Abstract
Sesame Sesamum indicum L. is a major oil-based seed crop that has been widely cultivated and consumed in Pakistan. Unfortunately, sesame is highly prone to Aspergillus fungal growth in the field, and under inappropriate storage conditions can become contaminated with aflatoxins, the most potent carcinogen found in nature. Here, we have isolated a high number of Aspergillus isolates from sesame seeds in fresh and stored conditions obtained from rainfed and irrigated zones of Punjab, Pakistan, and characterized them for aflatoxigenic potentials. Using morphological identification techniques, 260 isolates were grouped as potential Aspergillus section Flavi, with 126 and 134 originating from the rainfed and irrigated zones, respectively. Out of 260 in total, 188 isolates were confirmed to produce aflatoxins. There were no significant differences in potential aflatoxigenic isolates with respect to the rainfed and irrigated zones. However, the number of potential aflatoxigenic isolates was significantly higher (p < 0.05) in stored samples than that of those from fresh sesame seeds in the rainfed and irrigated zone. Whole genome sequencing and comparative analyses of 12 select isolates have revealed that one of the A. flavus isolates, which produced very low aflatoxins (AFP10), has an elevated missense variant rate, numerous high impact mutations, and a 600 base pair deletion in the norB gene. In summary, our study provides insights into aflatoxigenic potential and the associated genetic diversity of indigenous Aspergillus section Flavi isolates and potential management strategies for reducing aflatoxin contamination levels in a major crop consumed in Punjab, Pakistan.
Key Contribution:
This study reports the distribution of aflatoxigenic and non-aflatoxigenic isolates of Aspergillus section Flavi in the rainfed and irrigated zones of Punjab, Pakistan and the genomic diversity of 12 indigenous Aspergillus flavus isolates from sesame seeds.
1. Introduction
Aflatoxins are a group of naturally occurring carcinogens and toxic secondary metabolites of the fungal genus Aspergillus [1]. Numerous Aspergillus spp. produce aflatoxins but Aspergillus flavus is the most predominant one responsible for producing aflatoxins in fields and storage conditions [2]. The four major aflatoxins commonly isolated from foods and feeds are aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), aflatoxin G1 (AFG1), and aflatoxin G2 (AFG2), which are categorized as group 1 human carcinogens by the International Agency for Research on Cancer (IARC) [3]. AFB1 is the most toxic, common, and widespread food and feed contaminant and is responsible for 75% of all aflatoxin contamination [4]. Over half of the global population (some four billion people), mostly in developing countries, are at threat of chronic exposure to unknown levels of aflatoxins, which can be associated with decreased growth rate and feeding efficiency, reduced liver and kidney function, and immune system suppression [5,6]. The most important illness related to aflatoxin consumption is hepatocellular carcinoma, which is the third leading cause of cancer-related mortality worldwide [7]. In the developing countries, 40% of human productivity is lost as a result of disorders associated with aflatoxin contamination [8].
Sesame, Sesamum indicum L., is a major oilseed crop that is cultivated in about 9,398,770 hectares (ha) of tropical, sub-tropical, and temperate regions around the world. Pakistan ranks 14th among the major sesame-producing countries in the world with an annual production of about 35,699 tons, grown over 83,372 ha [9]. Sesame is grown in both rainfed and irrigated zones of the Punjab [10]. Sesame represents itself as a rich food source due to its extraordinary oil content ranging from 34% to 59% and its nutritious, medicinal, and cosmetics qualities [11]. Due to the presence of various bioactive compounds, sesame seeds have been shown to exhibit anti-inflammatory, antioxidant, antihypertensive, wound healing, anticancer, and neuroprotective activities [12,13,14,15,16,17]. In Pakistan, sesame seed is a regular part of the local cuisine and is commonly used in the food industry and in Unani herbal medicines. Sesame seeds and sesame oil are used for cooking, salad dressing, garnish, snacks, flavoring agents, as well as for use in the manufacturing of margarine and as a raw ingredient for paints, varnishes, soaps, perfumes and insecticides [18]. Sesame consumption is steadily increasing in Pakistan as well as globally mainly due to changing consumer consumption patterns and increasing health awareness [19]. Sesame is the only oilseed crop exported by Pakistan. During 2019–2020, 32,838 tons of sesame seeds were exported from Pakistan with huge foreign revenue [20]. Sesame is important in Pakistan because of its multidimensional uses. However, contamination from fungal agricultural pests is a major concern [21]. A. flavus is the most prevalent aflatoxin producer [22]. The warm and humid conditions of Pakistan, especially in the province of the Punjab, facilitate the thriving of A. flavus, and consequently, aflatoxin contamination poses serious threats to the health of the Pakistani people [23,24]. Additionally, many Pakistani farms do not utilize proper handling practices or have proper food storage facilities. As a consequence, economic losses due to A. flavus contamination can reach up to 100% when the presence of aflatoxins is beyond acceptable levels [25,26,27].
Aflatoxin production in A. flavus is polymorphic, and the collective species are relatively genetically diverse [28,29,30,31,32]. The ability of A. flavus to produce aflatoxins is strain-specific and it is associated with the intactness of the aflatoxin biosynthesis gene cluster, which consists of 26 genes responsible for the biosynthesis and transport of aflatoxins [33,34]. Currently, there is increasing awareness of aflatoxin contamination in foods and feeds, and previous studies have reported the presence of A. flavus in sesame from Pakistan. However, no systemic studies have been carried out for the aflatoxigenic potential and genetic intactness of the aflatoxin gene cluster in the indigenous A. flavus population isolated from sesame seeds in Pakistan. Here, we assessed the contamination rates of A. flavus in sesame seeds, determined the aflatoxigenic and non-aflatoxigenic potential of 260 isolates, and carried out comparative genomic analyses of 12 select indigenous A. flavus isolates from sesame seeds produced in two Agro-ecological zones of the Punjab, Pakistan.
2. Results
2.1. Sesame Seed Samples and Isolation of Fungi Belonging to Aspergillus Section Flavi
One hundred sesame seeds samples were collected directly from the fields in two Agro-ecological zones of Punjab, Pakistan i.e., the rainfed zone (Rawalpindi, Attock and, Chakwal) and the irrigated zone (Hafizabad, Gujranwala, Gujrat, Sargodha, and Bahawalpur: Figure 1A). Representative fresh and stored sesame seed samples used for Aspergillus isolation are shown in Figure 1B.
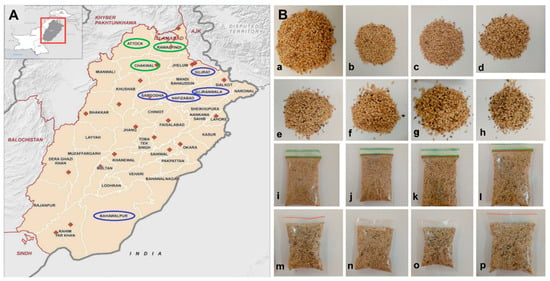
Figure 1.
Sample collection regions in Punjab and sesame seed samples. (A) A map of Punjab, Pakistan, showing the regions for collection of the sesame seed samples. Green circles indicate the districts of the rainfed, and blue circles indicates the districts of the irrigated zone. (B) Fresh sesame seeds collected from (a) Rawalpindi, (b) Attock, (c) Chakwal, (d) Hafizabad, (e) Gujranwala, (f) Gujrat, (g) Sargodha, (h) Bahawalpur, and (i–p) the sesame seeds from (a) Rawalpindi ~ (h) Bahawalpur being stored in polythene bags at room temperature.
Isolates of Aspergillus section Flavi show rapid growth on Potato Dextrose Agar (PDA). Initially, the isolates exhibited the white color of mycelia followed by the production of yellow green to olive-colored conidia within three days of incubation. The reverse side of the colonies was slightly pale. Microscopic examinations showed that the conidiophores were thick walled, non-septate, uncolored and unevenly pitted or roughened vesicles. The length of the conidiophore was about 600–800 µm and the diameter was 15–20 µm. The vesicles were sub-globose in a few isolates and globose in others. Phialides were either uniseriate, biseriate, or both. The conidia were globose with thin, slightly roughened walls. The length of the conidial head was about 20–45 µm yellow/greyish green and the diameter ranged from 2–6 µm (Figure 2A,B).
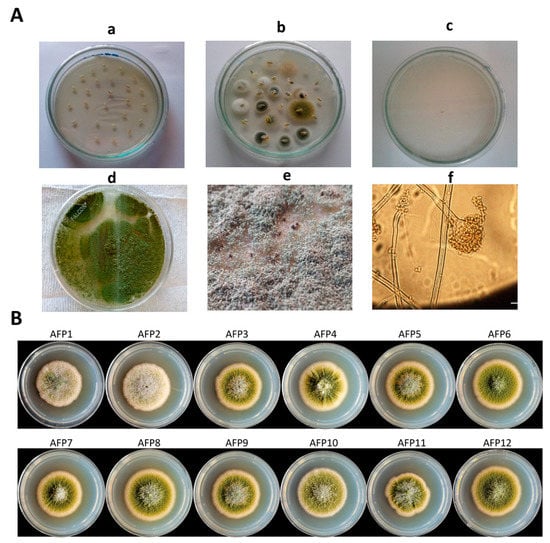
Figure 2.
Isolation and identification of fungal isolates belonging to Aspergillus section Flavi. (A) Procedure for isolation of Aspergillus section Flavi from sesame seeds (a) sesame seeds were placed on PDA, (b) fungal colonies emerging from the seeds after 3–5 days at 28 ± 2 °C under alternative cycle of darkness and light in a versatile environmental test chamber, (c) isolation of potential Aspergillus section Flavi on the basis of colony characteristics, (d) Growth of Aspergillus section Flavi on PDA, (e) Morphological appearance of Aspergillus section Flavi and brown exudates, and (f) A microscopic image of conidiophore of Aspergillus section Flavi. (B) Colonies of the 12 isolates of Aspergillus section Flavi selected for whole genome sequencing. Each isolate was point inoculated on the center of PDA and the colony photographs were taken at four days post incubation at 30 °C. Initially, the isolates exhibited the white color of mycelia followed by production of yellow green to olive-colored conidia within three days.
Out of all the molds in fresh and stored sesame seeds, 260 isolates were identified as potential Aspergillus section Flavi based on macro-morphological and micro-morphological characteristics. The present study tested the hypothesis that differences exist between fresh and stored sesame seeds derived from rainfed and irrigated zones in terms of contamination with Aspergillus section Flavi. Isolation frequency (%) and relative density (%) of Aspergillus section Flavi were calculated and are presented in Table 1.

Table 1.
Isolation frequency (%) and relative density (%) of Aspergillus section Flavi isolates in fresh and stored sesame seeds from two Agro-ecological (rain-fed and irrigated) zones of the Punjab, Pakistan.
In the rainfed zone, 126 isolates were identified as potential Aspergillus section Flavi including 34 isolates from fresh and 92 isolates from stored sesame seeds. It appears that stored samples were more contaminated with Aspergillus than fresh samples with a percentage incidence of about 100% and 80%, respectively. Stored samples from all the collection sites showed 100% occurrence of Aspergillus section Flavi while in fresh sesame seeds, and the highest incidence was reported from Rawalpindi (100%) followed by Attock (80%), and Chakwal (60%) (Table 1).
In the irrigated zone, 134 isolates were identified as potential Aspergillus section Flavi, which include 56 from fresh and 78 from stored sesame seeds. Stored samples were more contaminated than fresh samples, with the percentage incidence of about 93.32% and 71.84%, respectively. Stored samples from all the collection sites showed 100% occurrence except for Bahawalpur with 66.6% incidence. In the case of fresh samples, the highest incidence was reported from Sargodha and Gujranwala with 100% occurrence and the least was reported from Gujrat with 40% incidence (Table 1).
2.2. Examination of the Aflatoxigenic Potential of 260 Aspergillus Section Flavi Isolates
We then tested all 260 potential Aspergillus section Flavi isolates for their ability to produce aflatoxins in vitro employing HPLC. As shown in Table 2, 188 (72.31%) isolates produced aflatoxins whereas no aflatoxins were detectable from cultures of 72 (27.69%) isolates (Figure 3A,B). From aflatoxigenic isolates, 88.3% isolates were reported to produce AFB1, 39.89% isolates produced AFB2, 5.85% isolates produced AFG1, and 3.19% isolates were reported to produce AFG2 (Figure 3C). The average and range of AFB1, AFB2, AFG1, and AFG2 are presented in Table 3. HPLC chromatograms of select A. flavus isolates are shown in Supplementary Figure S1.

Table 2.
Aflatoxigenic potential of 260 isolates of Aspergillus section Flavi.
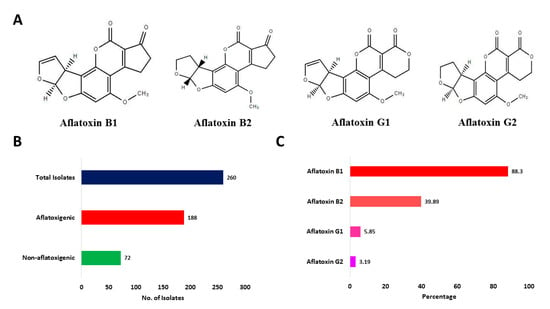
Figure 3.
Aflatoxin production potential of Aspergillus section Flavi isolates. (A) Chemical structures of AFB1, AFB2, AFG1, and AFG2. (B) Total number of aflatoxigenic and non-aflatoxigenic isolates of Aspergillus section Flavi isolated from fresh and stored sesame seeds. (C) Percentage of aflatoxigenic isolates producing AFB1, AFB2, AFG1, and AFG2.

Table 3.
Average and range of AFB1, AFB2, AFG1 and AFG2 of aflatoxigenic isolates of Aspergillus section Flavi in semi-solid (slant) culture condition.
In the rainfed zone, 92 (73.02%) isolates were found to be aflatoxigenic (20 isolates from fresh and 72 isolates from stored sesame seeds). However, no aflatoxins were detected in the cultures of 34 (26.98%: 14 isolates from fresh and 20 isolates from stored sesame seeds). The chances of getting aflatoxigenic isolates of Aspergillus section Flavi were significantly (p < 0.05) higher in stored samples as compared to fresh samples with frequencies of 78.26% and 58.82%, respectively. Moreover, the occurrence of aflatoxigenic Aspergillus section Flavi isolates was significantly (p < 0.05) higher in stored samples compared to fresh samples in all collection sites including Rawalpindi, Attock, and Chakwal (Table 2).
In the irrigated zone, 96 (71.64%) isolates were found to be aflatoxigenic (32 isolates from fresh and 64 isolates from stored sesame seeds) and 38 (28.36%) isolates were reported as non-aflatoxigenic (24 isolates from fresh and 14 isolates from stored sesame seeds). The occurrence of aflatoxigenic Aspergillus section Flavi isolates was significantly (p < 0.05) higher in stored samples as compared to fresh samples with the frequency of 82.05% and 57.14%, respectively. Moreover, the chances of getting aflatoxigenic Aspergillus section Flavi isolates was significantly (p < 0.05) higher in stored samples compared to fresh samples in Gujranwala, Gujrat, Sargodha, and Bahawalpur, except for Hafizabad (p ˃ 0.05) (Table 2).
2.3. Whole Genome Sequencing and Comparative Analyses of 12 Pakistani Isolates
To understand the genetic and genomic bases of the differences in their aflatoxigenic potential, 12 isolates (see Figure 2B) of Aspergillus section Flavi were chosen for the whole-genome sequencing analyses. As shown in Table 4, we selected six high producers of aflatoxins (AFP1~AFP6), and three were medium aflatoxin producers (AFP7~AFP9) and three were very low aflatoxin producers (AFP10~AFP12). Whole-genome sequencing data were deposited on the NCBI SRA under “Aspergillus flavus, Pakistani isolates Genome sequencing” (Accession No. PRJNA682409). Each genome was sequenced to a coverage of 25x~37x. The genome sequences of all 12 isolates highly matched to that of the reference A. flavus NRRL3357, confirming that all these isolates belong to A. flavus species (Supplementary Table S1). As shown in Figure 2, the 12 isolates have different developmental phenotypes.

Table 4.
Molecular identification and accession numbers of the genomes of the 12 isolates.
2.3.1. Phylogenetic Analysis
We inferred the evolutionary history of the 12 Pakistani A. flavus isolates along with 94 isolates of A. flavus strains from the United States that had been previously sequenced by Drott et al. [31]. Neighbor-net phylogenetic network analysis of 3581 SNPs spaced by a minimum of 10 kb revealed three major populations, corroborating the findings of Drott et al. [31]. All 12 Pakistani isolates are nested within population A (Figure 4). Branch lengths leading to terminal taxa are longer in population A compared with populations B and C, suggesting that population A has higher levels of genetic diversity (Figure 4).
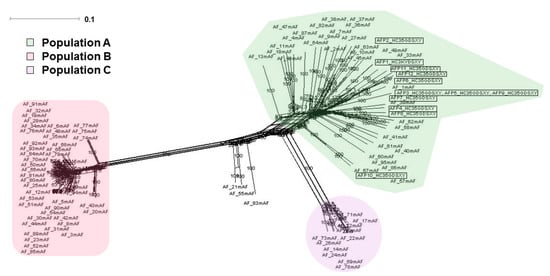
Figure 4.
Phylogenetic network analysis of the 12 Pakistani A. flavus isolates with 1000 bootstraps along with 94 isolates of A. flavus. Neighbor-net phylogenetic network analysis of 3581 SNPs spaced by a minimum of 10 kb revealed three major populations. All Pakistani isolates (marked by box) are nested within population A. Branch lengths suggest that population A has higher levels of genetic diversity.
2.3.2. Nucleotide Variation Analysis
To gain insights into associations between nucleotide variation and aflatoxin production, we calculated nucleotide diversity (π) within the Pakistani isolates across each gene in the aflatoxin gene cluster (norB − hypA) and two housekeeping genes encoding beta-tubulin and calmodulin. The highest levels of nucleotide diversity were present in avfA (π = 0.022), omtB (π = 0.022), and verB (π = 0.021). The lowest levels of nucleotide variation were observed in beta-tubulin (π = 0.0002), norB (π = 0.0007), aflT (π = 0.0009), pksA (π = 0.0009) and fas-1 (π = 0.001) (Figure 5A). Additionally, we quantified the missense variant rate for each isolate in each gene in the aflatoxin cluster as well as the housekeeping genes beta-tubulin and calmodulin. Only the isolates AFP8, AFP4, and AFP10 had missense variant rates greater than 0.6 (AFP8 = 1.98, AFP4 = 2.01, and AFP10 = 4.13). These isolates all possessed missense variant rates greater than 5 in avfA, verB, omtB and verA. AFP10 also displayed missense variant rates higher than 5 in pksA, fas2, hypA, omtA, and fas1 (Figure 5B).
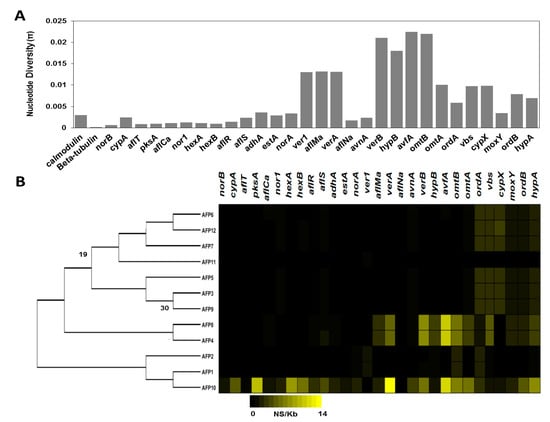
Figure 5.
Nucleotide diversity and missense variant rate of 12 isolates. (A) Nucleotide diversity (π) in the 12 Pakistani A. flavus isolates across the aflatoxin cluster genes and the housekeeping genes calmodulin and beta-tubulin. (B) Missense variant rate across genes in the aflatoxin gene cluster. Black represents a missense variant rate of 0, while bright yellow represents a missense variant rate of 14. A. flavus isolates are depicted based on their phylogenetic relationship from a maximum likelihood tree. The tree is rooted at the midpoint, and bootstrap values are shown when below 80.
2.3.3. Prediction of High Impact Mutations
Furthermore, we predicted putative high impact mutations as defined by SnpEff [35] in each isolate in all genes in the aflatoxin gene cluster. Interestingly, only AFP10 possessed SNP-based high impact mutations. AFP10 contained a premature stop codon in hypB and cypA, as well as a splice donor variant in cypA (Table 5). Additionally, AFP1, AFP10, AFP11, AFP2, AFP4 and AFP8 contained INDEL-based high impact mutations in aflatoxin genes. AFP10 possessed five high impact INDEL mutations (in hypA, avfA, verA, aflT, and cypA) (Table 5).

Table 5.
High impact mutations in all genes in the aflatoxin gene cluster in 12 isolates.
2.3.4. Copy Number Variation in the Aflatoxin Cluster Locus
Lastly, we predicted copy number variation across non-overlapping 100 bp bins in all isolates across the aflatoxin cluster locus using a read depth approach. This analysis allowed us to identify large-scale deletions. We observed a ~600 bp deletion in norB and a smaller deletion in aflT in AFP10 (Figure 6). The high missense variant rate, presence of numerous high impact mutations, and presence of large deletions in the aflatoxin gene cluster in AFP10 coincides with very low aflatoxin production.
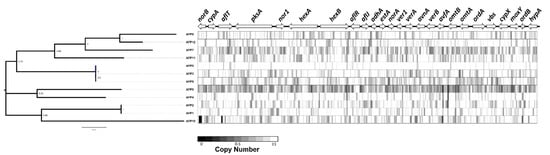
Figure 6.
Copy number variation and deletions across the aflatoxin cluster locus of 12 isolates.
3. Discussions
Microorganisms can grow on different commodities in fields, during harvesting, transportation, and storage. As filamentous fungi (molds) are ubiquitous and their spores can survive for several years in commodities, therefore, some careful measurements should be taken during storage [36]. In the present study, we have demonstrated that the Pakistani sesame seeds derived from Punjab were highly contaminated with the aflatoxigenic isolates of Aspergillus section Flavi in fresh and stored conditions. The primary reason for this high incidence of Aspergillus section Flavi is that the samples were collected from the areas where average temperature was 23~30 °C, an optimum range for A. flavus proliferation [37]. The hot and humid climatic conditions of rainfed and irrigated zones of Punjab, Pakistan, temperature ideal for fungal contamination, coupled with other factors including agronomic practices, post-harvest treatment (processing, drying, storage), and duration of storage may have contributed to the fungal growth, development, and secondary metabolite formation in agricultural products [38,39,40,41,42].
In Pakistan, limited studies have been carried out on the incidence of A. flavus in sesame seeds. In agreement with the present study, previous studies reported the aflatoxigenic potential of A. flavus and found that Aspergillus section Flavi members were common colonizers of all types of sesame seeds during post-harvest storage, [43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67]. In one study, 17 sesame samples were collected from Plateau State, Nigeria and were investigated for frequency of fungal species, especially for the Aspergillus species which revealed the highest frequency of A. flavus [68]. Ninety-one isolates of A. flavus derived from 63 samples of sesame collected from North Algeria were tested for their ability to produce aflatoxin and 23.52% of strains were aflatoxigenic [69]. A. flavus isolated from sesame seeds were aflatoxigenic, as reported by Sabry et al. [70]. In one study, sesame seeds were purchased from local markets of Nigeria and analyzed for fungal and mycotoxin contamination. The molecular analysis reported the presence of Aspergillus candidus, Aspergillus flavus, Aspergillus niger, Cladosporium spp., Fusarium fujikuroi, Penicillium spp., and Pleosporales/Didymellaceae spp. in the sesame seeds. The most frequent mycotoxin in the sesame seeds was AFB1 with the occurrence of 76% [71]. In the study by Abbas et al. [72], four sesame varieties were planted in the Mississippi Delta at four nitrogen fertilizer application rates from 44.8 to 112 kg N/ha for the evaluation of grain yields and contamination of mycotoxins, which revealed that N fertilizer application rate had no effect on yield or mycotoxin contamination in 2014, but significantly increased yield in 2015. Some studies from Pakistan reported the presence of A. flavus in sesame. Sesame seeds were collected from the National Agriculture Research Council and tested for the presence of mycoflora, which revealed that A. flavus, A. niger, and Fusarium oxysporum were predominant [73]. Sesame seeds from various areas of Sialkot, Pakistan was studied for mycoflora. A total number of 36 species belonging to 10 genera of fungi were isolated. The prevalent genera were Fusarium, Penicillium, Cercospora, Alternaria, and Cladosporium, followed by Aspergillus [74]. All the above-mentioned studies undoubtedly revealed that sesame is a highly threatened commodity with A. flavus.
In the present study, whole genome analyses have revealed that one isolate (AFP10; a very low aflatoxin producing isolate) has SNP-based high impact mutations, five high impact INDEL mutations (in hypA, avfA, verA, aflT, and cypA), and a 600 bp deletion in norB. Previous studies reported deletions in the norB–cypA region and several other large deletions in the aflatoxin-biosynthesis gene cluster in A. flavus [75,76,77,78,79]. The present study was also supported by previous reports in which 281 isolates of A. flavus were tested for aflatoxin production. The population was subdivided into two genetically different populations (A and B) which differ in allelic and genotypic diversity. The less diverse population was more abundant and may represent a clonal lineage derived from more diverse populations [80]. In a study, 94 isolates of A. flavus were sampled in the eastern and central latitudinal transects from seven states of the United States. The total population was divided into three genetically distinct populations (A, B, and C), which vary greatly in their ability to generate recombination, diversity, and to produce aflatoxin. Population B is sympatric with population A but produces substantially less aflatoxin and is the only population where multiple gene deletions have clarified the inability of non-aflatoxigenic isolates to produce aflatoxin. Population C is predominantly non-aflatoxigenic [31]. The present study was also confirmed by Adhikari et al. [81] In this study, clusters of aflatoxin genes from 35 genotypes were analyzed, showing a high degree of variations in terms of the amount and size of the gene deletions. Genotypes varied from those with a complete aflatoxin gene cluster to those with no genes at all, with most deletions occurring at the left end of the cluster or towards the telomeric end, depending on the size and type of deletions. In one study, isolates of A. flavus were tested for the presence/intactness of the aflD and aflQ genes. Any mutation or variation in these genes results in low aflatoxin production [82].
4. Conclusions
This study provides information on the aflatoxigenic potential and genetic diversity of certain isolates of Aspergillus section Flavi obtained from fresh and stored sesame seeds grown in Punjab, Pakistan. Punjab is a major sesame seed producing area of Pakistan and the world. Whole genome analyses of 12 aflatoxigenic isolates have revealed that the highest levels of nucleotide diversity were present in avfA, followed by omtB and verb. AFP10 (very low aflatoxin producing isolate) possessed SNP-based high impact mutations, five high impact INDEL mutations (in hypA, avfA, verA, aflT, and cypA), and a 600 bp deletion in norB and a smaller deletion in aflT. The high missense variant rate, the presence of numerous high impact mutations and large deletions in the aflatoxin gene cluster in AFP10 explains very low aflatoxin production. The identification of mutation and deletion in genes significantly affecting the resistance to aflatoxin accumulation would accelerate the development of resistant strains native to local agricultural areas, thus requiring more investigations.
5. Materials and Methods
5.1. Study Area and Sesame Seeds Samples Collection
The study was carried out in two Agro-ecological zones (rainfed and irrigated) of Punjab, Pakistan. One hundred sesame seeds samples were collected directly from the field in major sesame producing areas of rainfed (Rawalpindi, Attock and, Chakwal) and irrigated (Hafizabad, Gujranwala, Gujrat, Sargodha, and Bahawalpur) zones during the harvest season (Figure 1A). Samples were taken in the Mycology Laboratory, Department of Botany, Pir Mehr Ali Shah Arid Agriculture University Rawalpindi, Pakistan and subjected to the Agar Plate Method for the isolation of Aspergillus section Flavi. After that, sesame seeds samples were stored for 12 months at room temperature (Figure 1B). After 12 months, seeds were again analyzed for Aspergillus section Flavi contamination.
5.2. Isolation and Morphological Identification of Aspergillus Section Flavi
Seeds for isolating fungi were surface sterilized with 2% aqueous sodium hypochlorite (NaOCl) for two minutes. Potato Dextrose Agar (PDA, Oxoid, UK) (containing 4 g potato starch, 20 g glucose and 15 g agar) was prepared, 0.1 mg/mL streptomycin was added, and the culture media was poured in to 9 cm diameter petri dishes [83]. About 25 sesame seeds were placed on PDA and incubated for 3–5 days at 28 ± 2 °C under alternative cycles of darkness and light in a versatile environmental test chamber (Sanyo, Japan), with illumination provided by 55 W fluorescent tubes (125–130 μmolm−2s−1) [84]. The experiment was performed in triplicate. After 3–5 days, seeds were investigated under a light microscope (Eclips 80i, NIKON, Tokyo, Japan), and isolates of Aspergillus section Flavi arising from the seeds were counted. Isolates were maintained on fresh PDA and identified based on macro-morphological and micro-morphological characteristics (color and texture of colonies and morphology of conidial head, stipes, hyphal color, conidia size, color, length, shape of vesicle and metula produced on PDA) based on published procedure [46,47,85,86,87].
5.3. Aflatoxigenic and Non-Aflatoxigenic Potential of Aspergillus Section Flavi
All the isolates of Aspergillus section Flavi isolated from fresh and stored sesame seeds were analyzed for their aflatoxigenic and non-aflatoxigenic potential using the methodology described by Alshannaq et al. [88]. Individual aflatoxin standards for AFB1, AFB2, AFG1 and AFG2 were purchased from Sigma Chemical Co (St. Louis, MO, USA). As a positive control, the aflatoxigenic strain A. flavus NRRL 3357 and as a negative, A. oryzea NRRL 2999 was used.
5.3.1. Fungal Culture Preparations and Aflatoxin Extraction
Isolates of Aspergillus section Flavi were cultured into semi-solid (slant), solid and submerged conditions. For the semi-solid (slant) culture, 2 mL of Potato Dextrose Broth (PDB, Difco Lab, Sparks, MD, USA) was added in 25 mL glass test tubes, and fungal isolates (5 × 105 conidia/tube) were inoculated with the help of an inoculating loop. The tubes were put in a rack at a 45°angle and placed in an incubator at 28 ± 2 °C for seven days. For solid culture, fungal isolates (5 × 105 conidia/petri dish) were inoculated in petri dishes containing 25 mL of PDA (Difco Lab, Sparks, MD, USA) and placed in an incubator at 28 ± 2 °C for seven days. For submerged culture, fungal isolates at 5 × 105 conidia/flask were inoculated in 250 mL Erlenmeyer flasks containing 100 mL of PDB. All the flasks were incubated for seven days at 28 ± 2 °C with shaking at 220 rpm. After 7 days of incubation, aflatoxins were extracted from semi-solid, solid, and submerged cultures of fungal isolates as described in Alshannaq et al. [88]. Prior to HPLC analysis, all samples were filtered (0.45 mm with a diameter of 47 mm) (Thermo Fisher Science, Rockwood, TN, USA) into HPLC vials via a PTFE 0.45 μm syringe filter (0.45 mm with a diameter of 17 mm) (Thermo Fisher Science, Rockwood, TN, USA).
5.3.2. HPLC Analysis of Aflatoxins
The samples for AFB1, AFB2, AFG1, and AFG2 were analyzed using a Model 1100 HPLC device containing degasser, an autosampler, a quaternary pump fitted with a 1260 Infinity diode array (DAD), and a 1260 Infinity II fluorescence detector (FLD) connected in series (Agilent Technologies, Santa Clara, CA, USA). Samples were monitored for UV detection at a 365 nm wavelength and for FLD detection at 365 nm excitation and 450 nm emission. The samples were eluted with a mobile phase of water (H2O)/methanol (CH3OH)/acetonitrile (CH3CN) (50:40:10) at a flow rate of 0.8 mL/min. Before use, the mobile phase was degassed and purified through a membrane filter (47 mm, 0.45 µm). The injection volume was 100 μL. By using the Chem Station software, peak areas of aflatoxins were obtained and integrated (Agilent Technologies, Santa Clara, CA, USA).
5.4. Molecular Characterization
Twelve isolates of A. flavus were selected for molecular characterization based on aflatoxin producing ability. Six isolates were high aflatoxin producers, three isolates were medium producers and three were very low aflatoxin producers. The molecular characterization of selected isolates of A. flavus was performed by Whole Genome Sequencing (WGS) and their phylogenetic analysis was performed.
5.4.1. DNA Extraction
DNA of A. flavus isolates was extracted as described in Lee et al. [89]. DNA was extracted by using a commercially available DNA extraction kit (DNeasy® Plant Mini column kit, Qiagen, UK) by following the manufacturer’s instructions. The presence and quality of pure genomic DNA were confirmed through a 1% (w/v) agarose gel electrophoresis, and analysis was conducted at 110 V for 35 min. The gel was then viewed under a UV Transilluminator (Biorad Gel DocTM XR + Philadelphia, PA, USA) to confirm the presence of high molecular weight genomic DNA. The eluted DNA was stored at −80 °C until further molecular identification assays.
5.4.2. Genome Sequencing and Quality Control
Illumina libraries were constructed and sequenced by Novogene USA. Illumina sequencing was conducted in a paired-end 150 bp format with a 350 bp insert size. Raw reads were deduplicated using Tally with the “with-quality” and “pair-by-offset” parameters to remove exact duplicates [90]. The number of filtered read pairs and the % of reads that mapped against the A. flavus NRRL 3357 reference genome were presented in (supplementary Table S1). Next, deduplicated reads were trimmed with Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ accessed on 2 November 2020) using the “stringency 1”, “quality 30” and “length 50” parameters to remove residual adaptor sequences and low-quality positions.
5.4.3. Variant Calling and Annotation
Deduplicated, adapter and quality trimmed paired end reads from the 12 A. flavus isolates from Pakistan and the 94 publicly available A. flavus genomes previously sequenced by Drott et al. [31] were mapped against the A. flavus NRRL 3357 reference genome using BWA-MEM v0.7.15 [91,92]. SNPs and INDELs were called using freebayes v1.3.1 with the default settings with the exception of setting ploidy to haploid (--ploidy = 1) [93]. vcftools v0.1.14 was then used to filter variants with the following parameters: “--remove-filtered-all”, “--minQ 20”, “--recode” and “--recode-INFO-all” [94]. SNPs and INDELS from the filtered VCF file were annotated with SnpEff v4.3t using “Aspergillus flavus” as the genome database [35].
5.4.4. Phylogenetic Analysis
We conducted phylogenetic network analysis to infer the evolutionary relationship of the 12 A. flavus isolates from Pakistan in relation to the 94 A. flavus isolates previously sequenced by Drott et al. [31]. First, we used vcftools to ensure SNP markers were spaced by a minimum of 10 Kb in an effort to limit bias introduced by linkage. After filtering, 3581 SNPs remained, and an alignment of these sites were used to construct the neighbor-net phylogenetic network using Splitstree v4.16.1 [95] with 1000 bootstrap replicates. Additionally, a maximum likelihood phylogenetic tree was constructed in MEGA [96] with the 12 A. flavus isolates from Pakistan, using the Tamura-Nei model with 100 bootstrap replicates.
5.4.5. Genomic Analysis of Aflatoxin Loci
First, we used vcftools in haploid mode (“--haploid” option) to calculate nucleotide diversity (π) for each gene in the aflatoxin cluster. Next, using the SnpEff output, we calculated the missense variant rates for each gene in the aflatoxin cluster to identify genes with relatively elevated occurrences of missense variants. The er gene missense variant rate was calculated as:
Finally, we used the samtools depth function on sorted bam files to generate coverage values for each site in the aflatoxin locus as described previously by Alshannaq et al. [97]. Average coverage values for each non-overlapping 100 bp portion of the aflatoxin gene cluster were divided by the average coverage across the entire genome to estimate the copy number. Per genome average coverage was estimated by summing the samtools depth output and dividing by the A. flavus NRRL 3357 genome size (36,892,344 bp). Bins with coverage values of 0 represent deletions.
5.5. Statistical Analysis
Data were summarized and analyzed by using SPSS (version 16.0; SPSS Inc., Chicago, IL, USA). The isolation frequency and relative density of Aspergillus section Flavi were calculated by using the following formula.
where ns is the number of samples on which a fungus (Aspergillus section Flavi) occurred, N is the total number of seed sampled, ni is the number of isolates of a fungal genus/species (Aspergillus section Flavi), and Ni is the total number of fungal isolates obtained. Two-way Analysis of Variance (ANOVA) was performed with significant (p < 0.05) to determine the significant difference of aflatoxigenic potential of Aspergillus section Flavi isolates among the fresh and stored sesame seed samples obtained from rainfed and irrigated zones of the Punjab, Pakistan.
Supplementary Materials
The following are available online at www.mdpi.com/article/10.3390/toxins14020117/s1, Figure S1: High-performance liquid chromatography (HPLC) chromatograms of Aflatoxin B1 (AFB1). (A) Combined HPLC chromatograms for high producers AFP1 ~ AFP6. (B) HPLC chromatograms for high producers AFP1 ~ AFP6. (C) HPLC chromatograms for select medium and low producers, Table S1: Read mapping statistics for 12 AFP genomes with the A. flavus NRRL 3357 reference genome.
Author Contributions
Conceptualization, A.A. and M.A.; Methodology, J.-H.Y., A.F.A., B.G.N. and A.A.; Performed the experiments, M.A.; Supervision, J.-H.Y. and A.F.A.; Genomic Analysis, J.G.G. and H.M.; writing—original draft preparation, M.A.; writing—review and editing, M.A., J.-H.Y. and D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research is funded by Higher Education Commission (HEC) Pakistan under International Research Support Initiative Program IRSIP No. 1-8/HEC/HRD/2019/8790. The work is based upon work support by the National Institute of Food and Agriculture, United States Department of Agriculture, Hatch project 7000326. JGGs (U-Mass) contribution to this work is supported by the National Science Foundation under Grant No. 1942681.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We highly acknowledge United States Department of Agriculture (USDA), Animal and Plant Health Inspection Service Plant Protection & Quarantine for providing the import license for 260 isolates of Aspergillus section Flavi from Pakistan to USA under permit number P526P-20-00119.
Conflicts of Interest
The authors declare that they have no conflicts of interests.
References
- Diedhiou, P.M.; Bandyopadhyay, R.; Atehnkeng, J.; Ojiambo, P.S. Aspergillus colonization and aflatoxin contamination of maize and sesame kernels in two Agro-ecological zones in Senegal. J. Phytopathol. 2011, 159, 268–275. [Google Scholar] [CrossRef]
- Enyiukwu, D.N.; Awurum, A.N.; Nwaneri, J.A. Mycotoxins in stored agricultural products: Implications to food safety and health and prospects of plant–derived pesticides as novel approach to their management. Greener J. Microbiol. Antimicrob. 2014, 2, 32–48. [Google Scholar] [CrossRef]
- Richard, J.L. Some major mycotoxins and their mycotoxicosis—An overview. Int. J. Food Microbiol. 2007, 119, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Shivachandra, S.B.; Sah, R.L.; Singh, S.D.; Kataria, J.M.; Manimaran, K. Immunosuppression in broiler chicks fed aflatoxin and inoculated with fowl adenovirus serotype-4 (FAV-4) associated with hydropericardium syndrome. Vet. Res. Commun. 2003, 27, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Shuaib, F.M.; Jolly, P.E.; Ehiri, J.E.; Yatich, N.; Jiang, Y.; Funkhouser, E.; Person, S.D.; Wilson, C.; Ellis, W.O.; Wang, J.S.; et al. Association between birth outcomes and aflatoxin B1 biomarker blood levels in pregnant women in Kumasi, Ghana. Trop. Med. Int. Health. 2010, 15, 160–167. [Google Scholar] [CrossRef]
- Quezada, T.; Cuellar, H.; Jaramillo-Juarez, F.; Valdivia, A.G.; Reyes, J.L. Effects of aflatoxin B1 on the liver and kidney of broiler chickens during development. Comp. Biochem. Physiol. C-Pharmacol. Toxicol. Endocrinol. 2000, 125, 265–272. [Google Scholar] [CrossRef]
- Henry, S.H.; Bosch, X.F.; Bower, J.C. Mycotoxins and food safety. Adv. Exp. Med. Biol. 2002, 504, 229–233. [Google Scholar]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food. Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef]
- Food and Agriculture Organization Statistical Databases (FAOSTAT). 2019. Available online: http://faostat.fao.org (accessed on 20 October 2020).
- Amjad, M. Oilseed crops of Pakistan. Plant Sciences Division, Pakistan Agricultural 3 Research Council, Islamabad. 2014. Available online: http://www.parc.gov.pk/files/parc_pk/January–415/Status%20Papers/Status%20Paper%20(Oilseed%20Crops)%202014.pdf.5 (accessed on 4 August 2015).
- Morris, J.B. Characterization of sesame (Sesamum indicum L.) germplasm regenerated in Georgia, USA. Genet. Resour. Crop. Evol. 2009, 56, 925–936. [Google Scholar] [CrossRef]
- Khan, M.M.; Ishrat, T.; Ahmad, A.; Hoda, M.N.; Khan, M.B.; Khuwaja, G. Sesamin attenuates behavioral, biochemical and histological alterations induced by reversible middle cerebral artery occlusion in the rats. Chem. Biol. Interact. 2010, 183, 255–263. [Google Scholar] [CrossRef]
- Miyawaki, T.; Aono, H.; Toyoda-Ono, Y.; Maeda, H.; Kiso, Y.; Moriyama, K. Antihypertensive effects of sesamin in humans. J. Nutr. Sci. Vitaminol. 2009, 55, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Kiran, K.; Asad, M. Wound healing activity of Sesamum indicum L seed and oil in rats. Ind. J. Experi. Biol. 2008, 46, 777–782. [Google Scholar]
- Yokota, T.; Matsuzaki, Y.; Koyama, M.; Hitomi, T.; Kawanaka, M.; Enoki-Konishi, M. Sesamin, a Lignan of Sesame, Down-Regulates Cyclin D1 Protein Expression in Human Tumor Cells. Cancer Sci. 2007, 98, 1447–1453. [Google Scholar] [CrossRef] [PubMed]
- Jeng, K.C.; Hou, R.C.; Wang, J.C.; Ping, L.I. Sesamin inhibits lipopolysaccharide-induced cytokine production by suppression of p38 Nitrogen-activated protein Kinase and Nuclear factor-KappaB. Immunol. Lett. 2005, 97, 101–106. [Google Scholar] [CrossRef]
- Nakano, D.; Itoh, C.; Ishii, F.; Kawanishi, H.; Takaoka, M.; Kiso, Y.; Tsuruoka, N.; Tanaka, T.; Matsumura, Y. Effects of Effects of sesamin on aortic oxidative stress and endothelial dysfunction in deoxycorticosterone acetate-salt hypertensive rats. Biol. Pharm. Bull. 2003, 26, 1701–1705. [Google Scholar] [CrossRef]
- Quasem, J.M.; Mazahreh, A.S.; Abu-Alruz, K. Development of vegetable-based milk from decorticated sesame (Sesamum indicum). Am. J. Appl. Sci. 2009, 6, 888–896. [Google Scholar] [CrossRef]
- Mordor Intelligence. Global Sesame Seeds Market-Segmented by Geography-Growth, Trends, and Forescast. 2019. Available online: https://www.mordorintellignece.com/industry-reports (accessed on 11 November 2019).
- Food and Agriculture Organization (FAOSTAT) 2019–2020. Available online: https://www.fao.org/faostat/en/#home (accessed on 31 January 2022).
- Ojiambo, P.S.; Mibey, R.K.; Narla, R.D.; Ayiecho, P.O. Field transmission efficiency of Alternaria sesame in sesame from infected seed. Crop. Protec. 2003, 22, 1107–1115. [Google Scholar] [CrossRef]
- Hathout, A.S.; Aly, E.S. Biological detoxification of mycotoxins: A review. Annu. Microbiol. 2014, 64, 905–919. [Google Scholar] [CrossRef]
- Mobeen, A.K.; Aftab, A.; Asghar, A.; Zuzzer, A.S. Aflatoxins B1 and B2 Contamination of Peanut and Peanut Products and Subsequent Microwave Detoxification. J. Pharm. Nutr. Sci. 2011, 1, 1–3. [Google Scholar] [CrossRef]
- Nizami, H.M.; Zuberi, S.J. Aflatoxin and cancer in Karachi, a preliminary survey. J. Pak. Med. Assoc. 2004, 54, 351–352. [Google Scholar]
- Kumar, P.; Mahato, D.K.; Kamle, M.; Mohanta, T.K.; Kang, S.G. Aflatoxins: A global concern for food safety, human health and their management. Front. Microbiol. 2017, 7, 2170. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar-Mathur, P.; Sunkara, S.; Bhatnagar-Panwar, M.; Waliyar, F.; Sharma, K.K. Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 2015, 234, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.W.; Greene, R.L.; Dorner, J.W.; Sobolev, V.S.; Powell, J.H.; Layton, R.C. Association of morphology and mycotoxin production with vegetative compatibility groups in Aspergillus flavus, Aspergillus parasiticus and Aspergillus tamari. Mycologia 1996, 88, 574–587. [Google Scholar] [CrossRef]
- Bayman, P.; Cotty, P.J. Genetic diversity in the Aspergillus flavus: Association with aflatoxin production and morphology. Can. J. Bot. 1993, 71, 23–31. [Google Scholar] [CrossRef]
- Gibbons, J.G.; Salichos, L.; Slot, J.C.; Rinker, D.C.; McGary, K.L.; King, J.G.; Klich, M.A.; Tabb, D.L.; McDonald, W.H.; Rokas, A. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr. Biol. 2012, 22, 1403–1409. [Google Scholar] [CrossRef] [PubMed]
- Drott, M.T.; Satterlee, T.R.; Skerker, J.M.; Pfannenstiel, B.T.; Glass, N.L.; Keller, N.P.; Milgroom, M.G. The frequency of sex: Population genomics reveals differences in recombination and population structure of the aflatoxin-producing fungus Aspergillus flavus. Mbio 2020, 11, e00963-20. [Google Scholar] [CrossRef]
- Taghizadeh-Armaki, M.; Hedayati, M.T.; Ansari, S.; Omran, S.M.; Saber, S.; Rafati, H.; Zoll, J.; Van Der Lee, H.A.; Melchers, W.J.; Verweij, P.E.; et al. Genetic diversity and in vitro antifungal susceptibility of 200 clinical and environmental Aspergillus flavus isolates. Antimicrob. Agents Chemother. 2017, 61, e00004-17. [Google Scholar] [CrossRef]
- Payne, G.A.; Yu, J.; Nierman, W.C.; Machida, M.; Bhatnagar, D.; Cleveland, T.E.; Dean, R.A. A first glance into the genome sequence of Aspergillus flavus. In The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods; Osmani, S.A., Goldman, G.H., Eds.; CRC Press: Boca Raton, FL, USA, 2008; pp. 15–23. [Google Scholar]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.Y.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Klich, M.A. Identification of Common Aspergillus Species; Centraal Bureau Voor Schimmel Cultures: Utrecht, The Netherlands, 2002; 116p. [Google Scholar]
- Cotty, P.J.; Jaime-Garcia, R. Influences of cli-mate on aflatoxin producing fungi and aflatoxin contamination. Int. J. Food Microbiol. 2017, 119, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Khamees, M.A.F.; Schlosser, E. Seed-borne fungi on sesame in the Sudan. Mededelingen van de Faculteit Landbouwwetenschappen. Rijksuniversiteit Gent. 1990, 55, 877–887. [Google Scholar]
- Mbah, M.C.; Akueshi, C.D. Effect of Seed Borne Fungi Aspergillus flavus and A. niger on the germinability of sesame seeds. Niger J. Hort. Soc. 2000, 4, 57–64. [Google Scholar]
- Mbah, M.C.; Akueshi, C.O. Aflatoxin in Mould Infested Sesame Seeds. Afr. J. Biotechnol. 2009, 8, 391–394. [Google Scholar]
- Makun, H.A.; Gbodi, T.A.; Tijani, A.S.; Abai, A.; Kadir, G.U. Toxicological screening of fungi isolated from millet (Pennisetum spp.) during the rainy and dry Harmattan seasons in Niger State, Nigeria. Afr. J. Biotech. 2007, 6, 34–40. [Google Scholar]
- Amadi, J.E.; Adeniyi, D.O. Mycotoxin production by fungi isolated from stored grains. Afr. J. Biotech. 2009, 8, 1219–1221. [Google Scholar]
- Alwakeel, S. Molecular identification of isolated fungi from stored apples in Riyadh, Saudi Arabia. Saudi. J. Biol. Sci. 2013, 20, 311–317. [Google Scholar] [CrossRef]
- Bandh, S.; Kamili, A.; Ganai, B. Identification of some Aspergillus species isolated from Dal Lake, Kashmir by traditional approach of morphological observation and culture. Afr. J. Microbiol. Res. 2012, 6, 5824–5827. [Google Scholar]
- Morya, V.K.; Yadav, D. Diversity of indigenously isolated Aspergilli from soil of monoculture teak forest. Res. J. Soil Biol. 2009, 1, 77–83. [Google Scholar]
- Diba, K.; Kordbacheh, P.; Mirhendi, H.; Rezaie, S.; Mahmoud, M. Identification of Aspergillus species using morphological characteristics. Pak. J. Med. Sci. 2007, 23, 867–872. [Google Scholar]
- Mcclenny, N. Laboratory detection and identification of Aspergillus species by microscopic observation and culture: The traditional approach. Med. Mycol. Supplement. 2005, 43, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Curtis, L.; Cali, S.; Conroy, L.; Baker, K.; Ou, C.H.; Hershow, R.; Norlock-Cruz, F.; Scheff, P. Aspergillus surveillance project at a large tertiary-care hospital. J. Hosp. Infect. 2005, 59, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Jonsyn, F.E. Seedborne fungi of sesame (Sesamum indicum L.) in Sierra Leone and their potential aflatoxin/mycotoxin production. Mycopathologia 1988, 104, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Hassane, A.; El-Shanawany, A.; Abo-Dahab, N.; Abdel-Hadi, A.; Abdul-Raouf, U.; Mwanza, M. Influence of different moisture contents and temperature on growth and production of aflatoxin B1 by a toxigenic Aspergillus flavus isolate in wheat flour. J. Ecol. Health Environ. 2017, 5, 77–83. [Google Scholar] [CrossRef]
- Nikolic, M.; Stankovic, S.; Savic, I. Comparison of methods for determination of the toxigenic potential of Aspergillus parasiticus Speare and Aspergillus flavus Link isolated from maize. In Proceedings of the 6th International Scientific Meeting: Mycology, Mycotoxicology and Mycoses, Novi Sad, Serbia, 27–29 September 2017. [Google Scholar]
- Thathana, M.G.; Murage, H.; Abia, A.L.K.; Pillay, M. Morphological characterization and determination of aflatoxin production potentials of Aspergillus flavus isolated from maize and soil in Kenya. Agriculture 2017, 7, 80. [Google Scholar] [CrossRef]
- Donner, M.; Lichtemberg, P.S.F.; Doster, M.; Picot, A.; Cotty, P.J.; Puckett, R.D.; Michailides, T.J. Community structure of Aspergillus flavus and Aspergillus parasiticus in major almond producing areas of California, United States. Plant Dis. 2015, 99, 1161–1169. [Google Scholar] [CrossRef]
- Yassin, M.A.; Moslem, M.A.; El-Samawaty, A.M.A.; El-Shikh, M.S. Effectiveness of Allium sativum in controlling Sorghum grain molding Fungi. J. Pure Appl. Microbiol. 2013, 7, 101–107. [Google Scholar]
- Guezlane-Tebibel, N.; Bouras, N.; Mokrane, S.; Benayad, T.; Mathieu, F. Aflatoxigenic strains of Aspergillus section Flavi isolated from marketed peanuts (Arachis hypogaea) in Algiers (Algeria). Ann. Microbiol. 2012, 63, 295–305. [Google Scholar] [CrossRef]
- Sourabie, P.B.; Nikiema, P.; Barro, N.; Savadogo, A. Aflatoxigenic potential of Aspergillus spp. isolated from groundnut seeds, in Burkina Faso, West Africa. Afr. J. Microbiol. Res. 2012, 6, 2603–2609. [Google Scholar]
- Yazdani, D.; Abidin, M.; Tan, Y.; Kamaruzaman, S. Evaluation of the detection techniques of toxigenic Aspergillus isolates. Afr. J. Biotechnol. 2010, 9, 7654–7659. [Google Scholar]
- Ahsan, S.; Bhatti, I.A.; Asi, M.R.; Bhatti, H.N.; Sheikh, M.A. Occurrence of aflatoxins in maize grains from central areas of Punjab, Pakistan. Int. J. Agric. Biol. 2010, 12, 571–575. [Google Scholar]
- Martins, H.; Marques, M.; Fernando, B. Interaction of Wild Strains of Aspergilla with Aspergillus parasiticus ATCC15517 and Aflatoxin Production. Int. J. Mol. Sci. 2008, 9, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Cotty, P.J.; Antilla, L.; Wakelyn, P.J. Competitive exclusion of aflatoxin producers: Farmer-driven research and development. In Biological Control: A Global Perspective; Vincent, C., Goettel, M.S., Lazarovits, G., Eds.; CABI: Oxfordshire, UK, 2007; pp. 241–253. [Google Scholar] [CrossRef]
- Cotty, P.J. Bio competitive exclusion of Toxigenic fungi. In The Mycotoxin Factbook: Food and Feed Topics; Barug, D., Bhatnagar, D., van Egdmond, H.P., van der Kamp, J.W., van Osenbruggen, W.A., Visconti, A., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2006; pp. 179–197. ISBN 978-90-8686-006-7. [Google Scholar]
- Cotty, P.J. Influence of field application of an atoxigenic strain of Aspergillus flavus on the populations of A. flavus infecting cotton bolls and on the aflatoxin content of cottonseed. Phytopathology 1994, 84, 1270–1277. [Google Scholar] [CrossRef]
- Placinta, C.M.; D’Mello, J.P.F.; Macdonald, A.M.C. A review of worldwide contamination of cereal grains and animal feed with Fusarium mycotoxins. Anim. Feed Sci. Technol. 1999, 78, 21–37. [Google Scholar] [CrossRef]
- Cole, R.J.; Cotty, P.J. Biocontrol of aflatoxin production by using bio competitive agents. In A Perspective on Aflatoxin in Field Crops and Animal Food Products in the United States: A Symposium; USDA–ARS: Washington, DC, USA, 1990; pp. 62–66. [Google Scholar]
- Degola, F.; Dall-Asta, C.; Restivo, F. Development of a simple and high-throughput method for detecting aflatoxins production in culture media. Lett. Appl. Microbiol. 2012, 55, 82–89. [Google Scholar] [CrossRef]
- Ezekiel, C.N.; Sulyok, M.; Warth, B.; Krska, R. Multi-microbial metabolites in fonio millet (acha) and sesame seeds in Plateau State, Nigeria. Eur. Food Res. Technol. 2012, 235, 285–293. [Google Scholar] [CrossRef]
- Asadi, M.; Behest, H.R.; Feizy, J. A survey of aflatoxins in sesame in Iran. Mycotoxin Res. 2011, 27, 259–263. [Google Scholar] [CrossRef]
- Ezekiel, C.N.; Udom, I.E.; Frisvad, J.C.; Adetunji, M.C.; Houbraken, J.; Fapohunda, S.O.; Samson, R.A.; Atanda, O.O.; Agi-Otto, M.C.; Onashile, O.A. Assessment of aflatoxigenic Aspergillus and other fungi in millet and sesame from Plateau State, Nigeria. Mycology 2014, 5, 16–22. [Google Scholar] [CrossRef]
- Mimoune, N.A.; Riba, A.; Verheecke, C.; Mathieu, F.; Sabaou, N. Fungal contamination and mycotoxin production by Aspergillus Spp. in nuts and sesame seeds. J. Microbiol. Biotechnol. Food Sci. 2016, 5, 301–305. [Google Scholar] [CrossRef]
- Sabry, A.B.; Hathout, A.S.; Badr, A.N.; Aly, S.; Shehata, M.G. The prevalence of aflatoxin and Aspergillus parasiticus in Egyptian sesame seeds. Int. J. Chemtech. Res. 2016, 9, 308–319. [Google Scholar]
- Esan, A.O.; Fapohunda, S.O.; Ezekiel, C.N.; Sulyok, M.; Krska, R. Distribution of fungi and their toxic metabolites in melon and sesame seeds marketed in two major producing states in Nigeria. Mycotoxin Res. 2020, 36, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.K.; Ebelhar, M.W.; Bellaloui, N.; Mulvaney, M.J.; Stoner, G.R.D.; Kotowicz, J.K.; Little, N.S.; Accenelli, C.; Shier, W.T. Contamination of sesame seed grown in Mississippi with aflatoxin, fumonisin, and mycotoxin-producing fungi. World Mycotoxin J. 2019, 12, 123–132. [Google Scholar] [CrossRef]
- Altaf, N.; Khan, S.A.; Ahmad, M.; Asghar, R.; Ahmed, R.A.; Shaheen, S.; Zafar, M.; Saqib, M. Seed borne mycoflora of sesame (Sesamum indicum L.) and their effect on germination and seedling. Pak. J. Biol. Sci. 2004, 7, 243–245. [Google Scholar] [CrossRef][Green Version]
- Nayyar, B.G.; Akram, A.; Arshad, M.; Mughal, S.M.; Akhund, S.; Mushtaq, S. Mycoflora detected from seeds of (Sesamum indicum L.) in Sialkot, Pakistan. IOSR J. Pharm. Biol. Sci. 2013, 7, 99–103. [Google Scholar] [CrossRef]
- Callicot, K.A.; Cotty, P.J. Methods for monitoring deletions in the aflatoxin bio synthesis gene cluster of Aspergillus flavus with multiplex PCR. Lett. Appl. Microbiol. 2015, 60, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.; Castelle, C.; Singh, A.; Brown, C.; Anantharaman, K.; Sharon, I.; Hug, L.; Burstein, D.; Emerson, J.; Thomas, B.; et al. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ. Microbiol. 2014, 19, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Donner, M.; Atehnkeng, J.; Sikora, R.A.; Bandyopadhyay, R.; Cotty, P.J. Molecular characterization of atoxigenic strains for biological control of aflatoxins in Nigeria. Food Addit. Contam. Part A 2010, 27, 576–590. [Google Scholar] [CrossRef]
- Ehrlich, K.; Kobbeman, K.; Montalbano, B.; Cotty, P. Aflatoxin-producing Aspergillus species from Thailand. Int. J. Food Microbiol. 2007, 114, 153–159. [Google Scholar] [CrossRef]
- Chang, P.K.; Horn, B.W.; Yu, J.; Bhatnagar, D.; Cleveland, T.E. Genes differentially expressed by Aspergillus flavus strains after loss of aflatoxin production by serial transfers. Appl. Microbiol. Biotechnol. 2007, 77, 917–925. [Google Scholar] [CrossRef]
- Drott, M.T.; Lazzaro, B.P.; Brown, D.L.; Carbone, I.; Milgroom, M.G. Balancing selection for aflatoxin in Aspergillus flavus is maintained through interference competition with and fungivory by insects. Proc. R. Soc. B 2018, 284, 1–9. [Google Scholar] [CrossRef]
- Adhikari, K.; Fuentes-Guajardo, K.; Quinto-Sanchez, M.; Mendoza-Revilla, J.; Chacón-Duque, J.C.; Acuña-Alonzo, V.; Ruiz-Linares, A. A genome wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat. Commun. 2016, 7, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Okoth, S.; Nyongesa, B.; Ayugi, V.; Kangethe, E.; Korhonen, H.; Joutsjoki. V. Toxigenic Potential of Aspergillus Species Occurring on Maize Kernels from Two Agro-Ecological Zones in Kenya. Toxins 2012, 4, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Singleton, L.L.; Mihail, J.D.; Rush, C.M. Methods for Research on Soil Born Phytopathogenic Fungi; Phyto Pathological Society: St. Paul, MI, USA, 1993; 266p. [Google Scholar]
- ISTA. International rules for seed testing. Rules Amendments. Seed Sci. Technol. 2001, 29, 1–27. [Google Scholar]
- Samson, R.A.; Pitt, J. 1. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification; Harwood: Amsterdam, The Netherlands, 2000; 510p. [Google Scholar]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi; Academic Press: London, UK, 1980; Volume 1. [Google Scholar]
- Raper, K.B.; Fennell, D.I. The Genus Aspergillus; Williams & Wilkins: Baltimore, MD, USA, 1965; p. 686. [Google Scholar]
- Alshannaq, A.F.; Yu, J.H. A liquid chromatographic method for rapid and sensitive analysis of aflatoxins in laboratory fungal cultures. Toxins 2020, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Park, H.; Han, K.; Hong, S.; Yu, J. High molecular weight genomic DNA mini prep for filamentous fungi. Fungal Genet. Biol. 2017, 104, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.P.; Van Dongen, S.; Abreu-Goodger, C.; Bartonicek, N.; Enright, A.J. Kraken: A set of tools for quality control and analysis of high-throughput sequence data. Methods 2013, 63, 41–49. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907v2. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. Genomes Project Analysis, The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Huson, D.H. Splits Tree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Alshannaq, A.F.; Gibbons, J.G.; Lee, M.K.; Han, K.H.; Hong, S.B.; Yu, J.H. Controlling aflatoxin contamination and propagation of Aspergillus flavus by a soy-fermenting Aspergillus oryzae strain. Sci. Rep. 2018, 8, 16871. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).