Venomous Noodles: The Evolution of Toxins in Nemertea through Positive Selection and Gene Duplication
Abstract
1. Introduction
2. Results and Discussion
2.1. Transcriptome Assembly
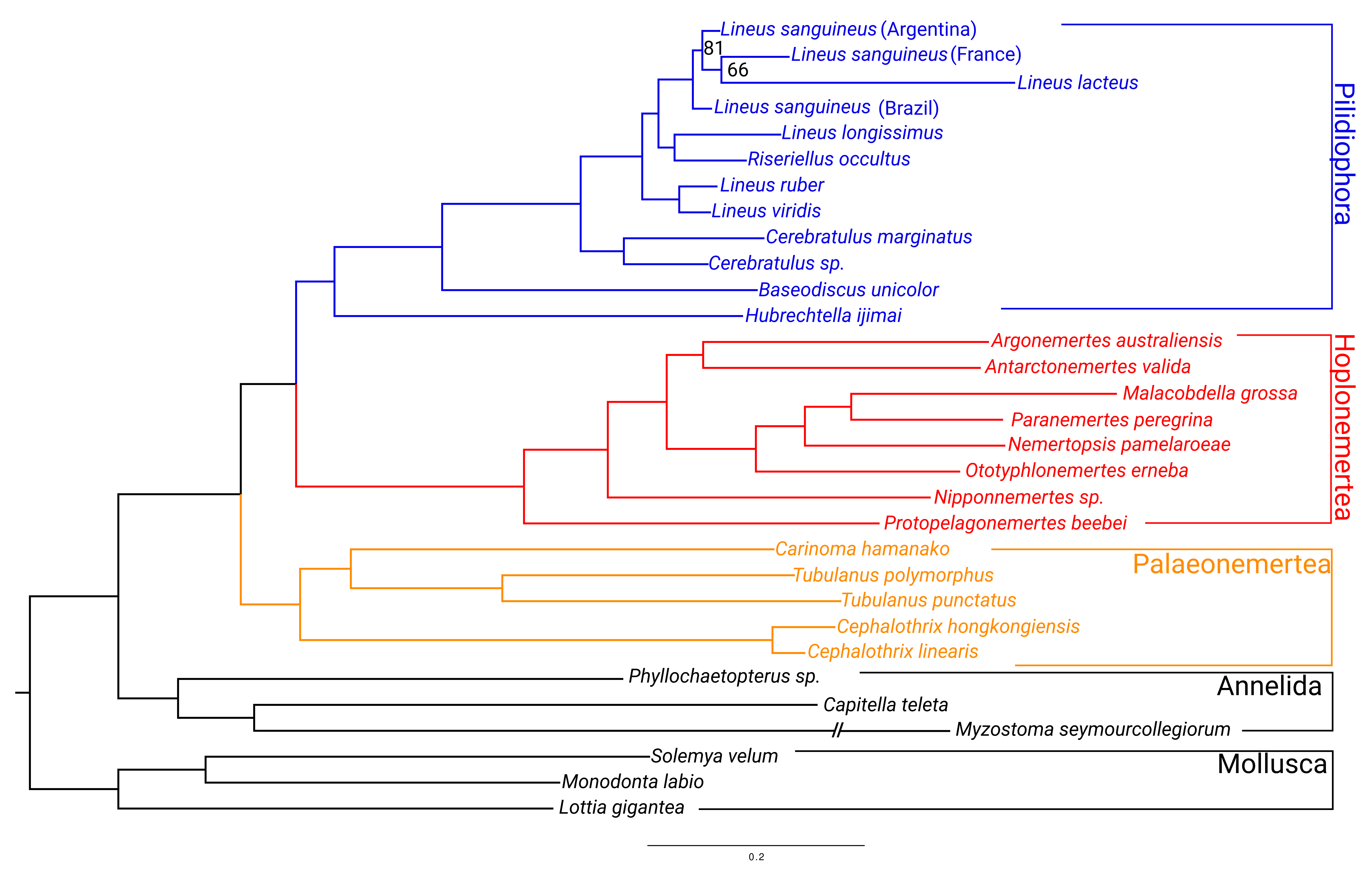
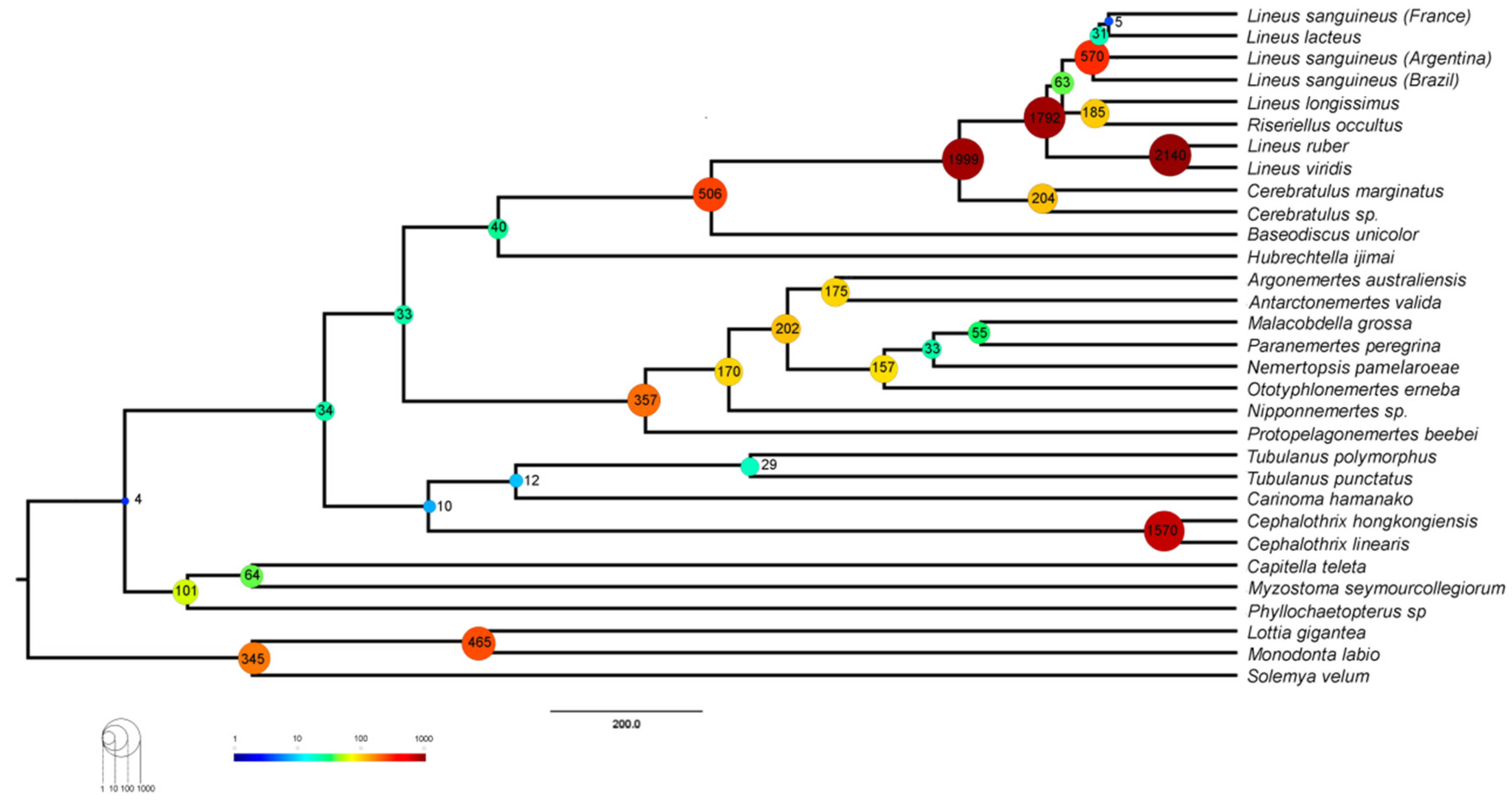
2.2. Phylogenomic Analysis
2.3. Identified Putative Toxins
2.4. Proteomic Experiments
2.5. Selected Toxin Gene Families
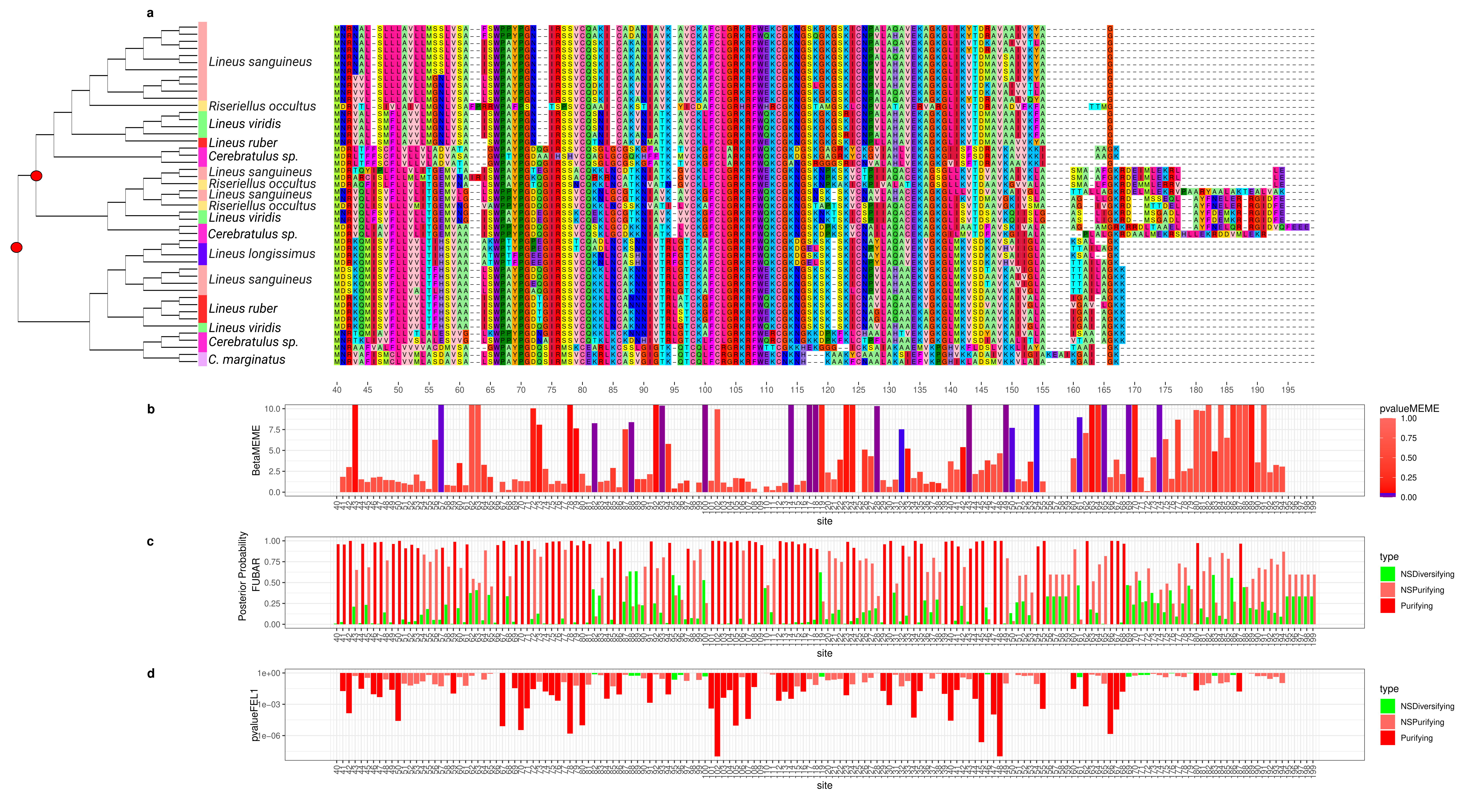
2.5.1. Cytotoxin-A
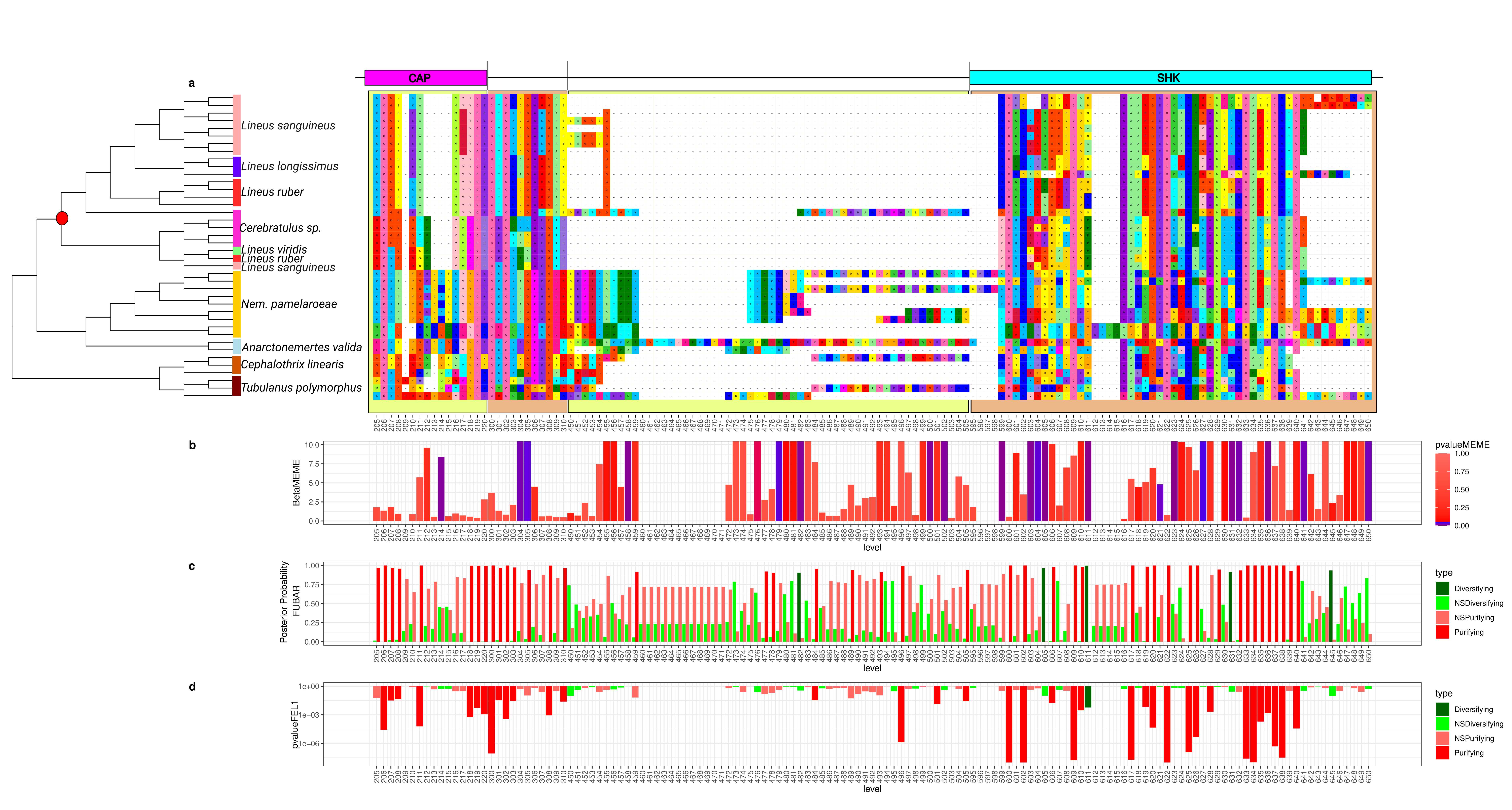
2.5.2. Scoloptoxin SD976-like Protein
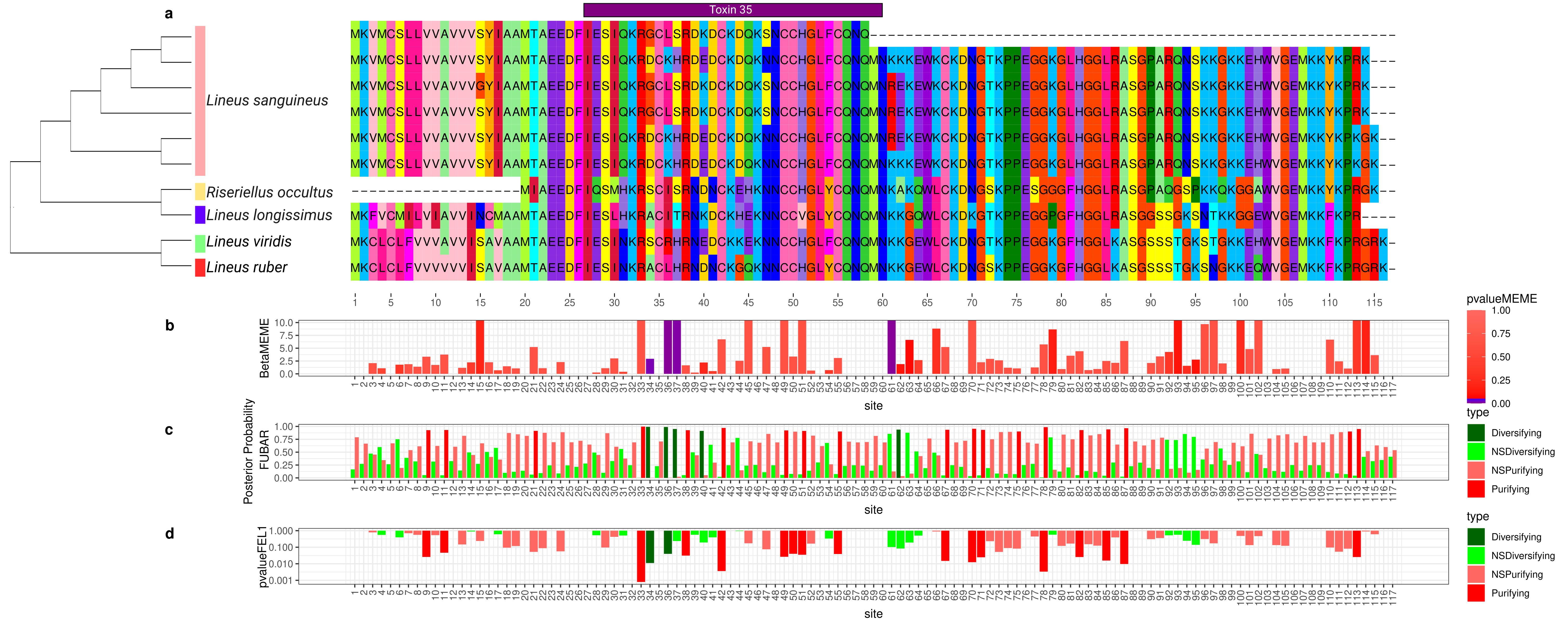
2.5.3. Alpha-Nemertide
2.5.4. Beta-Nemertide
2.5.5. The U-Nemertotoxin-3: Agelenin-like Proteins, a New Toxin?
2.5.6. Alpha-KTx-Like/Beta-Defensin-Like/Myticin-like
2.6. Selection Tests in Toxins
2.7. Gene Duplications in Nemertea
3. Conclusions
4. Materials and Methods
4.1. Sampling
4.2. RNA Extraction and Sequencing
4.3. Transcriptome Assembly and ORF Prediction
4.4. Phylogenomic Analysis
4.5. Toxin Identification
4.6. Proteomic of Mucus and Whole Individuals
4.7. Trees and Alignments for Toxin Orthogroups
4.8. Selection Test
4.9. Gene Duplications in Toxin Orthogroups
4.10. Nemertea Gene Duplication Events
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Norenburg, J.; Gibson, R.; Herrera Bachiller, A.; Strand, M. World Nemertea Database. Available online: https://www.Marinespecies.Org/Nemertea (accessed on 30 July 2023).
- Brusca, R.C.; Moore, W.; Shuster, S.M. Invertebrates, 3rd ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2016; ISBN 978-1-60535-375-3. [Google Scholar]
- Strand, M.; Norenburg, J.; Alfaya, J.E.; Fernández-Álvarez, F.Á.; Andersson, H.; Andrade, S.; Bartolomaeus, T.; Beckers, P.; Bigatti, G.; Cherneva, I.; et al. Nemertean Taxonomy Implementing Changes in the Higher Ranks, Dismissing Anopla and Enopla. Zool. Scr. 2019, 48, 118–119. [Google Scholar] [CrossRef]
- Andrade, S.C.; Montenegro, H.; Strand, M.; Schwartz, M.L.; Kajihara, H.; Norenburg, J.L.; Turbeville, J.M.; Sundberg, P.; Giribet, G. A Transcriptomic Approach to Ribbon Worm Systematics (Nemertea): Resolving the Pilidiophora Problem. Mol. Biol. Evol. 2014, 31, 3206–3215. [Google Scholar] [CrossRef] [PubMed]
- Göransson, U.; Jacobsson, E.; Strand, M.; Andersson, H.S. The Toxins of Nemertean Worms. Toxins 2019, 11, 120. [Google Scholar] [CrossRef]
- Bacq, Z.M. Les Poisons Des Nemertiens. Bull. De La Cl. Des Sci. 1936, 22, 1072–1079. [Google Scholar]
- Bacq, Z.M. L’Amphiporine et La Némertine Poisons Des Vers Némertiens. Arch. Int. De Physiol. 1937, 44, 190–204. [Google Scholar]
- Kem, W.R. A Study of the Occurrence of Anabaseine in Paranemertes and Other Nemertines. Toxicon 1971, 9, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Kem, W.R. Purification and Characterization of a New Family of Polypeptide Neurotoxins from the Heteronemertine Cerebratulus lacteus (Leidy). J. Biol. Chem. 1976, 251, 4184–4192. [Google Scholar] [CrossRef] [PubMed]
- Kem, W.R.; Blumenthal, K.M. Purification and Characterization of the Cytotoxic Cerebratulus A Toxins. J. Biol. Chem. 1978, 253, 5752–5757. [Google Scholar] [CrossRef]
- Kem, W.R. Structure and Action of Nemertine Toxins. Am. Zool. 1985, 25, 99–111. [Google Scholar] [CrossRef]
- Berne, S.; Sepčić, K.; Križaj, I.; Kem, W.R.; McClintock, J.B.; Turk, T. Isolation and Characterisation of a Cytolytic Protein from Mucus Secretions of the Antarctic Heteronemertine Parborlasia corrugatus. Toxicon 2003, 41, 483–491. [Google Scholar] [CrossRef]
- Jacobsson, E.; Andersson, H.S.; Strand, M.; Peigneur, S.; Eriksson, C.; Lodén, H.; Shariatgorji, M.; Andrén, P.E.; Lebbe, E.K.; Rosengren, K.J. Peptide Ion Channel Toxins from the Bootlace Worm, the Longest Animal on Earth. Sci. Rep. 2018, 8, 4596. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.J. Observations on the Food and Feeding Behavior of Estuarine Nemertean Worms Belonging to the Order Hoplonemertea. Biol. Bull. 1976, 150, 57–68. [Google Scholar] [CrossRef]
- Thiel, M.; Kruse, I. Status of the Nemertea as Predators in Marine Ecosystems. Hydrobiologia 2001, 456, 21–32. [Google Scholar] [CrossRef]
- Wexler, P.; Fonger, G.C.; White, J.; Weinstein, S. Toxinology: Taxonomy, Interpretation, and Information Resources. Sci. Technol. Libr. 2015, 34, 67–90. [Google Scholar] [CrossRef]
- McDermott, J.J. The Feeding Biology of Nipponnemertes pulcher (Johnston)(Hoplonemertea), with Some Ecological Implications. Ophelia 1984, 23, 1–21. [Google Scholar] [CrossRef]
- Christy, J.H.; Goshima, S.; Backwell, P.R.; Kreuter, T.J. Nemertean Predation on the Tropical Fiddler Crab Uca musica. Hydrobiologia 1997, 365, 233–239. [Google Scholar] [CrossRef]
- McDermott, J.J.; Roe, P. Food, Feeding Behavior and Feeding Ecology of Nemerteans. Am. Zool. 1985, 25, 113–125. [Google Scholar] [CrossRef]
- Stricker, S.A. The Stylet Apparatus of Monostiliferous Hoplonemerteans. Am. Zool. 1985, 25, 87–97. [Google Scholar] [CrossRef]
- Posner, P.; Kem, W.R. Cardiac Effects of Toxin A-III from the Heteronemertine Worm Cerebratulus Lacteus (Leidy). Toxicon 1978, 16, 343–349. [Google Scholar] [CrossRef]
- Kem, W.R.; Scott, K.N.; Duncan, J.H. Hoplonemertine Worms—A New Source of Pyridine Neurotoxins. Experientia 1976, 32, 684–686. [Google Scholar] [CrossRef]
- Kem, W.R. Pyridine alkaloid distribution in the hoplonemertines. Hydrobiologia 1988, 156, 145–151. [Google Scholar] [CrossRef]
- Kem, W.R.; Rocca, J.R.; Johnson, J.V.; Junoy, J. Discovery of the Nicotinic Receptor Toxin Anabaseine in a Polystiliferan Nemertean. Toxins 2023, 15, 46. [Google Scholar] [CrossRef]
- Ali, A.E.; Arakawa, O.; Noguchi, T.; Miyazawa, K.; Shida, Y.; Hashimoto, K. Tetrodotoxin and Related Substances in a Ribbon Worm Cephalothrix linearis (Nemertean). Toxicon 1990, 28, 1083–1093. [Google Scholar] [CrossRef]
- Hwang, D.; Noguchi, T. Tetrodotoxin Poisoning. In Advances in Food and Nutrition Research; Elsevier: Lincoln, NE, USA, 2007; Volume 52, pp. 141–236. ISBN 978-0-12-373711-3. [Google Scholar]
- Tanu, M.B.; Mahmud, Y.; Arakawa, O.; Takatani, T.; Kajihara, H.; Kawatsu, K.; Hamano, Y.; Asakawa, M.; Miyazawa, K.; Noguchi, T. Immunoenzymatic Visualization of Tetrodotoxin (TTX) in Cephalothrix species (Nemertea: Anopla: Palaeonemertea: Cephalotrichidae) and Planocera reticulata (Platyhelminthes: Turbellaria: Polycladida: Planoceridae). Toxicon 2004, 44, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Vlasenko, A.E.; Magarlamov, T.Y. Tetrodotoxin and Its Analogues in Cephalothrix cf. simula (Nemertea: Palaeonemertea) from the Sea of Japan (Peter the Great Gulf): Intrabody Distribution and Secretions. Toxins 2020, 12, 745. [Google Scholar] [CrossRef] [PubMed]
- Malykin, G.V.; Chernyshev, A.V.; Magarlamov, T.Y. Intrabody Tetrodotoxin Distribution and Possible Hypothesis for Its Migration in Ribbon Worms Cephalothrix c. simula (Palaeonemertea, Nemertea). Mar. Drugs 2021, 19, 494. [Google Scholar] [CrossRef]
- Beleneva, I.A.; Magarlamov, T.Y.; Kukhlevsky, A.D. Characterization, Identification, and Screening for Tetrodotoxin Production by Bacteria Associated with the Ribbon Worm (Nemertea) Cephalotrix simula (Ivata, 1952). Microbiology 2014, 83, 220–226. [Google Scholar] [CrossRef]
- Vlasenko, A.E.; Kuznetsov, V.G.; Malykin, G.V.; Pereverzeva, A.O.; Velansky, P.V.; Yakovlev, K.V.; Magarlamov, T.Y. Tetrodotoxins Secretion and Voltage-Gated Sodium Channel Adaptation in the Ribbon Worm Kulikovia alborostrata (Takakura, 1898) (Nemertea). Toxins 2021, 13, 606. [Google Scholar] [CrossRef] [PubMed]
- Whelan, N.V.; Kocot, K.M.; Santos, S.R.; Halanych, K.M. Nemertean Toxin Genes Revealed through Transcriptome Sequencing. Genome Biol. Evol. 2014, 6, 3314–3325. [Google Scholar] [CrossRef] [PubMed]
- Vlasenko, A.E.; Kuznetsov, V.G.; Magarlamov, T.Y. Investigation of Peptide Toxin Diversity in Ribbon Worms (Nemertea) Using a Transcriptomic Approach. Toxins 2022, 14, 542. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Lüddecke, T.; Timm, T.; Lochnit, G.; Vilcinskas, A.; von Döhren, J.; Nilsson, M.A. Proteo-Transcriptomic Analysis Identifies Potential Novel Toxins Secreted by the Predatory, Prey-Piercing Ribbon Worm Amphiporus lactifloreus. Mar. Drugs 2020, 18, 407. [Google Scholar] [CrossRef]
- Verdes, A.; Taboada, S.; Hamilton, B.R.; Undheim, E.A.B.; Sonoda, G.G.; Andrade, S.C.S.; Morato, E.; Marina, A.I.; Cárdenas, C.A.; Riesgo, A. Evolution, Expression Patterns, and Distribution of Novel Ribbon Worm Predatory and Defensive Toxins. Mol. Biol. Evol. 2022, 39, msac096. [Google Scholar] [CrossRef]
- von Reumont, B.; Campbell, L.; Jenner, R. Quo Vadis Venomics? A Roadmap to Neglected Venomous Invertebrates. Toxins 2014, 6, 3488–3551. [Google Scholar] [CrossRef]
- Kordiš, D.; Gubenšek, F. Adaptive Evolution of Animal Toxin Multigene Families. Gene 2000, 261, 43–52. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex Cocktails: The Evolutionary Novelty of Venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Duda, T.F.; Palumbi, S.R. Molecular Genetics of Ecological Diversification: Duplication and Rapid Evolution of Toxin Genes of the Venomous Gastropod Conus. Proc. Natl. Acad. Sci. USA 1999, 96, 6820–6823. [Google Scholar] [CrossRef] [PubMed]
- Pi, C.; Liu, J.; Peng, C.; Liu, Y.; Jiang, X.; Zhao, Y.; Tang, S.; Wang, L.; Dong, M.; Chen, S.; et al. Diversity and Evolution of Conotoxins Based on Gene Expression Profiling of Conus litteratus. Genomics 2006, 88, 809–819. [Google Scholar] [CrossRef]
- Duda, T.F., Jr.; Remigio, E.A. Variation and Evolution of Toxin Gene Expression Patterns of Six Closely Related Venomous Marine Snails. Mol. Ecol. 2008, 17, 3018–3032. [Google Scholar] [CrossRef]
- Chang, D.; Duda, T.F., Jr. Extensive and Continuous Duplication Facilitates Rapid Evolution and Diversification of Gene Families. Mol. Biol. Evol. 2012, 29, 2019–2029. [Google Scholar] [CrossRef] [PubMed]
- Binford, G.J.; Bodner, M.R.; Cordes, M.H.; Baldwin, K.L.; Rynerson, M.R.; Burns, S.N.; Zobel-Thropp, P.A. Molecular Evolution, Functional Variation, and Proposed Nomenclature of the Gene Family That Includes Sphingomyelinase D in Sicariid Spider Venoms. Mol. Biol. Evol. 2009, 26, 547–566. [Google Scholar] [CrossRef]
- Fry, B.G.; Wüster, W.; Kini, R.M.; Brusic, V.; Khan, A.; Venkataraman, D.; Rooney, A.P. Molecular Evolution and Phylogeny of Elapid Snake Venom Three-Finger Toxins. J. Mol. Evol. 2003, 57, 110–129. [Google Scholar] [CrossRef]
- Juarez, P.; Comas, I.; Gonzalez-Candelas, F.; Calvete, J.J. Evolution of Snake Venom Disintegrins by Positive Darwinian Selection. Mol. Biol. Evol. 2008, 25, 2391–2407. [Google Scholar] [CrossRef] [PubMed]
- Bayona-Serrano, J.D.; Viala, V.L.; Rautsaw, R.M.; Schramer, T.D.; Barros-Carvalho, G.A.; Nishiyama, M.Y., Jr.; Freitas-de-Sousa, L.A.; Moura-da-Silva, A.M.; Parkinson, C.L.; Grazziotin, F.G. Replacement and Parallel Simplification of Nonhomologous Proteinases Maintain Venom Phenotypes in Rear-Fanged Snakes. Mol. Biol. Evol. 2020, 37, 3563–3575. [Google Scholar] [CrossRef]
- Giorgianni, M.W.; Dowell, N.L.; Griffin, S.; Kassner, V.A.; Selegue, J.E.; Carroll, S.B. The Origin and Diversification of a Novel Protein Family in Venomous Snakes. Proc. Natl. Acad. Sci. USA 2020, 117, 10911–10920. [Google Scholar] [CrossRef]
- Ma, Y.; He, Y.; Zhao, R.; Wu, Y.; Li, W.; Cao, Z. Extreme Diversity of Scorpion Venom Peptides and Proteins Revealed by Transcriptomic Analysis: Implication for Proteome Evolution of Scorpion Venom Arsenal. J. Proteom. 2012, 75, 1563–1576. [Google Scholar] [CrossRef]
- Zhu, S.; Peigneur, S.; Gao, B.; Lu, X.; Cao, C.; Tytgat, J. Evolutionary Diversification of Mesobuthus α-Scorpion Toxins Affecting Sodium Channels. Mol. Cell. Proteom. 2012, 11, M111.012054. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Undheim, E.; Chan, A.; Koludarov, I.; Muñoz-Gómez, S.; Antunes, A.; Fry, B. Evolution Stings: The Origin and Diversification of Scorpion Toxin Peptide Scaffolds. Toxins 2013, 5, 2456–2487. [Google Scholar] [CrossRef]
- Cousin, X.; Bon, S.; Massoulié, J.; Bon, C. Identification of a Novel Type of Alternatively Spliced Exon from the Acetylcholinesterase Gene of Bungarus fasciatus: Molecular Forms of Acetylcholinesterase in the Snake Liver and Muscle. J. Biol. Chem. 1998, 273, 9812–9820. [Google Scholar] [CrossRef]
- Fry, B.G. From Genome to “Venome”: Molecular Origin and Evolution of the Snake Venom Proteome Inferred from Phylogenetic Analysis of Toxin Sequences and Related Body Proteins. Genome Res. 2005, 15, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, B.; Zhu, S. Target-Driven Evolution of Scorpion Toxins. Sci. Rep. 2015, 5, 14973. [Google Scholar] [CrossRef]
- Nakashima, K.; Ogawa, T.; Oda, N.; Hattori, M.; Sakaki, Y.; Kihara, H.; Ohno, M. Accelerated Evolution of Trimeresurus flavoviridis Venom Gland Phospholipase A2 Isozymes. Proc. Natl. Acad. Sci. USA 1993, 90, 5964–5968. [Google Scholar] [CrossRef] [PubMed]
- Van Valen, L. The Red Queen. Am. Nat. 1977, 111, 809–810. [Google Scholar] [CrossRef]
- Han, M.V.; Demuth, J.P.; McGrath, C.L.; Casola, C.; Hahn, M.W. Adaptive Evolution of Young Gene Duplicates in Mammals. Genome Res. 2009, 19, 859–867. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Rousselle, M.; Simion, P.; Tilak, M.-K.; Figuet, E.; Nabholz, B.; Galtier, N. Is Adaptation Limited by Mutation? A Timescale-Dependent Effect of Genetic Diversity on the Adaptive Substitution Rate in Animals. PLoS Genet. 2020, 16, e1008668. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving Fundamental Biases in Whole Genome Comparisons Dramatically Improves Orthogroup Inference Accuracy. Genome Biol. 2015, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Andrade, S.C.; Strand, M.; Schwartz, M.; Chen, H.; Kajihara, H.; von Doehren, J.; Sun, S.; Junoy, J.; Thiel, M.; Norenburg, J.L. Disentangling Ribbon Worm Relationships: Multi-locus Analysis Supports Traditional Classification of the Phylum Nemertea. Cladistics 2012, 28, 141–159. [Google Scholar] [CrossRef]
- Kvist, S.; Laumer, C.E.; Junoy, J.; Giribet, G. New Insights into the Phylogeny, Systematics and DNA Barcoding of Nemertea. Invert. Syst. 2014, 28, 287–308. [Google Scholar] [CrossRef]
- De Figueiredo, S.G.; De Lima, M.E.; Cordeiro, M.N.; Diniz, C.R.; Patten, D.; Halliwell, R.F.; Gilroy, J.; Richardson, M. Purification and Amino Acid Sequence of a Highly Insecticidal Toxin from the Venom of the Brazilian Spider Phoneutria nigriventer Which Inhibits NMDA-Evoked Currents in Rat Hippocampal Neurones. Toxicon 2001, 39, 309–317. [Google Scholar] [CrossRef]
- Gorokhova, S.; Bibert, S.; Geering, K.; Heintz, N. A Novel Family of Transmembrane Proteins Interacting with β Subunits of the Na, K-ATPase. Hum. Mol. Genet. 2007, 16, 2394–2410. [Google Scholar] [CrossRef]
- Corrêa, D.D. Ototyphlonemertes from the Brazilian Coast. Commun. Zool. Mus. Hist. Nat. Mont. 1948, 2, 1–12. [Google Scholar]
- Correa, D.D. Ecological Study of Brazilian Ototyphlonemertes. Commun. Zool. Mus. Hist. Nat. Mont. 1949, 3, 1–7. [Google Scholar]
- Envall, M.; Norenburg, J.L. Morphology and Systematics in Mesopsammic Nemerteans of the Genus Ototyphlonemertes (Nemertea, Hoplonemertea, Ototyphlonemertidae). Hydrobiologia 2001, 456, 145–163. [Google Scholar] [CrossRef]
- Kazandjian, T.D.; Petras, D.; Robinson, S.D.; van Thiel, J.; Greene, H.W.; Arbuckle, K.; Barlow, A.; Carter, D.A.; Wouters, R.M.; Whiteley, G. Convergent Evolution of Pain-Inducing Defensive Venom Components in Spitting Cobras. Science 2021, 371, 386–390. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J. Evolution of Separate Predation-and Defence-Evoked Venoms in Carnivorous Cone Snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef]
- Dunwiddie, C.; Thornberry, N.A.; Bull, H.G.; Sardana, M.; Friedman, P.A.; Jacobs, J.W.; Simpson, E. Antistasin, a Leech-Derived Inhibitor of Factor Xa: Kinetic Analysis of Enzyme Inhibition and Identification of the Reactive Site. J. Biol. Chem. 1989, 264, 16694–16699. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Iwama, R.E.; Tessler, M.; Siddall, M.E.; Kvist, S. The Origin and Evolution of Antistasin-like Proteins in Leeches (Hirudinida, Clitellata). Genome Biol. Evol. 2021, 13, evaa242. [Google Scholar] [CrossRef] [PubMed]
- Mourão, C.; Schwartz, E. Protease Inhibitors from Marine Venomous Animals and Their Counterparts in Terrestrial Venomous Animals. Mar. Drugs 2013, 11, 2069–2112. [Google Scholar] [CrossRef]
- Barzkar, N.; Khan, Z.; Jahromi, S.T.; Pourmozaffar, S.; Gozari, M.; Nahavandi, R. A Critical Review on Marine Serine Protease and Its Inhibitors: A New Wave of Drugs? Int. J. Biol. Macromol. 2021, 170, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-C.; Zhang, R.; Zhao, F.; Chen, Z.-M.; Liu, H.-W.; Wang, Y.-J.; Jiang, P.; Zhang, Y.; Wu, Y.; Ding, J.-P.; et al. Venomic and Transcriptomic Analysis of Centipede Scolopendra subspinipes dehaani. J. Proteome Res. 2012, 11, 6197–6212. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, O.; Sotolongo, V.; Amor, A.M.; Stöcklin, R.; Anderson, A.J.; Harvey, A.L.; Engström, Å.; Wernstedt, C.; Karlsson, E. Characterization of a Potassium Channel Toxin from the Caribbean Sea Anemone Stichodactyla helianthus. Toxicon 1995, 33, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Möhrlen, F.; Hutter, H.; Zwilling, R. The Astacin Protein Family in Caenorhabditis elegans: Astacin Protein Family in C. elegans. Eur. J. Biochem. 2003, 270, 4909–4920. [Google Scholar] [CrossRef]
- Rangaraju, S.; Khoo, K.K.; Feng, Z.-P.; Crossley, G.; Nugent, D.; Khaytin, I.; Chi, V.; Pham, C.; Calabresi, P.; Pennington, M.W.; et al. Potassium Channel Modulation by a Toxin Domain in Matrix Metalloprotease. J. Biol. Chem. 2010, 285, 9124–9136. [Google Scholar] [CrossRef]
- Jacobsson, E.; Peigneur, S.; Andersson, H.S.; Laborde, Q.; Strand, M.; Tytgat, J.; Goransson, U. Functional Characterization of the Nemertide α Family of Peptide Toxins. J. Nat. Prod. 2021, 84, 2121–2128. [Google Scholar] [CrossRef]
- Gao, B.; Harvey, P.J.; Craik, D.J.; Ronjat, M.; De Waard, M.; Zhu, S. Functional Evolution of Scorpion Venom Peptides with an Inhibitor Cystine Knot Fold. Biosci. Rep. 2013, 33, 513–527. [Google Scholar] [CrossRef]
- Lavergne, V.; Harliwong, I.; Jones, A.; Miller, D.; Taft, R.J.; Alewood, P.F. Optimized Deep-Targeted Proteotranscriptomic Profiling Reveals Unexplored Conus Toxin Diversity and Novel Cysteine Frameworks. Proc. Natl. Acad. Sci. USA 2015, 112, E3782–E3791. [Google Scholar] [CrossRef]
- Cardoso, F.C.; Dekan, Z.; Smith, J.J.; Deuis, J.R.; Vetter, I.; Herzig, V.; Alewood, P.F.; King, G.F.; Lewis, R.J. Modulatory Features of the Novel Spider Toxin μ-TRTX-Df1a Isolated from the Venom of the Spider Davus fasciatus. Br. J. Pharmacol. 2017, 174, 2528–2544. [Google Scholar] [CrossRef]
- Inui, T.; Hagiwara, K.; Nakajima, K.; Kimura, T.; Nakajima, T.; Sakakibara, S. Synthesis and Disulfide Structure Determination of Agelenin: Identification of the Carboxy-Terminus as an Amide Form. Pept. Res. 1992, 5, 140–144. [Google Scholar]
- Yamaji, N.; Sugase, K.; Nakajima, T.; Miki, T.; Wakamori, M.; Mori, Y.; Iwashita, T. Solution Structure of Agelenin, an Insecticidal Peptide Isolated from the Spider Agelena Opulenta, and Its Structural Similarities to Insect-Specific Calcium Channel Inhibitors. FEBS Lett. 2007, 581, 3789–3794. [Google Scholar] [CrossRef]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A Rational Nomenclature for Naming Peptide Toxins from Spiders and Other Venomous Animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Mitta, G.; Hubert, F.; Noel, T.; Roch, P. Myticin, a Novel Cysteine-Rich Antimicrobial Peptide Isolated from Haemocytes and Plasma of the Mussel Mytilus galloprovincialis. Eur. J. Biochem. 1999, 265, 71–78. [Google Scholar] [CrossRef]
- Zhang, N.; Li, M.; Chen, X.; Wang, Y.; Wu, G.; Hu, G.; Wu, H. Solution Structure of BmKK2, a New Potassium Channel Blocker from the Venom of Chinese Scorpion Buthus martensi Karsch. Proteins 2004, 55, 835–845. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, A.; Veldhuizen, E.J.A.; Haagsman, H.P. Avian Defensins. Vet. Immunol. Immunopathol. 2008, 124, 1–18. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S. The Toxicogenomic Multiverse: Convergent Recruitment of Proteins into Animal Venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Poon, A.F.Y.; Velazquez, R.; Weaver, S.; Hepler, N.L.; Murrell, B.; Shank, S.D.; Magalis, B.R.; Bouvier, D.; Nekrutenko, A.; et al. HyPhy 2.5—A Customizable Platform for Evolutionary Hypothesis Testing Using Phylogenies. Mol. Biol. Evol. 2020, 37, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Wray, K.P.; Lemmon, A.R.; Lemmon, E.M.; Caudle, S.B. A High-Throughput Venom-Gland Transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and Evidence for Pervasive Positive Selection across Toxin Classes. Toxicon 2011, 57, 657–671. [Google Scholar] [CrossRef]
- Zhu, L.; Peigneur, S.; Gao, B.; Zhang, S.; Tytgat, J.; Zhu, S. Target-Driven Positive Selection at Hot Spots of Scorpion Toxins Uncovers Their Potential in Design of Insecticides. Mol. Biol. Evol. 2016, 33, 1907–1920. [Google Scholar] [CrossRef]
- Modahl, C.M.; Mrinalini; Frietze, S.; Mackessy, S.P. Adaptive Evolution of Distinct Prey-Specific Toxin Genes in Rear-Fanged Snake Venom. Proc. R. Soc. B 2018, 285, 20181003. [Google Scholar] [CrossRef]
- Grueber, C.E.; Wallis, G.P.; Jamieson, I.G. Episodic Positive Selection in the Evolution of Avian Toll-Like Receptor Innate Immunity Genes. PLoS ONE 2014, 9, e89632. [Google Scholar] [CrossRef] [PubMed]
- Jouiaei, M.; Sunagar, K.; Federman Gross, A.; Scheib, H.; Alewood, P.F.; Moran, Y.; Fry, B.G. Evolution of an Ancient Venom: Recognition of a Novel Family of Cnidarian Toxins and the Common Evolutionary Origin of Sodium and Potassium Neurotoxins in Sea Anemone. Mol. Biol. Evol. 2015, 32, 1598–1610. [Google Scholar] [CrossRef]
- Bhere, K.V.; Haney, R.A.; Ayoub, N.A.; Garb, J.E. Gene Structure, Regulatory Control, and Evolution of Black Widow Venom Latrotoxins. FEBS Lett. 2014, 588, 3891–3897. [Google Scholar] [CrossRef]
- Dowell, N.L.; Giorgianni, M.W.; Kassner, V.A.; Selegue, J.E.; Sanchez, E.E.; Carroll, S.B. The Deep Origin and Recent Loss of Venom Toxin Genes in Rattlesnakes. Curr. Biol. 2016, 26, 2434–2445. [Google Scholar] [CrossRef] [PubMed]
- Schield, D.R.; Card, D.C.; Hales, N.R.; Perry, B.W.; Pasquesi, G.M.; Blackmon, H.; Adams, R.H.; Corbin, A.B.; Smith, C.F.; Ramesh, B.; et al. The Origins and Evolution of Chromosomes, Dosage Compensation, and Mechanisms Underlying Venom Regulation in Snakes. Genome Res. 2019, 29, 590–601. [Google Scholar] [CrossRef]
- Suryamohan, K.; Krishnankutty, S.P.; Guillory, J.; Jevit, M.; Schröder, M.S.; Wu, M.; Kuriakose, B.; Mathew, O.K.; Perumal, R.C.; Koludarov, I.; et al. The Indian Cobra Reference Genome and Transcriptome Enables Comprehensive Identification of Venom Toxins. Nat. Genet. 2020, 52, 106–117. [Google Scholar] [CrossRef]
- Almeida, D.D.; Viala, V.L.; Nachtigall, P.G.; Broe, M.; Gibbs, H.L.; de Toledo Serrano, S.M.; Moura-da-Silva, A.M.; Ho, P.L.; Nishiyama, M.Y., Jr.; Junqueira-de-Azevedo, I.L. Tracking the Recruitment and Evolution of Snake Toxins Using the Evolutionary Context Provided by the Bothrops jararaca Genome. Proc. Natl. Acad. Sci. USA 2021, 118, e2015159118. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.-J.; Kanda, M.; Koyanagi, R.; Hisata, K.; Akiyama, T.; Sakamoto, H.; Sakamoto, T.; Satoh, N. Nemertean and Phoronid Genomes Reveal Lophotrochozoan Evolution and the Origin of Bilaterian Heads. Nat. Ecol. Evol. 2017, 2, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. Pseudoallelism and Gene Evolution. Cold Spring Harb. Symp. Quant. Biol. 1951, 16, 159–174. [Google Scholar] [CrossRef]
- Qian, W.; Zhang, J. Genomic Evidence for Adaptation by Gene Duplication. Genome Res. 2014, 24, 1356–1362. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yang, H. The Multiple Fates of Gene Duplications: Deletion, Hypofunctionalization, Subfunctionalization, Neofunctionalization, Dosage Balance Constraints, and Neutral Variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef]
- Casewell, N.R.; Jackson, T.N.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef]
- Caplins, S.A.; Norenburg, J.L.; Turbeville, J.M. Molecular and Morphological Variation in the Barnacle Predator Nemertopsis bivitatta (Nemertea, Hoplonemertea). Integr. Comp. Biol. 2012, 52, E24. [Google Scholar]
- Walker, A.A. The Evolutionary Dynamics of Venom Toxins Made by Insects and Other Animals. Biochem. Soc. Trans. 2020, 48, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.-L.; Arbuckle, K. Coevolution of Snake Venom Toxic Activities and Diet: Evidence That Ecological Generalism Favours Toxicological Diversity. Toxins 2019, 11, 711. [Google Scholar] [CrossRef]
- Lyons, K.; Dugon, M.M.; Healy, K. Diet Breadth Mediates the Prey Specificity of Venom Potency in Snakes. Toxins 2020, 12, 74. [Google Scholar] [CrossRef]
- Holding, M.L.; Strickland, J.L.; Rautsaw, R.M.; Hofmann, E.P.; Mason, A.J.; Hogan, M.P.; Nystrom, G.S.; Ellsworth, S.A.; Colston, T.J.; Borja, M. Phylogenetically Diverse Diets Favor More Complex Venoms in North American Pitvipers. Proc. Natl. Acad. Sci. USA 2021, 118, e2015579118. [Google Scholar] [CrossRef] [PubMed]
- Riesgo, A.; Pérez-Porro, A.R.; Carmona, S.; Leys, S.P.; Giribet, G. Optimization of Preservation and Storage Time of Sponge Tissues to Obtain Quality mRNA for Next-generation Sequencing. Mol. Ecol. Resour. 2012, 12, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.; Xiao, C.; Durbrow, K.; Kimelman, M.; Rodarmer, K.; Shumway, M.; Yaschenko, E. Ncbi Sra Toolkit Technology for next Generation Sequence Data. In Proceedings of the Plant and Animal Genome XX Conference, San Diego, CA, USA, 14–18 January 2012. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Haas, B.; Papanicolaou, A. TransDecoder (Find Coding Regions within Transcripts). 2016. Available online: https://github.com/TransDecoder/TransDecoder (accessed on 12 October 2023).
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kocot, K.M.; Citarella, M.R.; Moroz, L.L.; Halanych, K.M. PhyloTreePruner: A Phylogenetic Tree-Based Approach for Selection of Orthologous Sequences for Phylogenomics. Evol. Bioinform. 2013, 9. [Google Scholar] [CrossRef]
- Kück, P.; Struck, T.H. BaCoCa–A Heuristic Software Tool for the Parallel Assessment of Sequence Biases in Hundreds of Gene and Taxon Partitions. Mol. Phylogenetics Evol. 2014, 70, 94–98. [Google Scholar] [CrossRef]
- Zhong, M.; Hansen, B.; Nesnidal, M.; Golombek, A.; Halanych, K.M.; Struck, T.H. Detecting the Symplesiomorphy Trap: A Multigene Phylogenetic Analysis of Terebelliform Annelids. BMC Evol. Biol. 2011, 11, 369. [Google Scholar] [CrossRef]
- Laumer, C.E.; Fernández, R.; Lemer, S.; Combosch, D.; Kocot, K.M.; Riesgo, A.; Andrade, S.C.S.; Sterrer, W.; Sørensen, M.V.; Giribet, G. Revisiting Metazoan Phylogeny with Genomic Sampling of All Phyla. Proc. R. Soc. B 2019, 286, 20190831. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Jungo, F.; Bairoch, A. Tox-Prot, the Toxin Protein Annotation Program of the Swiss-Prot Protein Knowledgebase. Toxicon 2005, 45, 293–301. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Dreon, M.S.; Frassa, M.V.; Ceolín, M.; Ituarte, S.; Qiu, J.-W.; Sun, J.; Fernández, P.E.; Heras, H. Novel Animal Defenses against Predation: A Snail Egg Neurotoxin Combining Lectin and Pore-Forming Chains That Resembles Plant Defense and Bacteria Attack Toxins. PLoS ONE 2013, 8, e63782. [Google Scholar] [CrossRef]
- Cadierno, M.P.; Dreon, M.S.; Heras, H. Apple Snail Perivitellin Precursor Properties Help Explain Predators’ Feeding Behavior. Physiol. Biochem. Zool. 2017, 90, 461–470. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mendes, C.B.; Norenburg, J.L.; Andrade, S.C.S. Species Delimitation Integrative Approach Reveals Three New Species in the Nemertopsis bivittata Complex. Invert. Syst. 2021, 35, 637–654. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gouveia-Oliveira, R.; Sackett, P.W.; Pedersen, A.G. MaxAlign: Maximizing Usable Data in an Alignment. BMC Bioinform. 2007, 8, 312. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Wu, Y.-C.; Rasmussen, M.D.; Bansal, M.S.; Kellis, M. Most Parsimonious Reconciliation in the Presence of Gene Duplication, Loss, and Deep Coalescence Using Labeled Coalescent Trees. Genome Res. 2014, 24, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, D.; Blaxter, M.; Darwin Tree of Life Barcoding collective; Wellcome Sanger Institute Tree of Life programme; Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective; Tree of Life Core Informatics collective; Darwin Tree of Life Consortium. The Genome Sequence of the Bootlace Worm, Lineus longissimus (Gunnerus, 1770). Wellcome Open Res. 2021, 6, 272. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Hahne, F.; Ivanek, R. Visualizing Genomic Data Using Gviz and Bioconductor. In Statistical Genomics; Mathé, E., Davis, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1418, pp. 335–351. ISBN 978-1-4939-3576-5. [Google Scholar]
- Han, M.V.; Thomas, G.W.; Lugo-Martinez, J.; Hahn, M.W. Estimating Gene Gain and Loss Rates in the Presence of Error in Genome Assembly and Annotation Using CAFÉ. Mol. Biol. Evol. 2013, 30, 1987–1997. [Google Scholar] [CrossRef]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An Efficient Algorithm for Large-Scale Detection of Protein Families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef] [PubMed]



| Toxin | Positive Selection Detection Method | Sites under Selection |
|---|---|---|
| Cytotoxin | M8/M7 (BEB) | 146 |
| BsA/BsA1 (BEB) | 154 | |
| MEME | 57; 82; 88; 93; 100; 114; 117; 118; 128; 132; 143; 149; 150; 154; 161; 165; 169; 174 | |
| FEL | NS | |
| FUBAR | NS | |
| Common (at least 2) | 154 | |
| Scoloptoxin | M8/M7 | 481; 482; 604; 605; 611; 624; 631; 647; 649 |
| BsA/BsA1 | NS | |
| MEME | 26; 33; 36; 37; 38; 40; 58; 74; 123; 214; 277; 304; 458; 479; 482; 500; 502; 599; 603; 604; 605; 611; 621; 623; 627; 631; 632; 636; 641; 650 | |
| FEL | 611; | |
| FUBAR | 482; 605; 611; 631; 645 | |
| Common (at least 2) | 482; 604; 605; 611; 631 | |
| Alpha-Nemertide | M8/M7 | 77 |
| BsA/BsA1 | 63; 77 | |
| MEME | 27; 74; 77; 78; 95 | |
| FEL | NS | |
| FUBAR | 74 | |
| Common (at least 2) | 77,74 | |
| Beta-Nemertide | M8/M7 | 26; 31 |
| BsA/BsA1 | NA | |
| MEME | 3; 38; 48; 61; 65; 85 | |
| FEL | 38 | |
| FUBAR | 34; 38 | |
| Common (at least 2) | 38 | |
| U-Nemertotoxin-3 | M8/M7 | 36; 37 |
| BsA/BsA1 | NA | |
| MEME | 34; 36; 37; 61 | |
| FEL | 34; 36 | |
| FUBAR | 34; 36; 37; 40; 62 | |
| Common (at least 2) | 34; 36; 37 | |
| Myt-alpha-ktx-like | M8/M7 | 37 |
| BsA/BsA1 | NA | |
| MEME | 25; 36; 57 | |
| FEL | 36 | |
| FUBAR | 36 | |
| Common (at least 2) | 36 |
| Model | Global Lambda (or Remaining Species) | Pilidiophora Lambda | Lineus Lambda | lognL | (2 × (lnLglobal − lnLmulti)) | Lowest (2 × (lnLglobal − lnLsimulation)) (N= 194) |
|---|---|---|---|---|---|---|
| 1 ƛ (Global) | 7.23 × 10−4 | NA | NA | −1.03 × 106 | NA | −7.65 |
| 2 ƛ Heteronemertea Remaining | 7.23 × 10−4 | 1.44 × 10−3 | NA | −9.73 × 105 | −1.19 × 105 | |
| 3 ƛ Lineus sanguineus Heteronemertea Remaining | 7.23 × 10−4 | 1.44 × 10−3 | 4.11 × 10−3 | −9.44 × 105 | −1.76 × 105 | |
| 1 ƛ Global + Error model | 7.23 × 10−4 | NA | NA | −1.03 × 106 | NA | |
| 3 ƛ Lineus sanguineus Heteronemertea Remaining+ error model | 5.51 × 10−4 | 1.34 × 10−3 | 8.55 × 10−4 | −1.02 × 106 | −2.54 × 104 |
| Species | Locality | Latitude | Longitude | Substrate | Number of Individuals |
|---|---|---|---|---|---|
| L. sanguineus * | Praia do Cabelo Gordo—SP | −23.828088 | −45.4232 | Crassostrea sp. bank | 1 |
| L. sanguineus +& | Praia do Araçá—SP | −23.812840 | −45.408216 | Crassostrea sp. bank | 2 |
| L. sanguineus & | Peró—RJ | −22.822844 | −41.969951 | Crassostrea sp. bank | 2 |
| N. berthalutzae +& | Praia Grande—SP | −23.824663 | −45.412764 | Crassostrea sp. bank | 2 |
| N. pamelaroeae * | Praia do Jabaquara—RJ | −23.211346 | −44.714191 | Crassostrea sp. bank | 1 |
| O. erneba * | Praia da Vila—SP | −23.777234 | −45.358997 | Sand sediment | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonoda, G.G.; Tobaruela, E.d.C.; Norenburg, J.; Fabi, J.P.; Andrade, S.C.S. Venomous Noodles: The Evolution of Toxins in Nemertea through Positive Selection and Gene Duplication. Toxins 2023, 15, 650. https://doi.org/10.3390/toxins15110650
Sonoda GG, Tobaruela EdC, Norenburg J, Fabi JP, Andrade SCS. Venomous Noodles: The Evolution of Toxins in Nemertea through Positive Selection and Gene Duplication. Toxins. 2023; 15(11):650. https://doi.org/10.3390/toxins15110650
Chicago/Turabian StyleSonoda, Gabriel Gonzalez, Eric de Castro Tobaruela, Jon Norenburg, João Paulo Fabi, and Sónia C. S. Andrade. 2023. "Venomous Noodles: The Evolution of Toxins in Nemertea through Positive Selection and Gene Duplication" Toxins 15, no. 11: 650. https://doi.org/10.3390/toxins15110650
APA StyleSonoda, G. G., Tobaruela, E. d. C., Norenburg, J., Fabi, J. P., & Andrade, S. C. S. (2023). Venomous Noodles: The Evolution of Toxins in Nemertea through Positive Selection and Gene Duplication. Toxins, 15(11), 650. https://doi.org/10.3390/toxins15110650








