Hennion, M.C.; Sandra, P. Determination of okadaic acid by micellar electrokinetic chromatography with ultraviolet detection. Toxicon 1997, 35, 273–281. [Google Scholar] [CrossRef] [PubMed]

| Compound | Cross-Reactivity (%) |
|---|---|
| DA | 0.13 |
| STX | 0.02 |
| MC-LR | 0.07 |
| DTX-1 | 29.52 |
| DTX-2 | 35.87 |
| Sample | Spiked Concentration (ng/mL) | AlphaLIS | |
|---|---|---|---|
| Mean Recovery ± SD (%, n = 3) | RSD (%) | ||
| 1 | 0.5 | 95.33 ± 6.11 | 6.41 |
| 2 | 10 | 102.44 ± 3.65 | 3.56 |
| 3 | 50 | 104.98 ± 8.97 | 8.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Y.; Li, J.; Kuang, J.; Shen, S.; Zhou, X.; Zhao, X.; Huang, B.; Han, B. Okadaic Acid Detection through a Rapid and Sensitive Amplified Luminescent Proximity Homogeneous Assay. Toxins 2023, 15, 501. https://doi.org/10.3390/toxins15080501
Qin Y, Li J, Kuang J, Shen S, Zhou X, Zhao X, Huang B, Han B. Okadaic Acid Detection through a Rapid and Sensitive Amplified Luminescent Proximity Homogeneous Assay. Toxins. 2023; 15(8):501. https://doi.org/10.3390/toxins15080501
Chicago/Turabian StyleQin, Yuan, Jiayu Li, Jiani Kuang, Sicheng Shen, Xiumei Zhou, Xueqin Zhao, Biao Huang, and Bingnan Han. 2023. "Okadaic Acid Detection through a Rapid and Sensitive Amplified Luminescent Proximity Homogeneous Assay" Toxins 15, no. 8: 501. https://doi.org/10.3390/toxins15080501
APA StyleQin, Y., Li, J., Kuang, J., Shen, S., Zhou, X., Zhao, X., Huang, B., & Han, B. (2023). Okadaic Acid Detection through a Rapid and Sensitive Amplified Luminescent Proximity Homogeneous Assay. Toxins, 15(8), 501. https://doi.org/10.3390/toxins15080501






