Brown Spider Venom Phospholipase-D Activity upon Different Lipid Substrates
Abstract
1. Introduction
2. Results
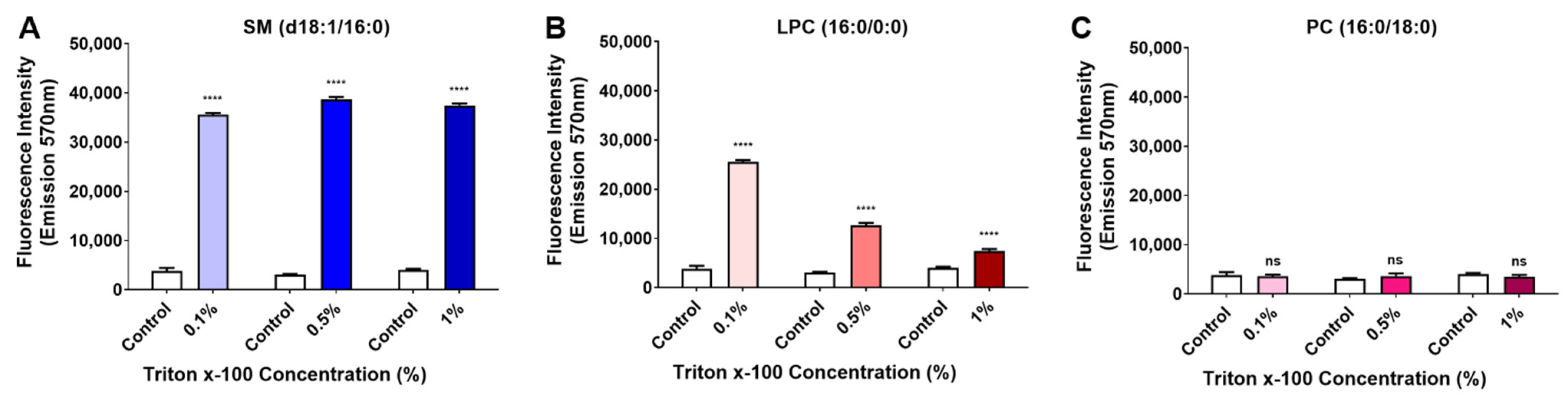
2.1. Evaluation of the Activity of Brown Spider Recombinant PLD on Different Sphingomyelins, Lysophosphatidylcholines, and Phosphatidylcholine
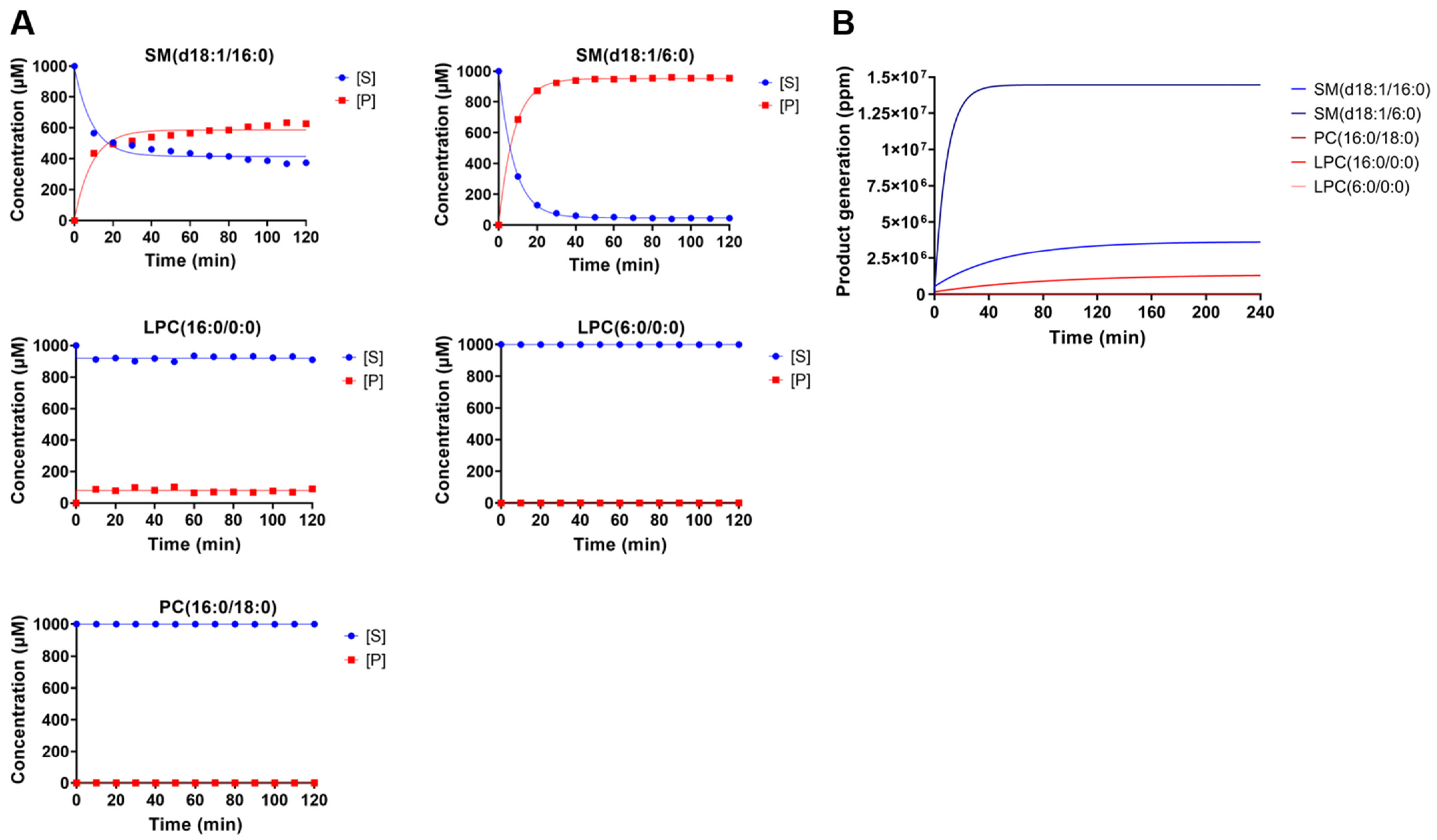
2.2. Kinetics Parameters of Degradation of Different Phospholipids by Brown Spider Recombinant PLD
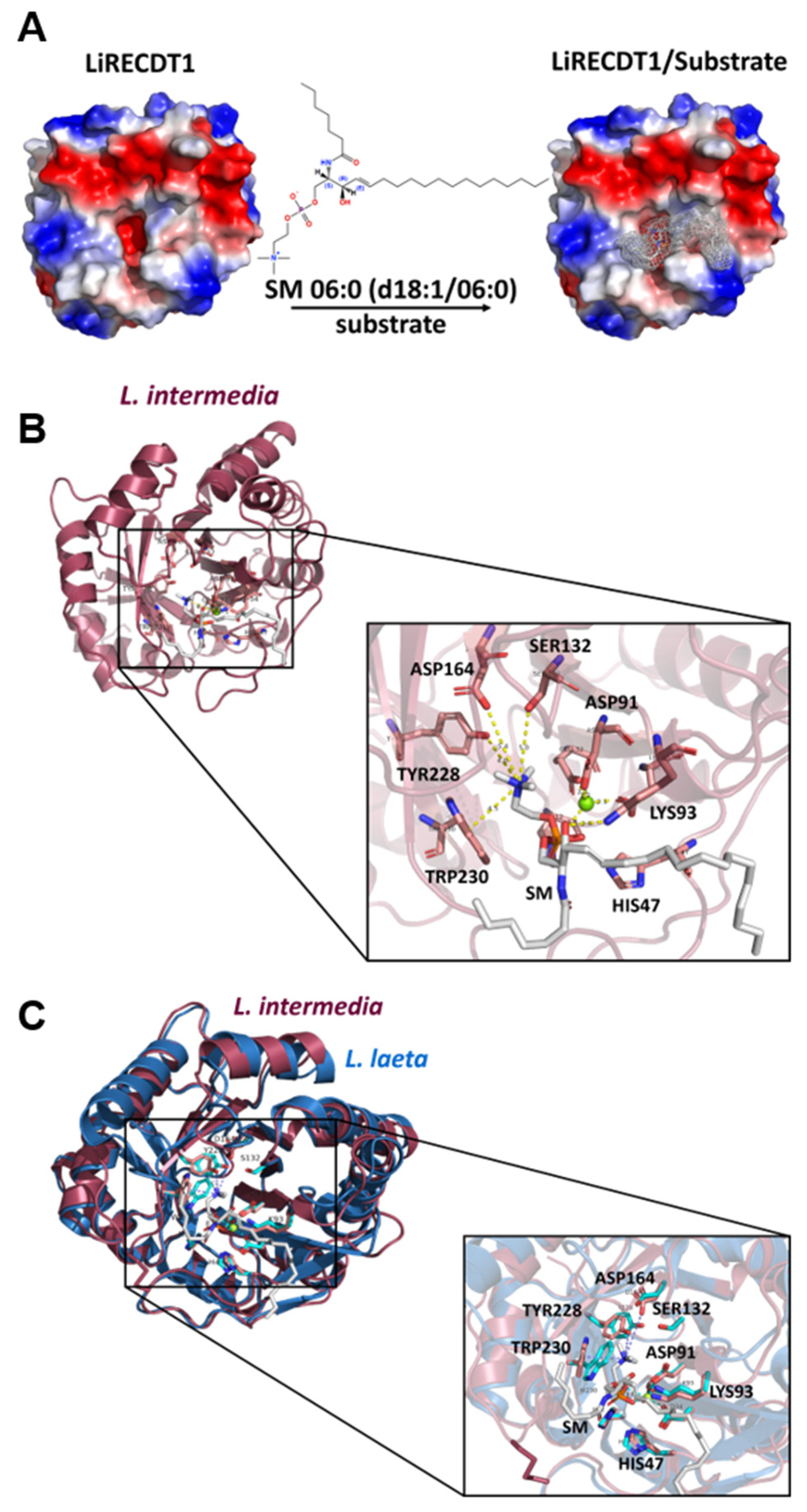
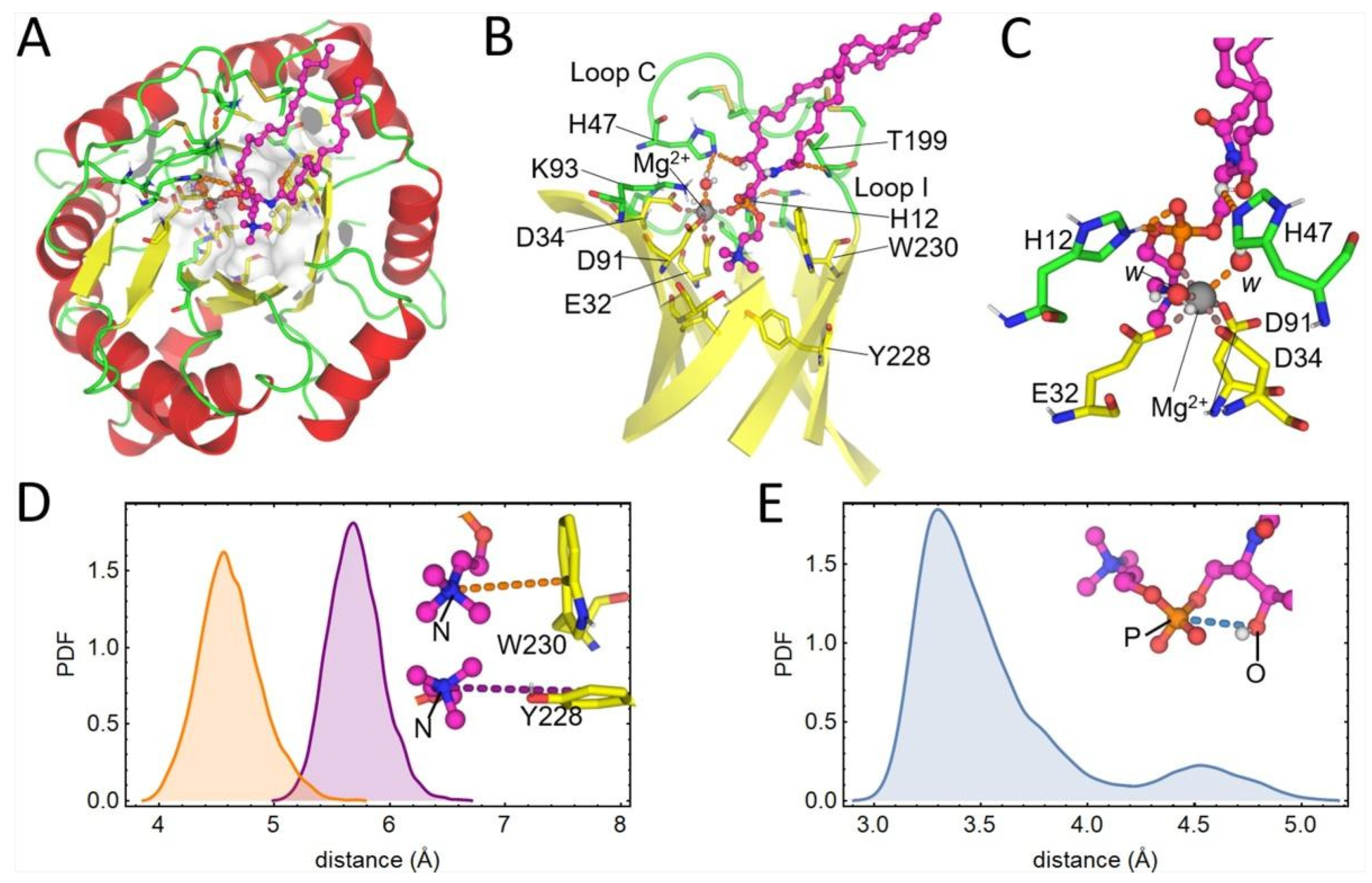
2.3. Docking Analysis and MD Simulations of the Brown Spider Recombinant PLD in the Presence of Substrates
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cloning, Expression, and Purification
4.3. PLD Activity
4.4. Nuclear Magnetic Resonance (NMR)
4.5. Molecular Docking
4.6. MD Simulations
4.7. Statistical Analysis
5. Associated Content
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Spider Catalog. Version 23.5. Available online: http://wsc.nmbe.ch (accessed on 14 September 2022).
- Vetter, R.S.; Swanson, D.L. Bites of Recluse Spiders. Available online: https://www.uptodate.com/contents/bites-of-recluse-spiders (accessed on 20 November 2022).
- Futrell, J.M. Loxoscelism. Am. J. Med. Sci. 1992, 304, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Malaque, C.M.S.; Santoro, M.L.; Cardoso, J.L.C.; Conde, M.R.; Novaes, C.T.G.; Risk, J.Y.; França, F.O.S.; de Medeiros, C.R.; Fan, H.W. Clinical Picture and Laboratorial Evaluation in Human Loxoscelism. Toxicon 2011, 58, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, P.L.M.M.; Tessarolo, L.D.; Menezes, F.H.; de Lima, T.B.; Paiva, J.H.H.G.L.; da Silva Júnior, G.B.; Martins, A.M.C.; de Francesco Daher, E. Acute Kidney Injury Due to Systemic Loxoscelism: A Cross Sectional Study in Northeast Brazil. Rev. Soc. Bras. Med. Trop. 2018, 51, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Isbister, G.K.; Fan, H.W. Spider Bite. Lancet 2011, 378, 2039–2047. [Google Scholar] [CrossRef] [PubMed]
- de Fernandes-Pedrosa, M.F.; Junqueira-de-Azevedo, I.L.M.; Gonçalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D.v. Transcriptome Analysis of Loxosceles Laeta (Araneae, Sicariidae) Spider Venomous Gland Using Expressed Sequence Tags. BMC Genomics 2008, 9, 279. [Google Scholar] [CrossRef]
- Gremski, L.H.; da Silveira, R.B.; Chaim, O.M.; Probst, C.M.A.; Ferrer, V.P.; Nowatzki, J.; Weinschutz, H.C.; Madeira, H.M.I.; Gremski, W.; Nader, H.B.; et al. A Novel Expression Profile of the Loxosceles Intermedia Spider Venomous Gland Revealed by Transcriptome Analysis. Mol. Biosyst. 2010, 6, 2403–2416. [Google Scholar] [CrossRef]
- Dantas, A.E.; Carmo, A.O.; Horta, C.C.R.; Leal, H.G.; Oliveira-Mendes, B.B.R.; Martins, A.P.V.; Chávez-Olórtegui, C.; Kalapothakis, E. Description of Loxtox Protein Family and Identification of a New Group of Phospholipases D from Loxosceles Similis Venom Gland. Toxicon 2016, 120, 97–106. [Google Scholar] [CrossRef]
- Medina-Santos, R.; Fernandes Costa, T.G.; Silva de Assis, T.C.; Kalapothakis, Y.; de Almeida Lima, S.; do Carmo, A.O.; Gonzalez-Kozlova, E.E.; Kalapothakis, E.; Chávez-Olórtegui, C.; Guerra-Duarte, C. Analysis of NGS Data from Peruvian Loxosceles Laeta Spider Venom Gland Reveals Toxin Diversity. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2022, 43, 101017. [Google Scholar] [CrossRef]
- Chaves-Moreira, D.; Senff-Ribeiro, A.; Wille, A.C.M.; Gremski, L.H.; Chaim, O.M.; Veiga, S.S. Highlights in the Knowledge of Brown Spider Toxins. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 1–12. [Google Scholar] [CrossRef]
- da Silva, M.S.; Lopes, P.H.; Elias, M.C.; Tambourgi, D.v. Cytotoxic and Genotoxic Effects on Human Keratinocytes Triggered by Sphingomyelinase D from Loxosceles Venom. Arch. Toxicol. 2020, 94, 3563–3577. [Google Scholar] [CrossRef]
- Dragulev, B.; Bao, Y.; Ramos-Cerrillo, B.; Vazquez, H.; Olvera, A.; Stock, R.; Algaron, A.; Fox, J.W. Upregulation of IL-6, IL-8, CXCL1, and CXCL2 Dominates Gene Expression in Human Fibroblast Cells Exposed to Loxosceles Reclusa Sphingomyelinase D: Insights into Spider Venom Dermonecrosis. J. Investig. Dermatol. 2007, 127, 1264–1266. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, D.M.; Roberts, S.A.; Zobel-Thropp, P.A.; Delahaye, J.L.; Bandarian, V.; Binford, G.J.; Cordes, M.H.J. Variable Substrate Preference among Phospholipase D Toxins from Sicariid Spiders. J. Biol. Chem. 2015, 290, 10994–11007. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, G.S.; Caporrino, M.C.; Della-Casa, M.S.; Kimura, L.F.; Prezotto-Neto, J.P.; Fukuda, D.A.; Portes-Junior, J.A.; Neves-Ferreira, A.G.C.; Santoro, M.L.; Barbaro, K.C. Cloning, Expression and Characterization of a Phospholipase D from Loxosceles Gaucho Venom Gland. Biochimie 2013, 95, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Catalán, A.; Cortes, W.; Sagua, H.; González, J.; Araya, J.E. Two New Phospholipase D Isoforms of Loxosceles Laeta: Cloning, Heterologous Expression, Functional Characterization, and Potential Biotechnological Application. J. Biochem. Mol. Toxicol. 2011, 25, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lynch, K.R. Brown Recluse Spider (Loxosceles Reclusa) Venom Phospholipase D (PLD) Generates Lysophosphatidic Acid (LPA). Biochem. J. 2005, 391, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, D.M.; Zobel-Thropp, P.A.; Kumirov, V.K.; Bandarian, V.; Binford, G.J.; Cordes, M.H.J. Phospholipase D Toxins of Brown Spider Venom Convert Lysophosphatidylcholine and Sphingomyelin to Cyclic Phosphates. PLoS ONE 2013, 8, e0072372. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.T.; Freitas Fernandes-Pedrosa, M.; de Andrade, S.A.; Gabdoulkhakov, A.; Betzel, C.; Tambourgi, D.V.; Arni, R.K. Structural Insights into the Catalytic Mechanism of Sphingomyelinases D and Evolutionary Relationship to Glycerophosphodiester Phosphodiesterases. Biochem. Biophys. Res. Commun. 2006, 342, 323–329. [Google Scholar] [CrossRef]
- de Giuseppe, P.O.; Ullah, A.; Silva, D.T.; Gremski, L.H.; Wille, A.C.M.; Chaves Moreira, D.; Ribeiro, A.S.; Chaim, O.M.; Murakami, M.T.; Veiga, S.S.; et al. Structure of a Novel Class II Phospholipase D: Catalytic Cleft Is Modified by a Disulphide Bridge. Biochem. Biophys. Res. Commun. 2011, 409, 622–627. [Google Scholar] [CrossRef]
- Coronado, M.A.; Ullah, A.; Silva, L.S.; Chaves-Moreira, D.; Vuitika, L.; Chaim, O.M.; Veiga, S.S.; Chahine, J.; Murakami, M.T.; Arni, R.K. Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D. Curr. Protein. Pept. Sci. 2015, 16, 768–774. [Google Scholar] [CrossRef]
- Vuitika, L.; Chaves-Moreira, D.; Caruso, I.; Lima, M.A.; Matsubara, F.H.; Murakami, M.T.; Takahashi, H.K.; Toledo, M.S.; Coronado, M.A.; Nader, H.B.; et al. Active Site Mapping of Loxosceles Phospholipases D: Biochemical and Biological Features. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 970–979. [Google Scholar] [CrossRef]
- Ramos-Cerrillo, B.; Olvera, A.; Odell, G.V.; Zamudio, F.; Paniagua-Solís, J.; Alagón, A.; Stock, R.P. Genetic and Enzymatic Characterization of Sphingomyelinase D Isoforms from the North American Fiddleback Spiders Loxosceles Boneti and Loxosceles Reclusa. Toxicon 2004, 44, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Moutoussamy, E.E.; Waheed, Q.; Binford, G.J.; Khan, H.M.; Moran, S.M.; Eitel, A.R.; Cordes, M.H.J.; Reuter, N. Specificity of Loxosceles α Clade Phospholipase D Enzymes for Choline-Containing Lipids: Role of a Conserved Aromatic Cage. PLoS Comput. Biol. 2022, 18, e1009871. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, D.M.; Cordes, M.H.J. Spider, Bacterial and Fungal Phospholipase D Toxins Make Cyclic Phosphate Products. Toxicon 2015, 108, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Birrell, G.B.; Zaikova, T.O.; Rukavishnikov, A.V.; Keana, J.F.W.; Griffith, O.H. Allosteric Interactions within Subsites of a Monomeric Enzyme: Kinetics of Fluorogenic Substrates of PI-Specific Phospholipase C. Biophys. J. 2003, 84, 3264–3275. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Goldstein, R.; Gershenson, A.; Stec, B.; Roberts, M.F. The Cation-π Box Is a Specific Phosphatidylcholine Membrane Targeting Motif. J. Biol. Chem. 2013, 288, 14863–14873. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, A.; Liu, F.; Illes, K.; Nagar, B. Crystal Structure of the Human Alkaline Sphingomyelinase Provides Insights into Substrate Recognition. J. Biol. Chem. 2017, 292, 7087–7094. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System, Version 2.5; Schrödinger LLC: New York, NY, USA, 2022.
- ben Yekhlef, R.; Felicori, L.; Santos, L.H.; Oliveira, C.F.B.; Fadhloun, R.; Torabi, E.; Shahbazzadeh, D.; Bagheri, K.P.; Ferreira, R.S.; Borchani, L. Antigenic and Substrate Preference Differences between Scorpion and Spider Dermonecrotic Toxins, a Comparative Investigation. Toxins 2020, 12, 631. [Google Scholar] [CrossRef]
- Futerman, A.H.; Hannun, Y.A. The Complex Life of Simple Sphingolipids. EMBO Rep. 2004, 5, 777–782. [Google Scholar] [CrossRef]
- Rojas, J.M.; Arán-Sekul, T.; Cortés, E.; Jaldín, R.; Ordenes, K.; Orrego, P.R.; González, J.; Araya, J.E.; Catalán, A. Phospholipase d from Loxosceles Laeta Spider Venom Induces IL-6, IL-8, CXCL1/GRO-α, and CCL2/MCP-1 Production in Human Skin Fibroblasts and Stimulates Monocytes Migration. Toxins 2017, 9, 125. [Google Scholar] [CrossRef]
- Horta, C.C.R.; Oliveira-Mendes, B.B.R.; do Carmo, A.O.; Siqueira, F.F.; Barroca, T.M.; dos Santos Nassif Lacerda, S.M.; de Almeida Campos, P.H.; de França, L.R.; Ferreira, R.L.; Kalapothakis, E. Lysophosphatidic Acid Mediates the Release of Cytokines and Chemokines by Human Fibroblasts Treated with Loxosceles Spider Venom. J. Investig. Dermatol. 2013, 133, 1682–1685. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Principles of Bioactive Lipid Signalling: Lessons from Sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Pettus, B.; Chalfant, C.; Hannun, Y. Sphingolipids in Inflammation: Roles and Implications. Curr. Mol. Med. 2005, 4, 405–418. [Google Scholar] [CrossRef] [PubMed]
- el Alwani, M.; Wu, B.X.; Obeid, L.M.; Hannun, Y.A. Bioactive Sphingolipids in the Modulation of the Inflammatory Response. Pharmacol. Ther. 2006, 112, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Bartke, N.; Hannun, Y.A. Bioactive Sphingolipids: Metabolism and Function. J. Lipid. Res. 2009, 50, 91–96. [Google Scholar] [CrossRef]
- Milhas, D.; Clarke, C.J.; Hannun, Y.A. Sphingomyelin Metabolism at the Plasma Membrane: Implications for Bioactive Sphingolipids. FEBS Lett. 2010, 584, 1887–1894. [Google Scholar] [CrossRef]
- Stock, R.P.; Brewer, J.; Wagner, K.; Ramos-Cerrillo, B.; Duelund, L.; Jernshøj, K.D.; Olsen, L.F.; Bagatolli, L.A. Sphingomyelinase D Activity in Model Membranes: Structural Effects of in Situ Generation of Ceramide-1-Phosphate. PLoS ONE 2012, 7, e36003. [Google Scholar] [CrossRef]
- Carvalho, M.; Sampaio, J.L.; Palm, W.; Brankatschk, M.; Eaton, S.; Shevchenko, A. Effects of Diet and Development on the Drosophila Lipidome. Mol. Syst. Biol. 2012, 8, 1–17. [Google Scholar] [CrossRef]
- Bednaski, A.V.; Trevisan-Silva, D.; Matsubara, F.H.; Boia-Ferreira, M.; Olivério, M.M.; Gremski, L.H.; Cavalheiro, R.P.; de Paula, D.M.B.; Paredes-Gamero, E.J.; Takahashi, H.K.; et al. Characterization of Brown Spider (Loxosceles Intermedia) Hemolymph: Cellular and Biochemical Analyses. Toxicon 2015, 98, 62–74. [Google Scholar] [CrossRef]
- Dougherty, D. A Cation-p Interactions Involving Aromatic Amino Acids. J. Nutr. 2007, 137, 1504–1508. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for Ligand-Receptor Docking. In Current Protocols in Bioinformatics; John Wiley & Sons: Hoboken, NJ, USA, 2008; pp. 1–40. ISBN 0471250953. [Google Scholar]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cisneros, G.A.; Cruzeiro, V.W.D.; et al. Amber 2022 Software, University of California: San Francisco, CA, USA, 2022.
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Dickson, C.J.; Walker, R.C.; Gould, I.R. Lipid21: Complex Lipid Membrane Simulations with AMBER. J. Chem. Theory Comput. 2022, 18, 1726–1736. [Google Scholar] [CrossRef] [PubMed]
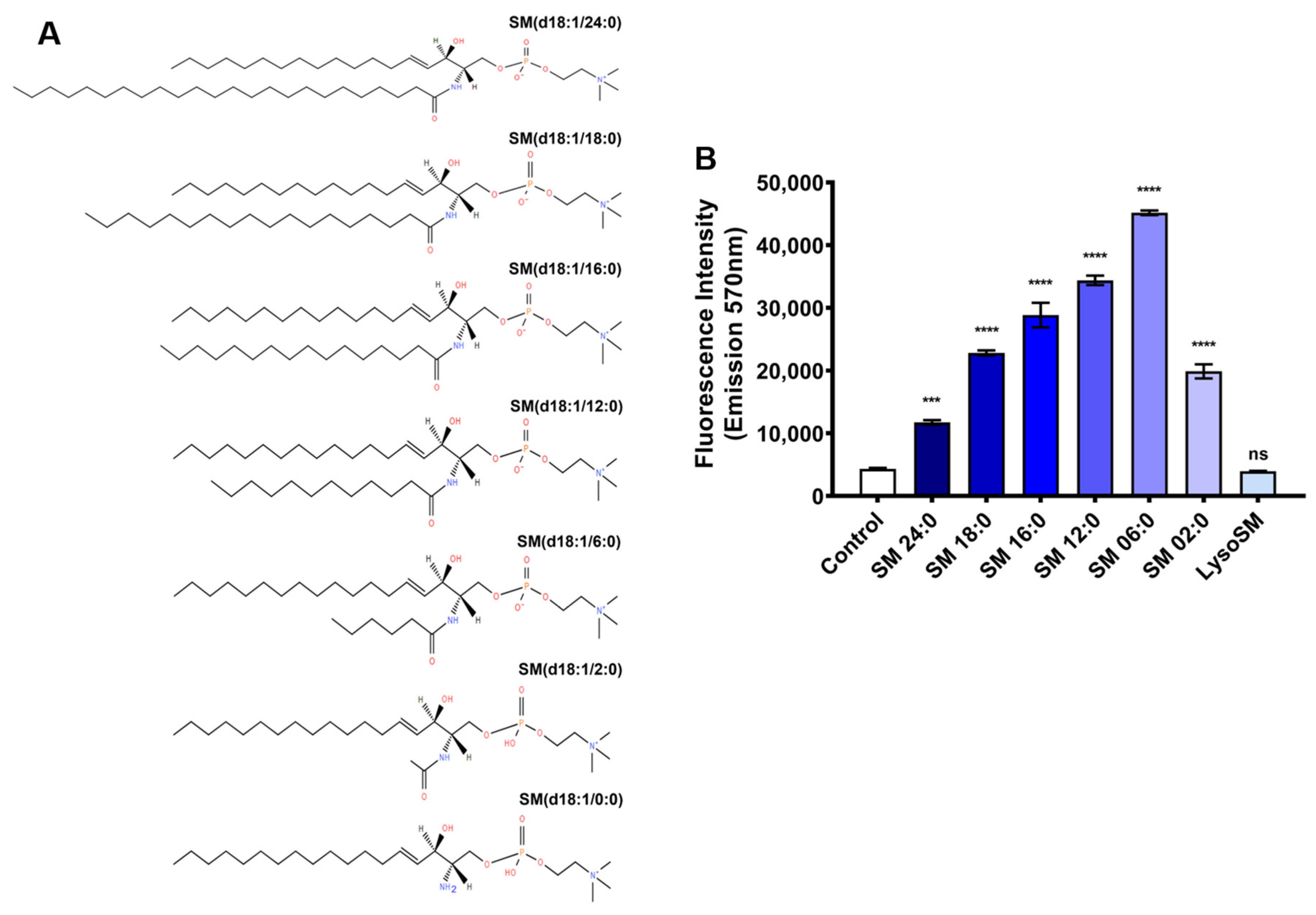

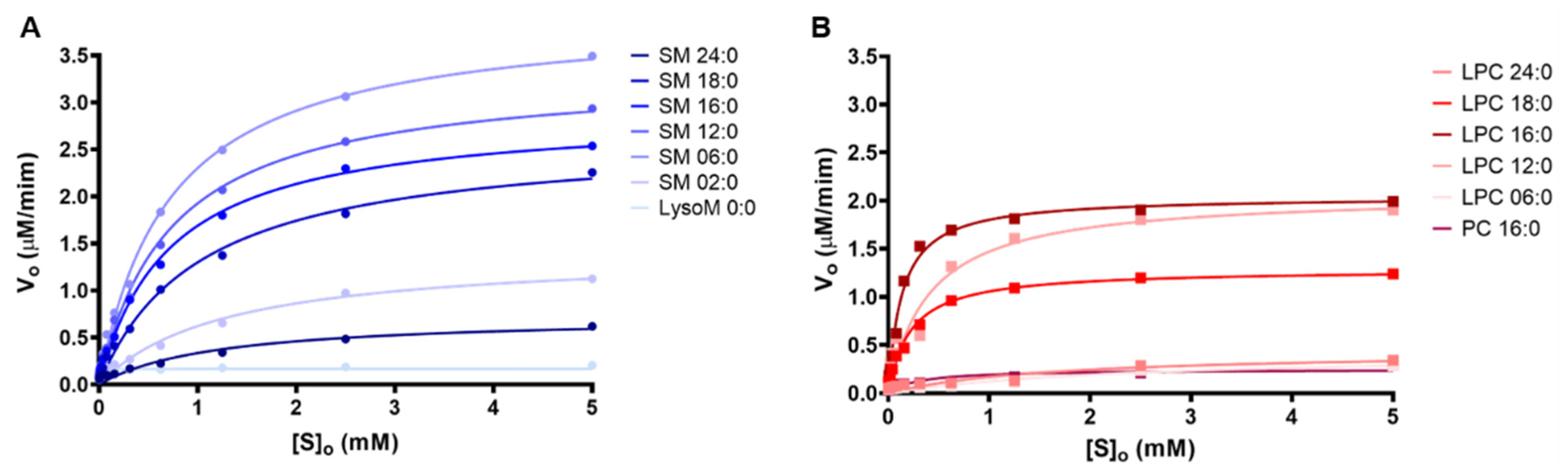
| Sphingomyelins V0 (μM.min−1) | Phosphatidylcholines V0 (μM.min−1) | ||
|---|---|---|---|
| SM 24:0 | 0.618 | PC 16:0 | 0.212 |
| SM 18:0 | 2.257 | LPC 24:0 | 0.287 |
| SM 16:0 | 2.537 | LPC 18:0 | 1.199 |
| SM 12:0 | 2.933 | LPC 16:0 | 1.906 |
| SM 06:0 | 3.493 | LPC 12:0 | 1.805 |
| SM 02:0 | 1.124 | LPC 06:0 | 0.238 |
| LSM 0:0 | 0.206 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaves-Moreira, D.; Gremski, L.H.; de Moraes, F.R.; Vuitika, L.; Wille, A.C.M.; Hernández González, J.E.; Chaim, O.M.; Senff-Ribeiro, A.; Arni, R.K.; Veiga, S.S. Brown Spider Venom Phospholipase-D Activity upon Different Lipid Substrates. Toxins 2023, 15, 109. https://doi.org/10.3390/toxins15020109
Chaves-Moreira D, Gremski LH, de Moraes FR, Vuitika L, Wille ACM, Hernández González JE, Chaim OM, Senff-Ribeiro A, Arni RK, Veiga SS. Brown Spider Venom Phospholipase-D Activity upon Different Lipid Substrates. Toxins. 2023; 15(2):109. https://doi.org/10.3390/toxins15020109
Chicago/Turabian StyleChaves-Moreira, Daniele, Luiza Helena Gremski, Fábio Rogério de Moraes, Larissa Vuitika, Ana Carolina Martins Wille, Jorge Enrique Hernández González, Olga Meiri Chaim, Andrea Senff-Ribeiro, Raghuvir Krishnaswamy Arni, and Silvio Sanches Veiga. 2023. "Brown Spider Venom Phospholipase-D Activity upon Different Lipid Substrates" Toxins 15, no. 2: 109. https://doi.org/10.3390/toxins15020109
APA StyleChaves-Moreira, D., Gremski, L. H., de Moraes, F. R., Vuitika, L., Wille, A. C. M., Hernández González, J. E., Chaim, O. M., Senff-Ribeiro, A., Arni, R. K., & Veiga, S. S. (2023). Brown Spider Venom Phospholipase-D Activity upon Different Lipid Substrates. Toxins, 15(2), 109. https://doi.org/10.3390/toxins15020109






