Charged Amino Acids Contribute to ZorO Toxicity
Abstract
1. Introduction
2. Results
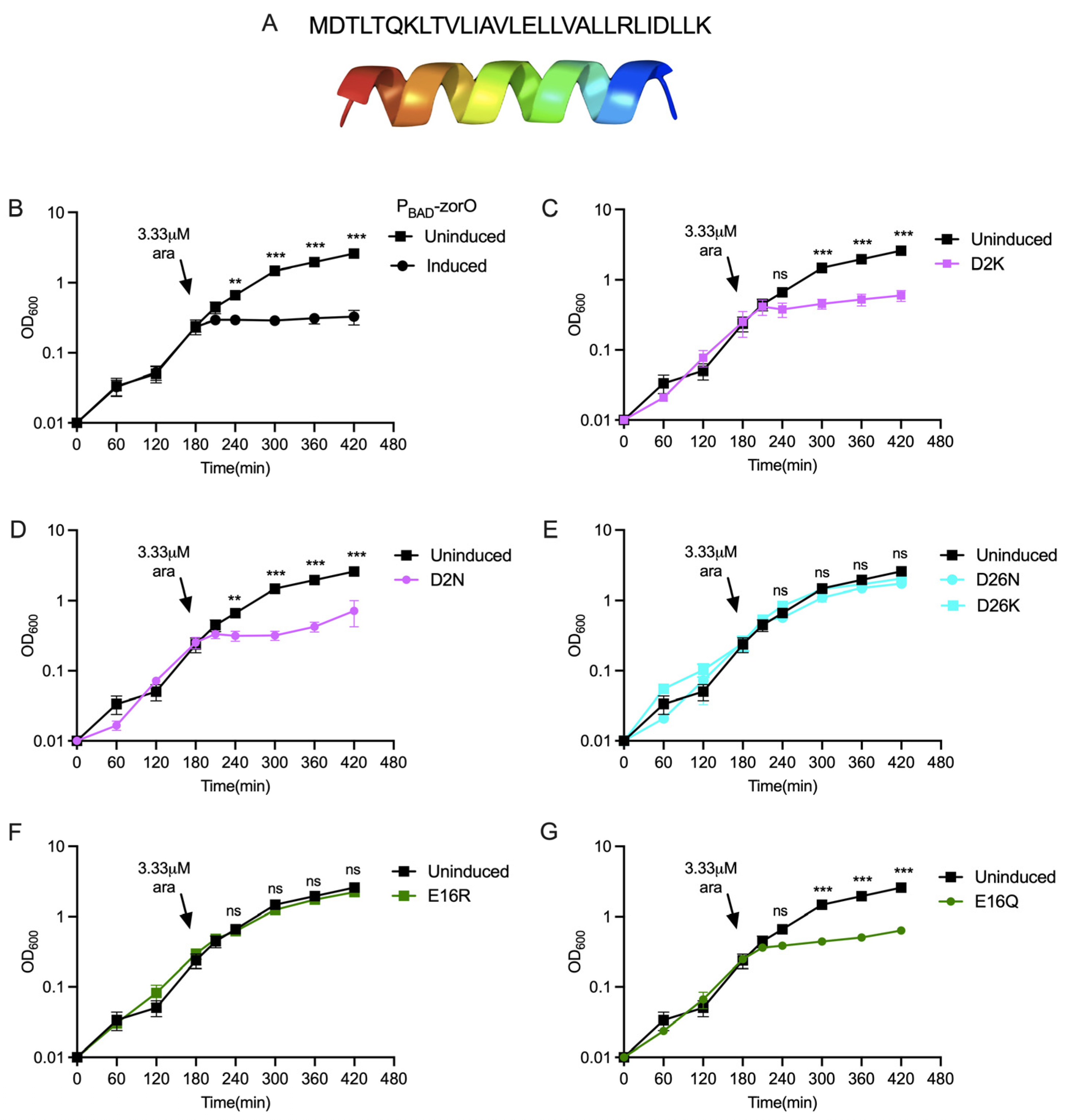
2.1. Alterations of Charged Residues of ZorO Do Not Impact Predicted Hydrophobicity
2.2. Specific Charged Amino Acids within the Predicted Transmembrane Domain Are Critical for ZorO Induced Growth Stasis
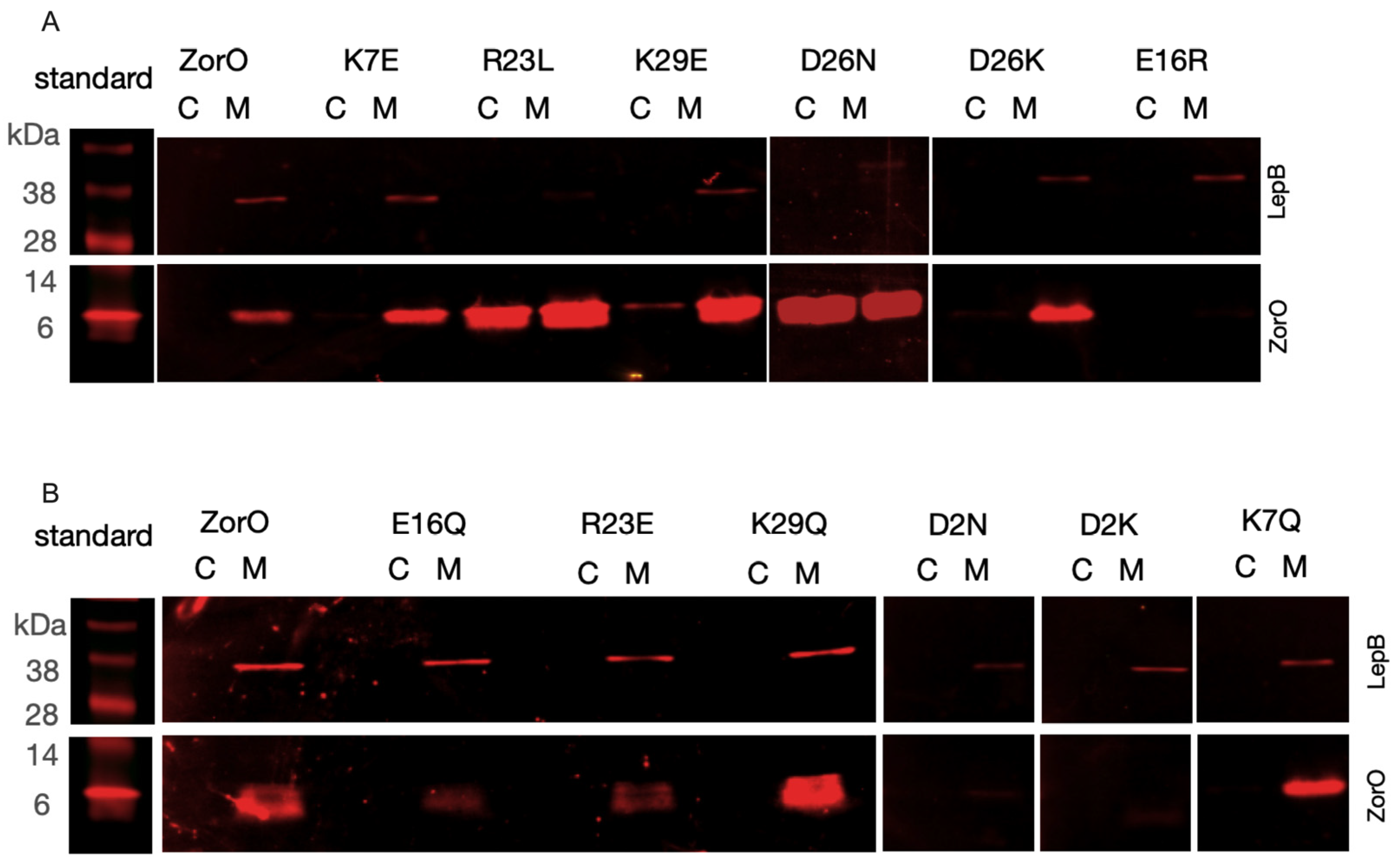
2.3. Localization of ZorO after Mutation of the Charged Residues
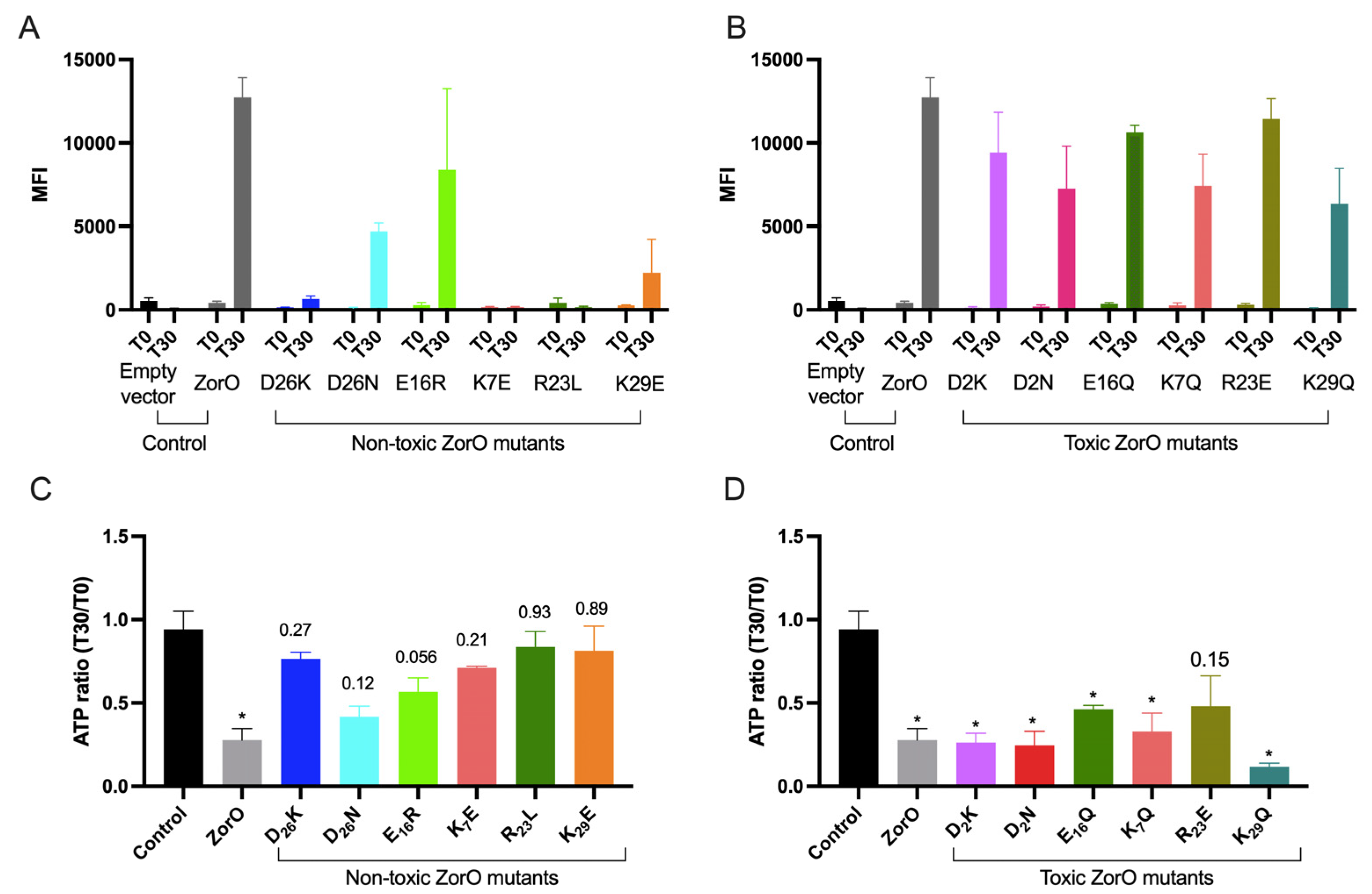
2.4. Impact of Charged Amino Acid Residues on ZorO Induced Membrane Depolarization and ATP Depletion
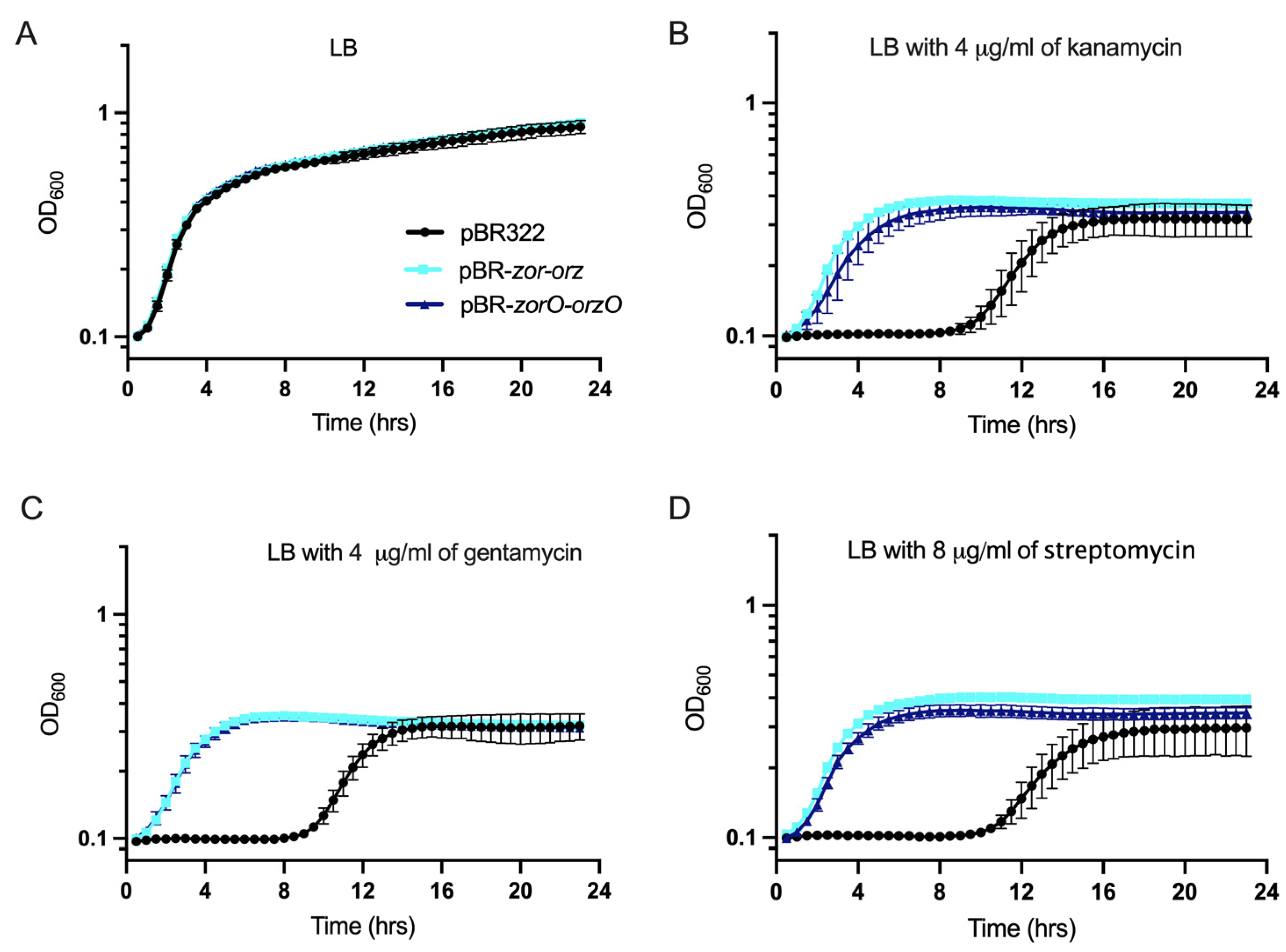
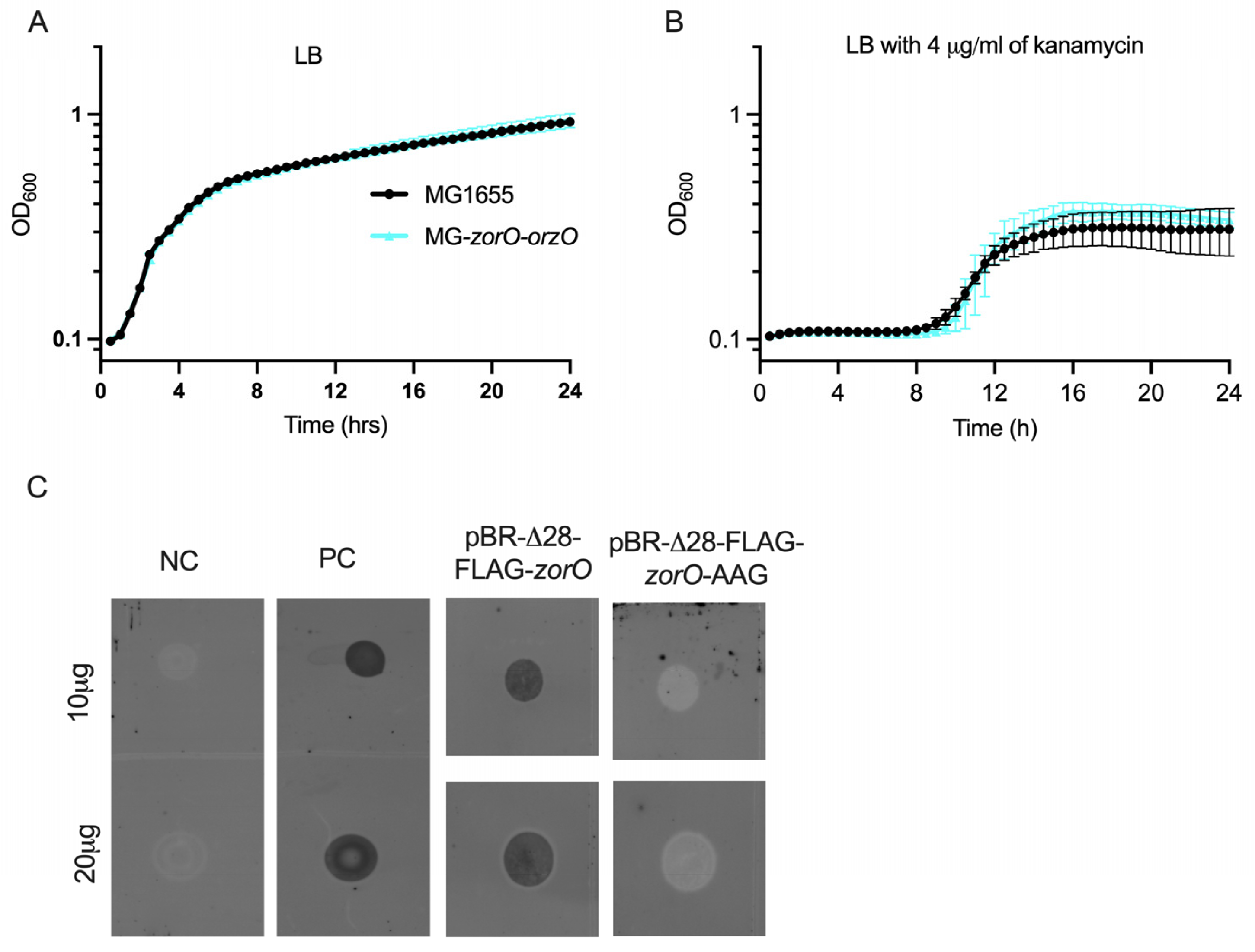
2.5. Multiple Copies of zorO-orzO Improve Growth in the Presence of Aminoglycosides but Not a Single Copy in the Chromosome
2.6. ZorO Translation from Its Native Promoter Can Be Detected when the Gene Copy Number Is Increased
2.7. The Non-Toxic ZorO Mutant R23L Improves Growth in Presence of Kanamycin
3. Discussion
3.1. Charged Residues in ZorO Are Important for ZorO Mediated Toxicity
3.2. ZorO Mutants Unable to Inhibit Growth Can Accumulate in the Cytoplasm
3.3. ZorO Translation under Native Promoter and the Role of zorO-orzO to Improve Bacterial Growth in Presence of Aminoglycoside
3.4. Decoupling ZorO Toxicity and zorO-orzO Mediated Improved Growth in Aminoglycoside
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Plasmids
5.2. Growth Conditions
5.3. Plasmid and Strain Construction
5.4. Membrane Depolarization Assay
5.5. ATP Measurements
5.6. Western Blot and Dot-Blot Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brantl, S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012, 9, 1488–1490. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Fozo, E.M. sRNA antitoxins: More than one way to repress a toxin. Toxins 2014, 6, 2310–2335. [Google Scholar] [CrossRef] [PubMed]
- Nonin-Lecomte, S.; Fermon, L.; Felden, B.; Pinel-Marie, M.-L. Bacterial type I toxins: Folding and membrane interactions. Toxins 2021, 13, 490. [Google Scholar] [CrossRef]
- Guo, Y.; Quiroga, C.; Chen, Q.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K.; Wang, X. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014, 42, 6448–6462. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Bayoumi, M.; Dewachter, L.; Knapen, W.; Mika, J.T.; Hofkens, J.; Dedecker, P.; Maglia, G.; Verstraeten, N.; Michiels, J. The persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. MBio 2018, 9, e00744-18. [Google Scholar] [CrossRef]
- Schneider, V.; Wadhwani, P.; Reichert, J.; Bürck, J.; Elstner, M.; Ulrich, A.S.; Kubař, T. Tetrameric Charge-Zipper Assembly of the TisB peptide in membranes-computer simulation and experiment. J. Phys. Chem. B 2019, 123, 1770–1779. [Google Scholar] [CrossRef]
- Unoson, C.; Wagner, E.G.H. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008, 70, 258–270. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Dewachter, L.; De Loose, P.-J.; Bollen, C.; Verstraeten, N.; Michiels, J. Hokb monomerization and membrane repolarization control persister awakening. Mol. Cell 2019, 75, 1031–1042.e4. [Google Scholar] [CrossRef]
- Verstraeten, N.; Knapen, W.J.; Kint, C.I.; Liebens, V.; Van den Bergh, B.; Dewachter, L.; Michiels, J.E.; Fu, Q.; David, C.C.; Fierro, A.C.; et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 2015, 59, 9–21. [Google Scholar] [CrossRef]
- Gurnev, P.A.; Ortenberg, R.; Dörr, T.; Lewis, K.; Bezrukov, S.M. Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 2012, 586, 2529–2534. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.H.; Unoson, C. The toxin-antitoxin system tisB-istR1: Expression, regulation, and biological role in persister phenotypes. RNA Biol. 2012, 9, 1513–1519. [Google Scholar] [CrossRef] [PubMed]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [PubMed]
- Berghoff, B.A.; Hoekzema, M.; Aulbach, L.; Wagner, E.G.H. Two regulatory RNA elements affect TisB-dependent depolarization and persister formation. Mol. Microbiol. 2017, 103, 1020–1033. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Argaman, L.; Wagner, E.G.H.; Altuvia, S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004, 14, 2271–2276. [Google Scholar] [CrossRef] [PubMed]
- Knopp, M.; Gudmundsdottir, J.S.; Nilsson, T.; König, F.; Warsi, O.; Rajer, F.; Ädelroth, P.; Andersson, D.I. De novo emergence of peptides that confer antibiotic resistance. MBio 2019, 10, e00837-19. [Google Scholar] [CrossRef]
- Kawano, M.; Oshima, T.; Kasai, H.; Mori, H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002, 45, 333–349. [Google Scholar] [CrossRef]
- Fozo, E.M.; Kawano, M.; Fontaine, F.; Kaya, Y.; Mendieta, K.S.; Jones, K.L.; Ocampo, A.; Rudd, K.E.; Storz, G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008, 70, 1076–1093. [Google Scholar] [CrossRef]
- Bogati, B.; Wadsworth, N.; Barrera, F.; Fozo, E.M. Improved growth of Escherichia coli in aminoglycoside antibiotics by the zor-orz toxin-antitoxin system. J. Bacteriol. 2021, 204, JB0040721. [Google Scholar] [CrossRef]
- Patel, S.; Weaver, K.E. Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis. J. Bacteriol. 2006, 188, 5374–5384. [Google Scholar] [CrossRef]
- Göbl, C.; Kosol, S.; Stockner, T.; Rückert, H.M.; Zangger, K. Solution structure and membrane binding of the toxin fst of the par addiction module. Biochemistry 2010, 49, 6567–6575. [Google Scholar] [CrossRef]
- Jahn, N.; Brantl, S.; Strahl, H. Against the mainstream: The membrane-associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 2015, 98, 651–666. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.; Sadlon, J.; Weaver, K. Charged residues flanking the transmembrane domain of two related toxin-antitoxin system toxins affect host response. Toxins 2021, 13, 329. [Google Scholar] [CrossRef] [PubMed]
- Mok, W.W.K.; Patel, N.H.; Li, Y. Decoding toxicity: Deducing the sequence requirements of IbsC, a type I toxin in Escherichia coli. J. Biol. Chem. 2010, 285, 41627–41636. [Google Scholar] [CrossRef]
- Wen, J.; Won, D.; Fozo, E.M. The ZorO-OrzO type I toxin-antitoxin locus: Repression by the OrzO antitoxin. Nucleic Acids Res. 2014, 42, 1930–1946. [Google Scholar] [CrossRef]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of type I toxin-antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Harp, J.R.; Fozo, E.M. The 5΄ UTR of the type I toxin ZorO can both inhibit and enhance translation. Nucleic Acids Res. 2017, 45, 4006–4020. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Otsuka, Y.; Ishikawa, T.; Takahashi, C.; Masuda, M. A short peptide derived from the ZorO toxin functions as an effective antimicrobial. Toxins 2019, 11, 392. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Wilmaerts, D.; De Loose, P.-J.; Vercauteren, S.; De Smedt, S.; Verstraeten, N.; Michiels, J. Functional analysis of cysteine residues of the Hok/Gef type I toxins in Escherichia coli. FEMS Microbiol. Lett. 2021, 368, fnab069. [Google Scholar] [CrossRef] [PubMed]
- Steinbrecher, T.; Prock, S.; Reichert, J.; Wadhwani, P.; Zimpfer, B.; Bürck, J.; Berditsch, M.; Elstner, M.; Ulrich, A.S. Peptide-lipid interactions of the stress-response peptide TisB that induces bacterial persistence. Biophys. J. 2012, 103, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef]
- Cheverton, A.M.; Gollan, B.; Przydacz, M.; Wong, C.T.; Mylona, A.; Hare, S.A.; Helaine, S. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell 2016, 63, 86–96. [Google Scholar] [CrossRef]
- Weaver, K.E.; Reddy, S.G.; Brinkman, C.L.; Patel, S.; Bayles, K.W.; Endres, J.L. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 2009, 155, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Loeffler, A.; Kadlec, K. Bacterial resistance to antimicrobial agents and its impact on veterinary and human medicine. Vet. Dermatol. 2017, 28, 82-e19. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef]
- Huang, X.; Chen, R.; Sun, M.; Peng, Y.; Pu, Q.; Yuan, Y.; Chen, G.; Dong, J.; Du, F.; Cui, X.; et al. Frame-shifted proteins of a given gene retain the same function. Nucleic Acids Res. 2020, 48, 4396–4404. [Google Scholar] [CrossRef]
- Bruni, G.N.; Kralj, J.M. Membrane voltage dysregulation driven by metabolic dysfunction underlies bactericidal activity of aminoglycosides. Elife 2020, 9, e58706. [Google Scholar] [CrossRef]
- Doublet, B.; Douard, G.; Targant, H.; Meunier, D.; Madec, J.-Y.; Cloeckaert, A. Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J. Microbiol. Methods 2008, 75, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, F.; Fuchs, R.T.; Storz, G. Membrane localization of small proteins in Escherichia coli. J. Biol. Chem. 2011, 286, 32464–32474. [Google Scholar] [CrossRef] [PubMed]
- Hemm, M.R.; Paul, B.J.; Miranda-Ríos, J.; Zhang, A.; Soltanzad, N.; Storz, G. Small stress response proteins in Escherichia coli: Proteins missed by classical proteomic studies. J. Bacteriol. 2010, 192, 46–58. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogati, B.; Shore, S.F.H.; Nipper, T.D.; Stoiculescu, O.; Fozo, E.M. Charged Amino Acids Contribute to ZorO Toxicity. Toxins 2023, 15, 32. https://doi.org/10.3390/toxins15010032
Bogati B, Shore SFH, Nipper TD, Stoiculescu O, Fozo EM. Charged Amino Acids Contribute to ZorO Toxicity. Toxins. 2023; 15(1):32. https://doi.org/10.3390/toxins15010032
Chicago/Turabian StyleBogati, Bikash, Selene F. H. Shore, Thomas D. Nipper, Oana Stoiculescu, and Elizabeth M. Fozo. 2023. "Charged Amino Acids Contribute to ZorO Toxicity" Toxins 15, no. 1: 32. https://doi.org/10.3390/toxins15010032
APA StyleBogati, B., Shore, S. F. H., Nipper, T. D., Stoiculescu, O., & Fozo, E. M. (2023). Charged Amino Acids Contribute to ZorO Toxicity. Toxins, 15(1), 32. https://doi.org/10.3390/toxins15010032




