Expression of the First Recombinant Anti-Tumoral Snake Venom Kunitz-Type Serine Protease Inhibitor
Abstract
:1. Introduction
2. Results and Discussion
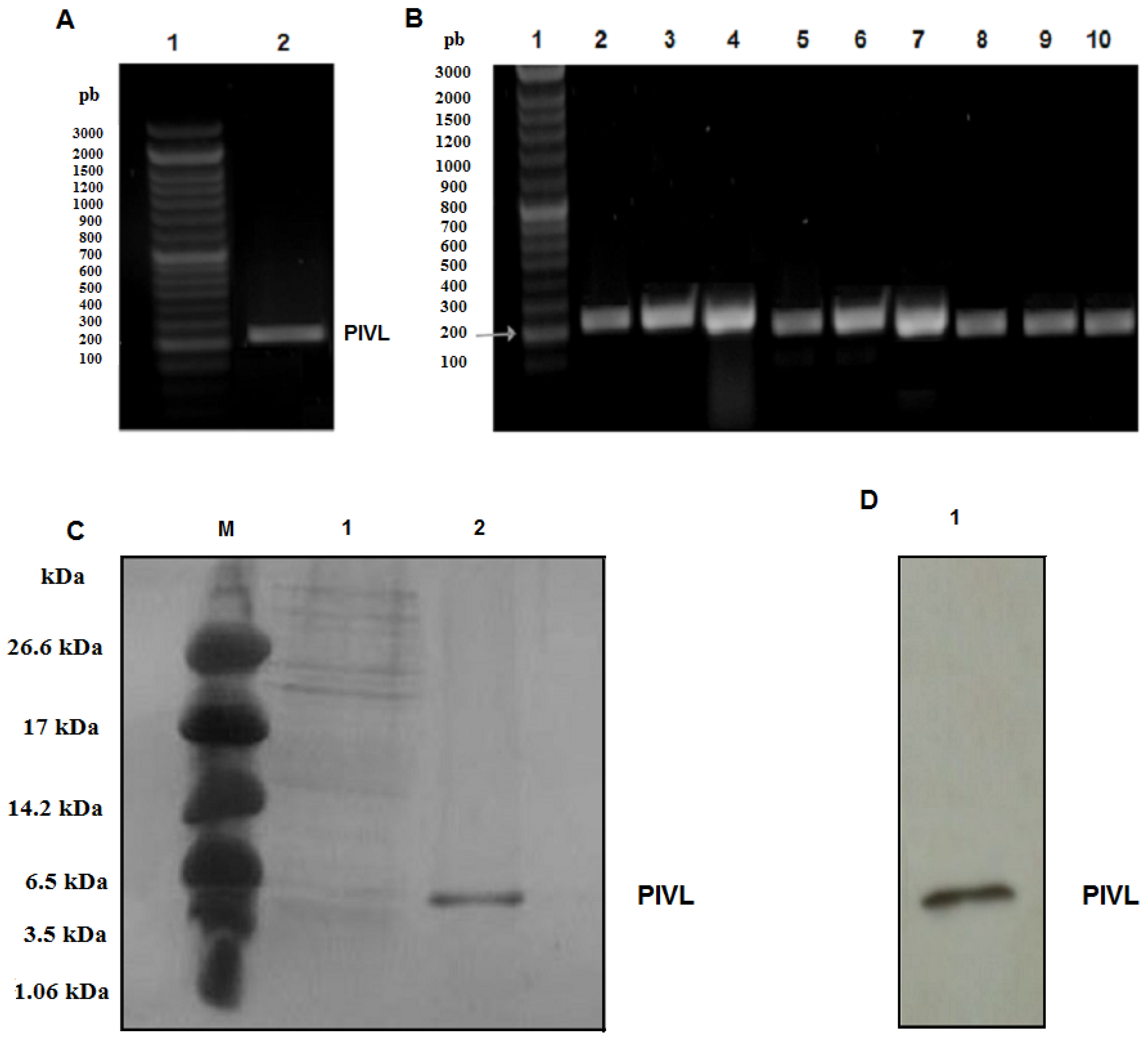
2.1. Amplification of the cDNA Encoding PIVL
2.2. Expression and Purification of Recombinant PIVL
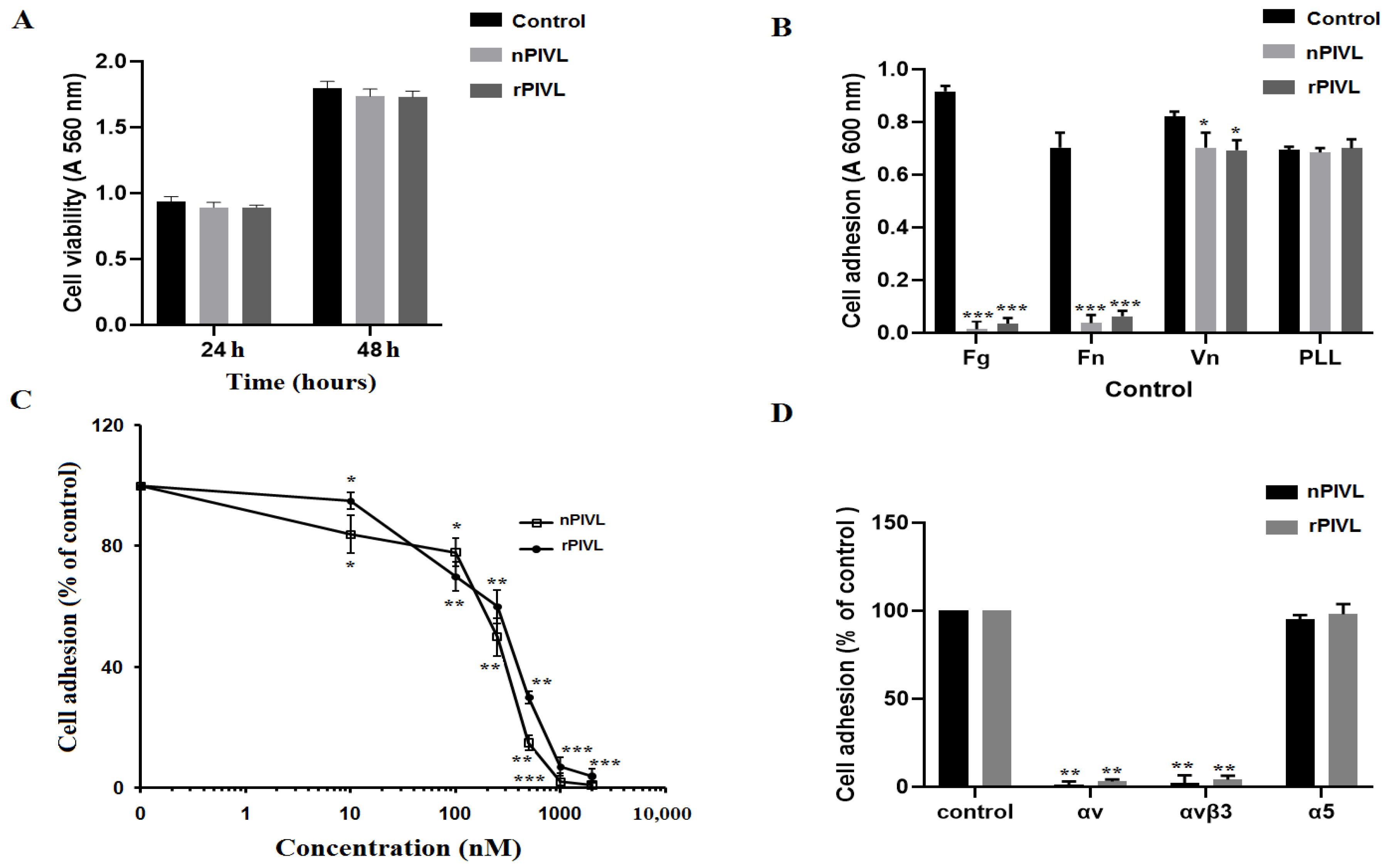
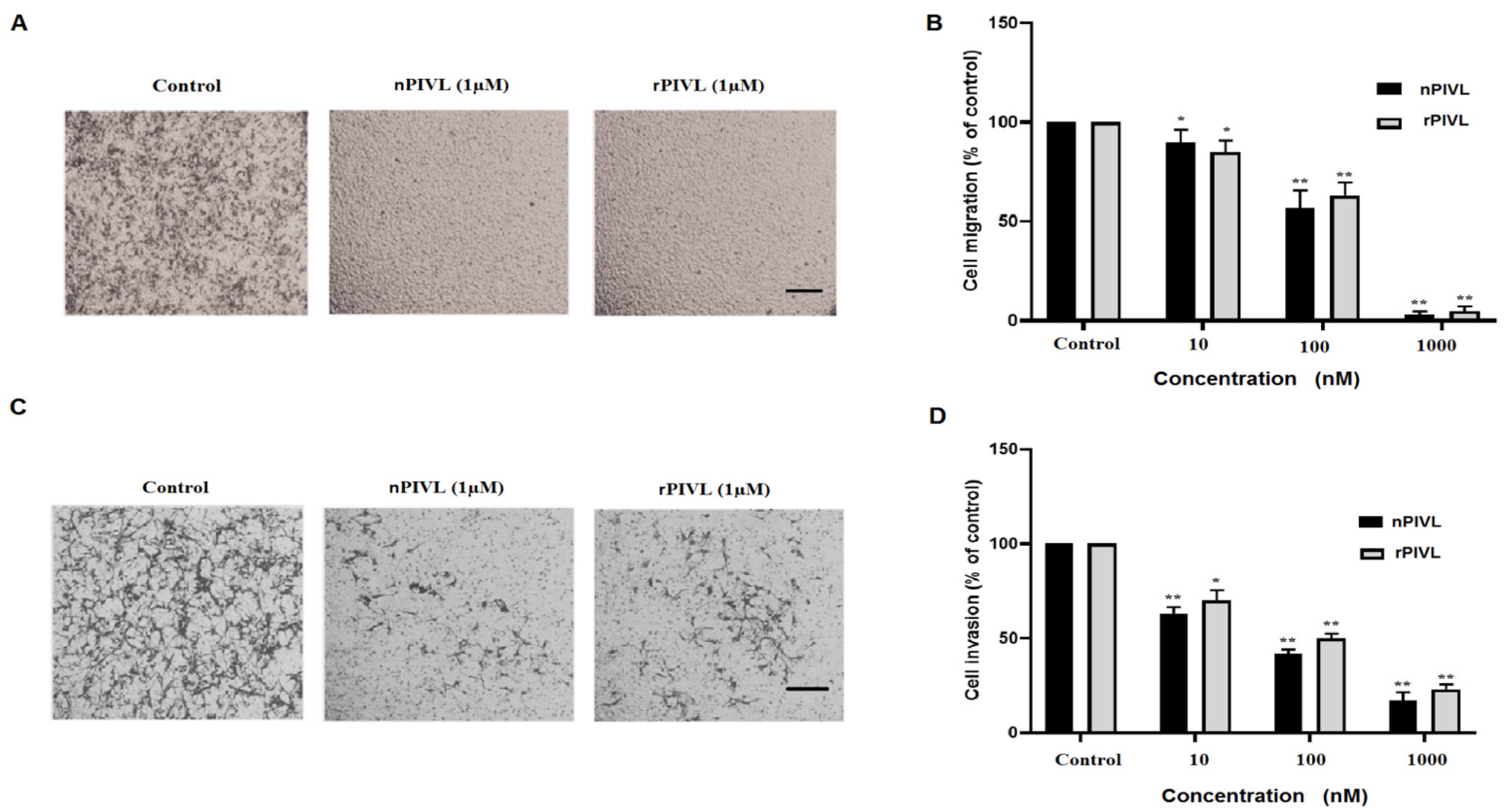
2.3. Recombinant PIVL Affects U87 Cells Adhesion and Motility
2.4. Recombinant PIVL Blocks Angiogenesis
2.5. Recombinant and Native PIVL Modulate PI3/AKT and MAPK Signaling Pathways in Glioblastoma Cells
3. Conclusions
4. Materials and Methods
4.1. Materials, Reagents, and Ethics Statement
4.2. Host Strains and Plasmids
4.3. Molecular Cloning Procedure of PIVL
4.4. Expression of Recombinant PIVL
4.5. Protein Recovery and Purification
4.6. Protein Electrophoresis and Western Blot Analysis
4.7. Cell Culture and Adhesion Assays
4.8. Cell Viability Assays
4.9. Cell Migration Assays
4.10. Cell Invasion Assays
4.11. Chicken Chorioallantoic Membrane Assay
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Desai, V.; Bhushan, A. Natural Bioactive Compounds: Alternative Approach to the Treatment of Glioblastoma Multiforme. Biomed. Res. Int. 2017, 2017, 9363040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paolillo, M.; Boselli, C.; Schinelli, S. Glioblastoma under Siege: An Overview of Current Therapeutic Strategies. Brain Sci. 2018, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamberlain, M.C. Temozolomide: Therapeutic limitations in the treatment of adult high-grade gliomas. Expert Rev. Neurother. 2010, 10, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Stevens, M.F.; Bradshaw, T.D. Temozolomide: Mechanisms of action, repair and resistance. Curr. Mol. Pharmacol. 2012, 5, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Abd El-Aziz, T.; Soares, A.G.; Stockand, J.D. Snake Venoms in Drug Discovery: Valuable Therapeutic Tools for Life Saving. Toxins 2019, 11, 564. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, M., Jr.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef]
- Ranasinghe, S.; McManus, D.P. Structure and function of invertebrate Kunitz serine protease inhibitors. Dev. Comp. Immunol. 2013, 39, 219–227. [Google Scholar] [CrossRef]
- Chou, W.M.; Liu, W.H.; Chen, K.C.; Chang, L.S. Structure-function studies on inhibitory activity of Bungarus multicinctus protease inhibitor-like protein on matrix metalloprotease-2, and invasion and migration of human neuroblastoma SK-N-SH cells. Toxicon 2010, 55, 353–360. [Google Scholar] [CrossRef]
- Morjen, M.; Kallech-Ziri, O.; Bazaa, A.; Othman, H.; Mabrouk, K.; Zouari-Kessentini, R.; Sanz, L.; Calvete, J.J.; Srairi-Abid, N.; El Ayeb, M.; et al. PIVL, a new serine protease inhibitor from Macrovipera lebetina transmediterranea venom, impairs motility of human glioblastoma cells. Matrix Biol. 2013, 32, 52–62. [Google Scholar] [CrossRef]
- Danen, E.H.J. Integrin Signaling as a Cancer Drug Target. Int. Sch. Res. Not. 2013, 2013, 135164. [Google Scholar] [CrossRef] [Green Version]
- Tucci, M.; Stucci, S.; Silvestris, F. Does cilengitide deserve another chance? Lancet Oncol. 2014, 15, e584–e585. [Google Scholar] [CrossRef]
- van der Horst, G.; van den Hoogen, C.; Buijs, J.T.; Cheung, H.; Bloys, H.; Pelger, R.C.; Lorenzon, G.; Heckmann, B.; Feyen, J.; Pujuguet, P.; et al. Targeting of αv-integrins in stem/progenitor cells and supportive microenvironment impairs bone metastasis in human prostate cancer. Neoplasia 2011, 13, 516–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morjen, M.; Honoré, S.; Bazaa, A.; Abdelkafi-Koubaa, Z.; Ellafi, A.; Mabrouk, K.; Kovacic, H.; El Ayeb, M.; Marrakchi, N.; Luis, J. PIVL, a snake venom Kunitz-type serine protease inhibitor, inhibits in vitro and in vivo angiogenesis. Microvasc. Res. 2014, 95, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Devi, A.; Saikia, B.; Doley, R. Expression and characterization of Flavikunin: A Kunitz-type serine protease inhibitor identified in the venom gland cDNA library of Bungarus flaviceps. J. Biochem. Mol. Toxicol. 2019, 33, e22273. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, S.L.; Fischer, K.; Gobert, G.N.; McManus, D.P. Functional expression of a novel Kunitz type protease inhibitor from the human blood fluke Schistosoma mansoni. Parasites Vectors 2015, 8, 408. [Google Scholar] [CrossRef] [Green Version]
- Okegawa, T.; Pong, R.C.; Li, Y.; Hsieh, J.T. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim. Pol. 2004, 51, 445–457. [Google Scholar] [CrossRef] [Green Version]
- Wank, M.; Schilling, D.; Schmid, T.E.; Meyer, B.; Gempt, J.; Barz, M.; Schlegel, J.; Liesche, F.; Kessel, K.A.; Wiestler, B.; et al. Human Glioma Migration and Infiltration Properties as a Target for Personalized Radiation Medicine. Cancers 2018, 10, 456. [Google Scholar] [CrossRef]
- Smith, L.A.; Lafaye, P.J.; LaPenotiere, H.F.; Spain, T.; Dolly, J.O. Cloning and functional expression of dendrotoxin K from black mamba, a K+ channel blocker. Biochemistry 1993, 32, 5692–5697. [Google Scholar] [CrossRef]
- Smith, L.A.; Reid, P.F.; Wang, F.C.; Parcej, D.N.; Schmidt, J.J.; Olson, M.A.; Dolly, J.O. Site-directed mutagenesis of dendrotoxin K reveals amino acids critical for its interaction with neuronal K+ channels. Biochemistry 1997, 36, 7690–7696. [Google Scholar] [CrossRef]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [Green Version]
- Zhi, Z.; Ouyang, Z.; Ren, Y.; Cheng, Y.; Liu, P.; Wen, Y.; Shao, Y. Non-canonical phosphorylation of Bmf by p38 MAPK promotes its apoptotic activity in anoikis. Cell Death Differ. 2021. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; New, L.; Pan, Z.; Han, J.; Nemerow, G.R. Urokinase plasminogen activator/urokinase-specific surface receptor expression and matrix invasion by breast cancer cells requires constitutive p38alpha mitogen-activated protein kinase activity. J. Biol. Chem. 2000, 275, 12266–12272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aikawa, R.; Nagai, T.; Kudoh, S.; Zou, Y.; Tanaka, M.; Tamura, M.; Akazawa, H.; Takano, H.; Nagai, R.; Komuro, I. Integrins play a critical role in mechanical stress-induced p38 MAPK activation. Hypertension 2002, 39, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Jebali, J.; Bazaa, A.; Sarray, S.; Benhaj, K.; Karboul, A.; El Ayeb, M.; Marrakchi, N.; Gargouri, A. C-type lectin protein isoforms of Macrovipera lebetina: cDNA cloning and genetic diversity. Toxicon 2009, 53, 228–237. [Google Scholar] [CrossRef]
- Kadi, A.; Pichard, V.; Lehmann, M.; Briand, C.; Braguer, D.; Marvaldi, J.; Rognoni, J.B.; Luis, J. Effect of microtubule disruption on cell adhesion and spreading. Biochem. Biophys. Res. Commun. 1998, 246, 690–695. [Google Scholar] [CrossRef]
- Defilles, C.; Lissitzky, J.C.; Montero, M.P.; André, F.; Prévot, C.; Delamarre, E.; Marrakchi, N.; Luis, J.; Rigot, V. Alphavbeta5/beta6 integrin suppression leads to a stimulation of alpha2beta1 dependent cell migration resistant to PI3K/Akt inhibition. Exp. Cell Res. 2009, 315, 1840–1849. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Sarray, S.; Delamarre, E.; Marvaldi, J.; El Ayeb, M.; Marrakchi, N.; Luis, J. Lebectin and lebecetin, two C-type lectins from snake venom, inhibit alpha5beta1 and alphaV-containing integrins. Matrix Biol. 2007, 26, 306–313. [Google Scholar] [CrossRef]
- Rigot, V.; Lehmann, M.; André, F.; Daemi, N.; Marvaldi, J.; Luis, J. Integrin ligation and PKC activation are required for migration of colon carcinoma cells. J. Cell Sci. 1998, 111 Pt 20, 3119–3127. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morjen, M.; Moslah, W.; Touihri-Baraketi, I.; Srairi-Abid, N.; Luis, J.; Marrakchi, N.; Jebali, J. Expression of the First Recombinant Anti-Tumoral Snake Venom Kunitz-Type Serine Protease Inhibitor. Toxins 2022, 14, 170. https://doi.org/10.3390/toxins14030170
Morjen M, Moslah W, Touihri-Baraketi I, Srairi-Abid N, Luis J, Marrakchi N, Jebali J. Expression of the First Recombinant Anti-Tumoral Snake Venom Kunitz-Type Serine Protease Inhibitor. Toxins. 2022; 14(3):170. https://doi.org/10.3390/toxins14030170
Chicago/Turabian StyleMorjen, Maram, Wassim Moslah, Imen Touihri-Baraketi, Najet Srairi-Abid, José Luis, Naziha Marrakchi, and Jed Jebali. 2022. "Expression of the First Recombinant Anti-Tumoral Snake Venom Kunitz-Type Serine Protease Inhibitor" Toxins 14, no. 3: 170. https://doi.org/10.3390/toxins14030170
APA StyleMorjen, M., Moslah, W., Touihri-Baraketi, I., Srairi-Abid, N., Luis, J., Marrakchi, N., & Jebali, J. (2022). Expression of the First Recombinant Anti-Tumoral Snake Venom Kunitz-Type Serine Protease Inhibitor. Toxins, 14(3), 170. https://doi.org/10.3390/toxins14030170





