Type A Trichothecene Metabolic Profile Differentiation, Mechanisms, Biosynthetic Pathways, and Evolution in Fusarium Species—A Mini Review
Abstract
1. Introduction
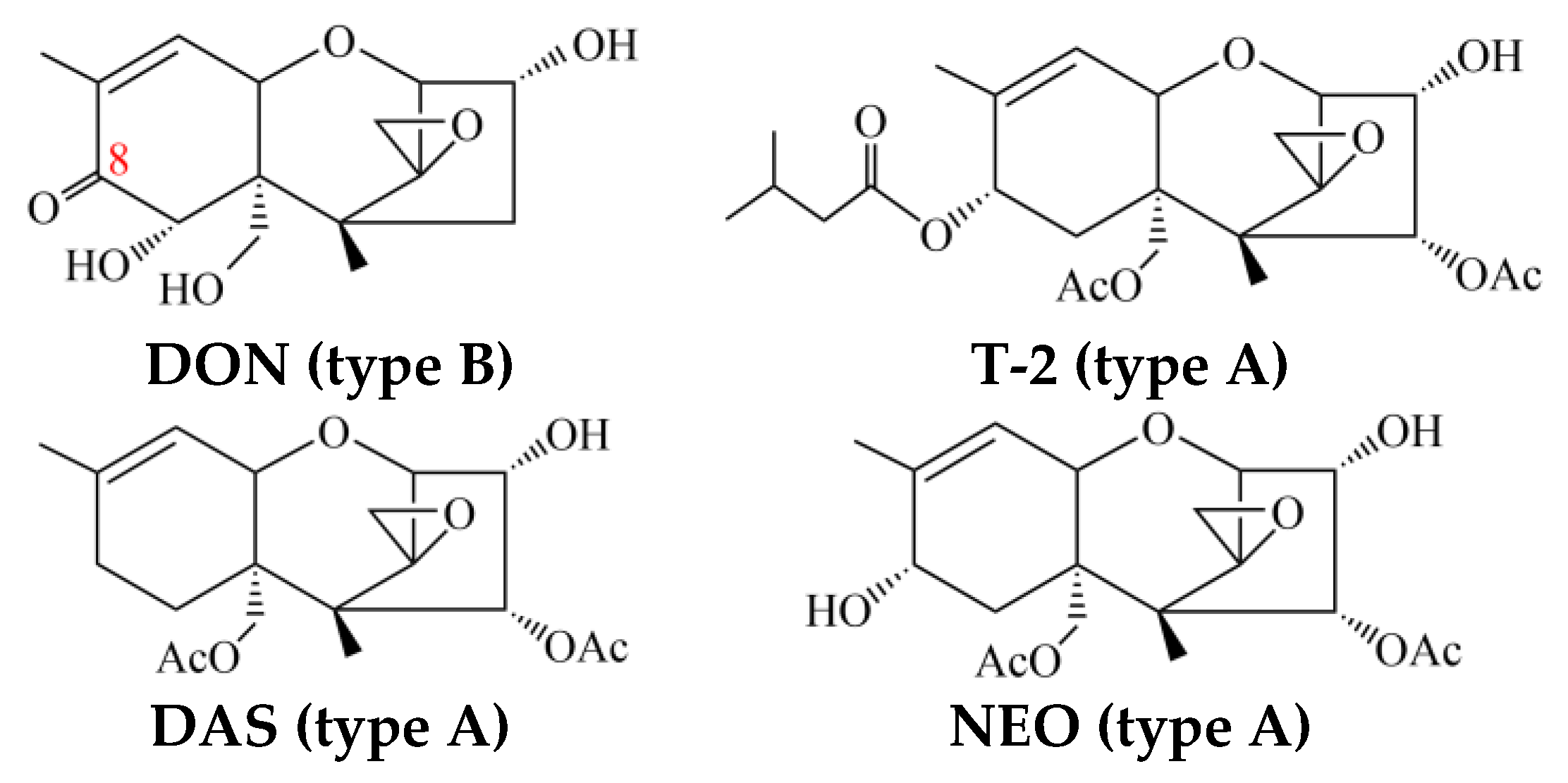
2. Metabolic Profile Differentiation in Type-A-Trichothecene-Producing Fusarium
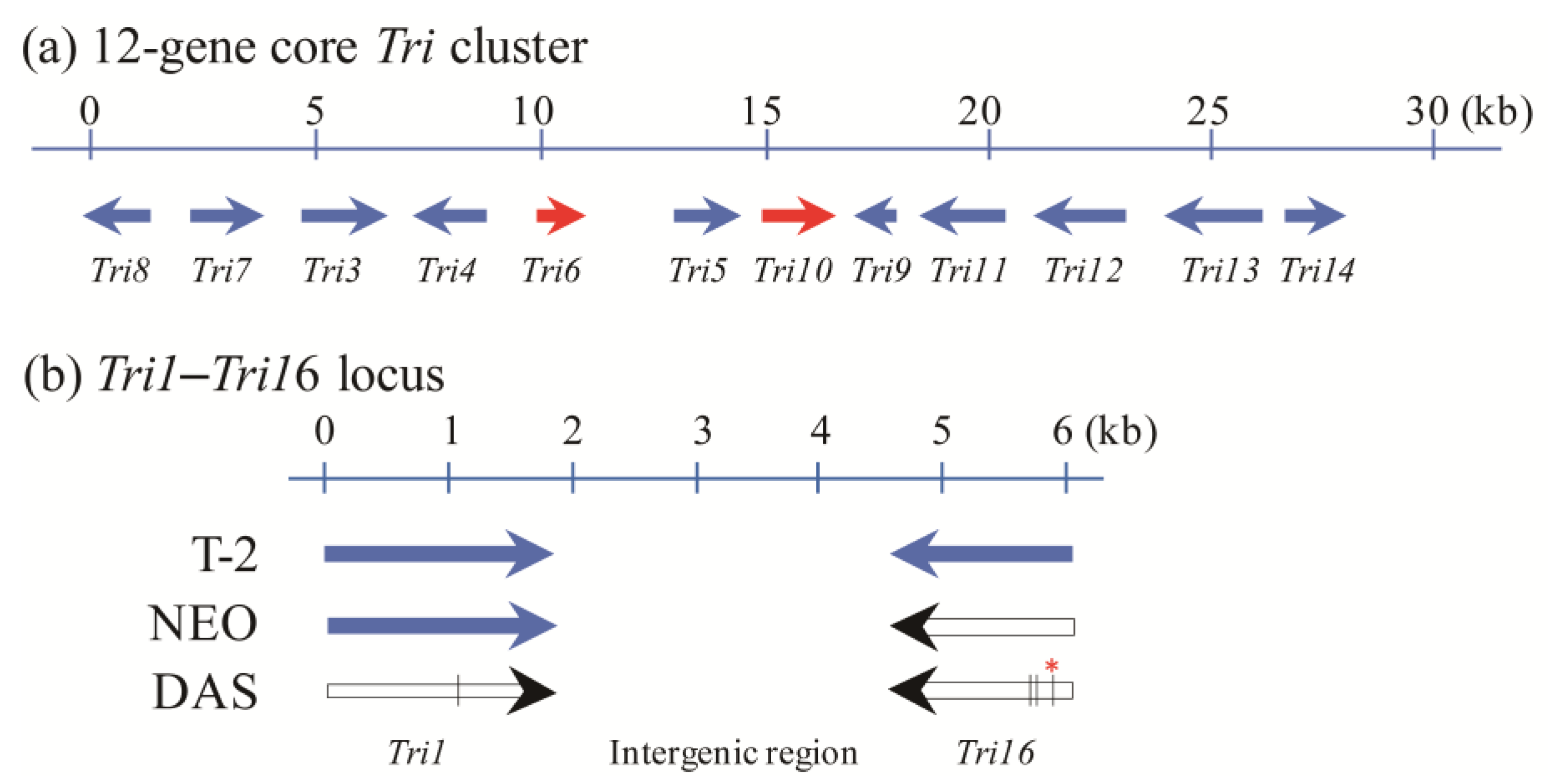
3. Tri1 and Tri16 Genes Are the Key Determinants of Trichothecene Profiles
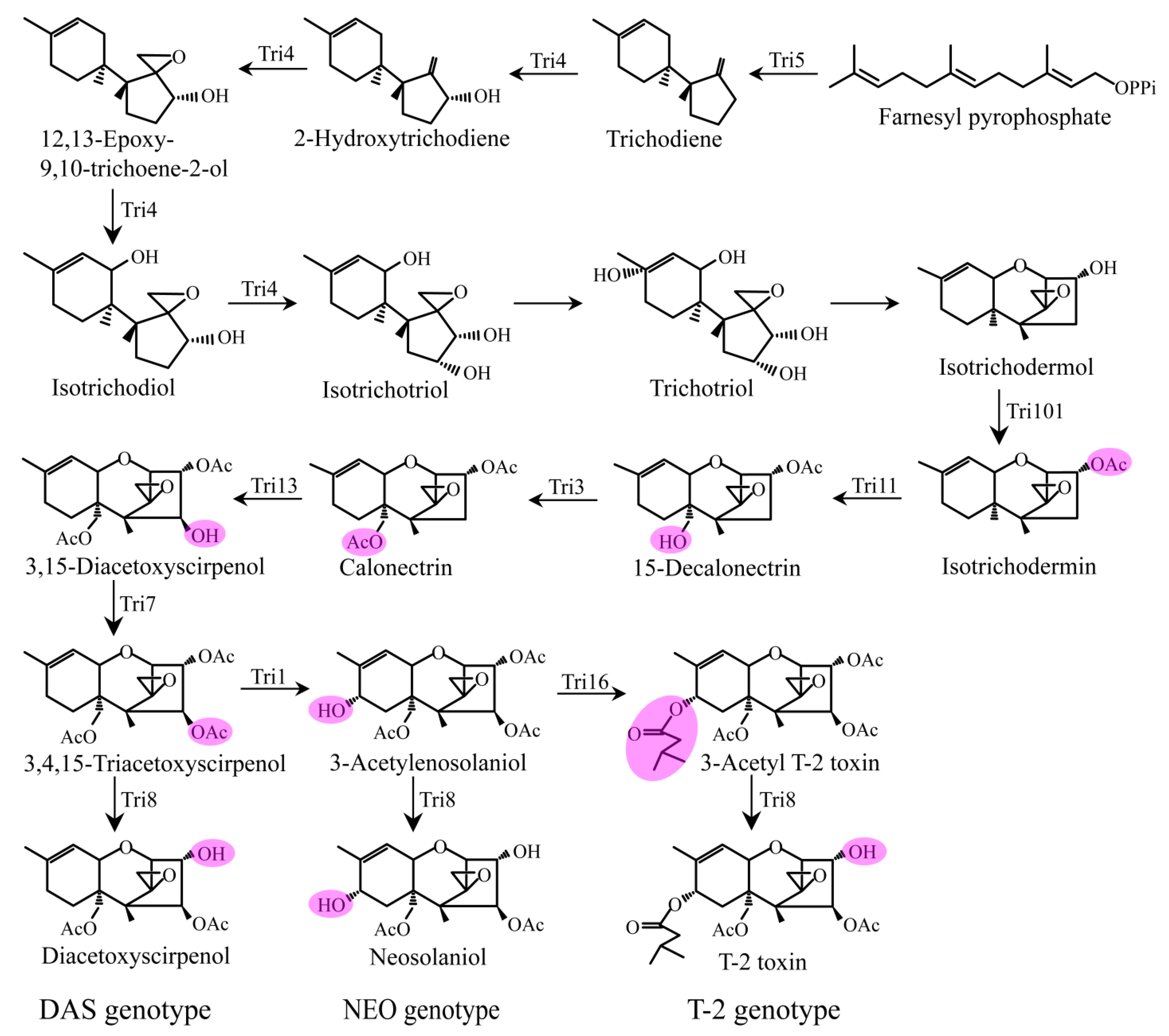
4. Proposed Biosynthetic Pathways of DAS and NEO and Comparisons with T-2
5. Evolution Potential of Type A Trichothecene Metabolic Profile Differentiation in Fusarium
6. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- D’Mello, J.P.F.; Placinta, C.M.; Macdonald, A.M.C. Fusarium mycotoxins: A review of global implications for animal health, welfare and productivity. Anim. Feed. Sci. Technol. 1999, 80, 183–205. [Google Scholar]
- Yazar, S.; Omurtag, G.Z. Fumonisins, trichothecenes and zearalenone in cereals. Int. J. Mol. Sci. 2008, 9, 2062–2090. [Google Scholar] [PubMed]
- Arunachalam, C.; Doohan, F.M. Trichothecene toxicity in eukaryotes: Cellular and molecular mechanisms in plants and animals. Mol. Toxicol. Lett. 2013, 217, 149–158. [Google Scholar]
- Wu, Q.; Wang, X.; Nepovimova, E.; Miron, A.; Liu, Q.; Wang, Y.; Su, D.; Yang, H.; Li, L.; Kuca, K. Trichothecenes: Immunomodulatory effects, mechanisms, and anticancer potential. Arch. Toxicol. 2017, 91, 3737–3785. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, Z.; Yang, X.; Yang, J.; Gong, A.; Zhang, J.; Chen, L.; Zhou, C. Fusarium graminearum species complex and trichothecene genotype. In Mycotoxins and Food Safety; Sabuncuoglu, S., Ed.; IntechOpen: London, UK, 2019. [Google Scholar]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From simple to complex mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar]
- Wu, J.; Jing, L.; Yuan, H.; Peng, S. T-2 toxin induces apoptosis in ovarian granulosa cells of rats through reactive oxygen species-mediated mitochondrial pathway. Toxicol. Lett. 2011, 202, 168–177. [Google Scholar]
- Rocha, O.; Ansari, K.; Doohan, F.M. Effects of trichothecene mycotoxins on eukaryotic cells: A review. Food Addit. Contam. 2015, 22, 369–378. [Google Scholar]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant Microbe Interact. 1995, 8, 593–601. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Proctor, R.H.; Bai, G.; McCormick, S.P.; Shaner, G.; Buechely, G.; Hohn, T.M. Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant Microbe Interact. 1996, 9, 775–781. [Google Scholar]
- Jansen, C.; Von Wettstein, D.; Schafer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Pro. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Greenhalgh, R.; Wang, Y.; Lu, M. Trichothecene chemotypes of three Fusarium species. Mycologia 1991, 83, 121–130. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Qu, B.; Zhang, J.; Huang, T.; Chen, F.; Liao, Y. Development of a generic PCR detection of 3-acetyldeoxynivalenol-, 15-acetyldeoxynivalenol- and nivalenol-chemotypes of Fusarium graminearum clade. Int. J. Mol. Sci. 2008, 9, 2495–2504. [Google Scholar] [PubMed]
- Zhang, J.; Li, H.; Dang, F.; Qu, B.; Xu, Y.; Zhao, C.; Liao, Y. Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycol. Res. 2007, 111, 967–975. [Google Scholar]
- Wang, J.H.; Ndoye, M.; Zhang, J.B.; Li, H.P.; Liao, Y.C. Population structure and genetic diversity of the Fusarium graminearum species complex. Toxins 2011, 3, 1020–1037. [Google Scholar]
- Edwards, S.G.; Imathiu, S.M.; Ray, R.V.; Back, M.; Hare, M.C. Molecular studies to identify the Fusarium species responsible for HT-2 and T-2 mycotoxins in UK oats. Int. J. Food Microbiol. 2012, 156, 168–175. [Google Scholar]
- Torp, M.; Nirenberg, H.I. Fusarium langsethiae sp. nov. on cereals in Europe. Int. J. Food Microbiol. 2004, 95, 247–256. [Google Scholar]
- Torp, M.; Langseth, W. Production of T-2 toxin by a Fusarium resembling Fusarium poae. Mycopathologia 1999, 147, 89–96. [Google Scholar] [CrossRef]
- Thrane, U.; Adler, A.; Clasen, P.; Galvano, F.; Langseth, W.; Lew, H.; Logrieco, A.; Nielsen, K.F.; Ritieni, A. Diversity in metabolite production by Fusarium langsethiae, Fusarium poae, and Fusarium sporotrichioides. Int. J. Food Microbiol. 2004, 95, 257–266. [Google Scholar]
- Covarelli, L.; Beccari, G.; Prodi, A.; Generotti, S.; Etruschi, F.; Juan, C.; Ferrer, E.; Mañes, J. Fusarium species, chemotypes characterization and trichothecene contamination of durum and soft wheat in an area of central Italy. J. Sci. Food Agr. 2015, 95, 540–551. [Google Scholar]
- Stenglein, S.A.; Dinolfo, M.I.; Barros, G.; Bongiorno, F.; Chulze, S.N.; Moreno, M.V. Fusarium poae pathogenicity and mycotoxin accumulation on selected wheat and barley genotypes at a single location in Argentina. Plant Dis. 2014, 98, 1733–1738. [Google Scholar] [CrossRef]
- Vogelgsang, S.; Sulyok, M.; Hecker, A.; Jenny, E.; Krska, R.; Schuhmacher, R.; Forrer, H.R. Toxigenicity and pathogenicity of Fusarium poae and Fusarium avenaceum on wheat. Eur. J. Plant Pathol. 2008, 122, 265–276. [Google Scholar]
- Vogelgsang, S.; Sulyok, M.; Bänziger, I.; Krska, R.; Schuhmacher, R.; Forrer, H.R. Effect of fungal strain and cereal substrate on in vitro mycotoxin production by Fusarium poae and Fusarium avenaceum. Food Addit. Contam. Part A 2008, 25, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Nichea, M.J.; Cendoya, E.; Zachetti, V.G.L.; Chiacchiera, S.M.; Sulyok, M.; Krska, R.; Torres, A.M.; Chulze, S.N.; Ramirez, M.L. Mycotoxin profile of Fusarium armeniacum isolated from natural grasses intended for cattle feed. World Mycotoxin J. 2014, 8, 451–457. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Ward, T.J.; O’Donnell, K.; Proctor, R.H.; Burkin, A.A.; Kononenko, G.P.; Gavrilova, O.P.; Aoki, T.; McCormick, S.P.; Gagkaeva, T.Y. Fusarium sibiricum sp. nov, a novel type A trichothecene-producing Fusarium from northern Asia closely related to F. sporotrichioides and F. langsethiae. Int. J. Food Microbiol. 2011, 147, 58–68. [Google Scholar]
- Elmer, W.H.; Marra, R.E. New species of Fusarium associated with dieback of Spartina alterniflora in Atlantic salt marshes. Mycologia 2011, 103, 806–819. [Google Scholar] [CrossRef]
- Laurence, M.H.; Walsh, J.L.; Shuttleworth, L.A.; Robinson, D.M.; Johansen, R.M.; Petrovic, T.; Vu, T.T.H.; Burgess, L.W.; Summerell, B.A.; Liew, E.C.Y. Six novel species of Fusarium from natural ecosystems in Australia. Fungal Divers. 2016, 77, 349–366. [Google Scholar]
- Rocha, L.O.; Laurence, M.H.; Proctor, R.H.; McCormick, S.P.; Summerell, B.A.; Liew, E.C.Y. Variation in type A trichothecene production and trichothecene biosynthetic genes in Fusarium goolgardi from natural ecosystems of Australia. Toxins 2015, 7, 4577–4594. [Google Scholar]
- Varga, E.; Wiesenberger, G.; Hametner, C.; Ward, T.J.; Dong, Y.; Schöfeck, D.; McCormick, S.; Broz, K.; Stückler, R.; Schuhmacher, R.; et al. New tricks of an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015, 17, 2588–2600. [Google Scholar] [PubMed]
- Lattanzio, V.M.T.; Ciasca, B.; Terzi, V.; Ghizzoni, R.; McCormick, S.P.; Pascale, M. Study of the natural occurrence of T-2 and HT-2 toxins and their glucosyl derivatives from field barley to malt by high-resolution Orbitrap mass spectrometry. Food Addit. Contam. Part A 2015, 32, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Kaneko, I.; Komiyama, M.; Takatsuki, A.; Koshino, H.; Yoneyama, K.; Yamaguchi, I. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. J. Biol. Chem. 1998, 273, 1654–1661. [Google Scholar] [PubMed]
- Meek, I.B.; Peplow, A.W.; Ake, C., Jr.; Phillips, T.D.; Beremand, M.N. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl. Environ. Microbiol. 2003, 69, 1607–1613. [Google Scholar]
- Brown, D.W.; Proctor, R.H.; Dyer, R.B.; Plattner, R.D. Characterization of a Fusarium 2-gene cluster involved in trichothecene C-8 modification. J. Agric. Food Chem. 2003, 51, 7936–7944. [Google Scholar] [CrossRef] [PubMed]
- Peplow, A.W.; Meek, I.B.; Wiles, M.C.; Phillips, T.D.; Beremand, M.N. Tri16 is required for esterification of position C-8 during trichothecene mycotoxin production by Fusarium sporotrichioides. Appl. Environ. Microbiol. 2003, 69, 5935–5940. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Alexander, N.J.; Desjardins, A.E. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 2009, 74, 1128–1142. [Google Scholar] [PubMed]
- Chen, L.; Yang, J.; Wang, H.; Yang, X.; Zhang, C.; Zhao, Z.; Wang, J. NX toxins: New threat posed by Fusarium graminearum species complex. Trends Food Sci. Technol. 2022, 119, 179–191. [Google Scholar]
- McCormic, S.P.; Alexander, N.J. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 2002, 68, 2959–2964. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Waalwijk, C.; van der Lee, T.; Proctor, R.H. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium graminearum. Fungal Genet. Biol. 2011, 48, 485–495. [Google Scholar]
- Brown, D.W.; McCormic, S.P.; Alexander, N.J.; Proctor, R.H.; Desjardins, A.E. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol. 2001, 32, 121–133. [Google Scholar]
- McCormic, S.P.; Hohn, T.M.; Desjardins, A.E. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 1996, 62, 353–359. [Google Scholar] [CrossRef]
- Hohn, T.M.; Desjardins, A.E.; McCormic, S.P. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 1995, 248, 95–102. [Google Scholar] [PubMed]
- Proctor, R.H.; Hohn, T.M.; McCormic, S.P.; Desjardins, A.E. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 1995, 61, 1923–1930. [Google Scholar] [PubMed]
- Hohn, T.M.; Vanmiddlesworth, F. Purification and characterization of the sesquiterpene cyclase trichodiene synthetase from Fusarium sporotrichioides. Arch. Biochem. Biophys. 1986, 251, 756–761. [Google Scholar] [PubMed]
- Tag, A.G.; Garifullina, G.F.; Peplow, A.W.; Ake, C., Jr.; Phillips, T.D.; Hohn, T.M.; Beremand, M.N. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 2001, 67, 5294–5302. [Google Scholar] [CrossRef]
- Alexander, N.J.; Hohn, T.M.; McCormic, S.P. The Tri11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 1998, 64, 221–225. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormic, S.P.; Hohn, T.M. Tri12, a trichothecene efflux pump from Fusarium sporotrichioides: Gene isolation and expression in yeast. Mol. Gen. Genet. 1999, 261, 977–984. [Google Scholar] [CrossRef]
- McCormic, S.P.; Alexander, N.J.; Trapp, S.E.; Hohn, T.M. Disruption of Tri101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl. Environ. Microbiol. 1999, 65, 5252–5256. [Google Scholar]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminerum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Gens, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Desjardins, A.E. From yellow rain to green wheat: 25 years of trichothecene biosynthesis research. J. Agr. Food Chem. 2009, 57, 4478–4484. [Google Scholar] [CrossRef]
- Foroud, N.A.; Eudes, F. Trichothecenes in cereal grains. Int. J. Mol. Sci. 2009, 10, 147–173. [Google Scholar] [PubMed]
- Proctor, R.H.; McCormick, S.P.; Kim, H.; Cardoza, R.E.; Stanley, A.M.; Lindo, L.; Kelly, A.; Brown, D.W.; Lee, T.; Vaughan, M.M.; et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog. 2018, 14, e1006946. [Google Scholar]
- Ward, T.J.; Bielawski, J.P.; Kistler, H.C.; Sullivan, E.; O’Donnell, K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 2002, 99, 9278–9283. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.; Proctor, R.H.; Belzile, F.; Chulze, S.N.; Clear, R.M.; Cowger, C.; Elmer, W.; Lee, T.; Obanor, F.; Waalwijk, C.; et al. The geographic distribution and complex evolutionary history of the NX-2 trichothecene chemotype from Fusarium graminearum. Fungal Genet. Biol. 2016, 95, 39–48. [Google Scholar] [PubMed]
| Tri Gene | Function | Mutant Phenotype |
|---|---|---|
| Tri8 | C-3 deacetylase | 3-acetyl T-2 |
| Tri7 | C-4 acetyltransferase | HT-2 |
| Tri3 | C-15 acetyltransferase | 15-decalonectrin, 3,15-didecalonectrin |
| Tri4 | multifunctional oxygenase | trichodiene |
| Tri6 | zinc finger transcription factor | low levels of trichodiene |
| Tri5 | trichodiene synthase | no trichothecenes |
| Tri10 | regulatory gene | no trichothecenes |
| Tri9 | unknown | not determined |
| Tri11 | C-15 hydroxylase | isotrichodermin |
| Tri12 | trichothecene efflux pump | no trichothecenes |
| Tri13 | C-4 hydroxylase | 8-hydroxycalonectrin, 4-deoxy T-2, 8-hydroxy-3-deacetylcalonectrin |
| Tri14 | virulence factor | T-2 |
| Tri1 | C-8 hydroxylase | DAS |
| Tri16 | C-8 acyltransferase | NEO, DAS |
| Tri101 | C-3 acetyltransferase | isotrichodermol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Zhang, M.; Yang, J.; Yang, X.; Zhang, J.; Zhao, Z. Type A Trichothecene Metabolic Profile Differentiation, Mechanisms, Biosynthetic Pathways, and Evolution in Fusarium Species—A Mini Review. Toxins 2023, 15, 446. https://doi.org/10.3390/toxins15070446
Wang J, Zhang M, Yang J, Yang X, Zhang J, Zhao Z. Type A Trichothecene Metabolic Profile Differentiation, Mechanisms, Biosynthetic Pathways, and Evolution in Fusarium Species—A Mini Review. Toxins. 2023; 15(7):446. https://doi.org/10.3390/toxins15070446
Chicago/Turabian StyleWang, Jianhua, Mengyuan Zhang, Junhua Yang, Xianli Yang, Jiahui Zhang, and Zhihui Zhao. 2023. "Type A Trichothecene Metabolic Profile Differentiation, Mechanisms, Biosynthetic Pathways, and Evolution in Fusarium Species—A Mini Review" Toxins 15, no. 7: 446. https://doi.org/10.3390/toxins15070446
APA StyleWang, J., Zhang, M., Yang, J., Yang, X., Zhang, J., & Zhao, Z. (2023). Type A Trichothecene Metabolic Profile Differentiation, Mechanisms, Biosynthetic Pathways, and Evolution in Fusarium Species—A Mini Review. Toxins, 15(7), 446. https://doi.org/10.3390/toxins15070446






