Development of a Method for Detecting Alexandrium pacificum Based on the Quantification of sxtA4 by Chip-Based Digital PCR
Abstract
:1. Introduction
2. Results
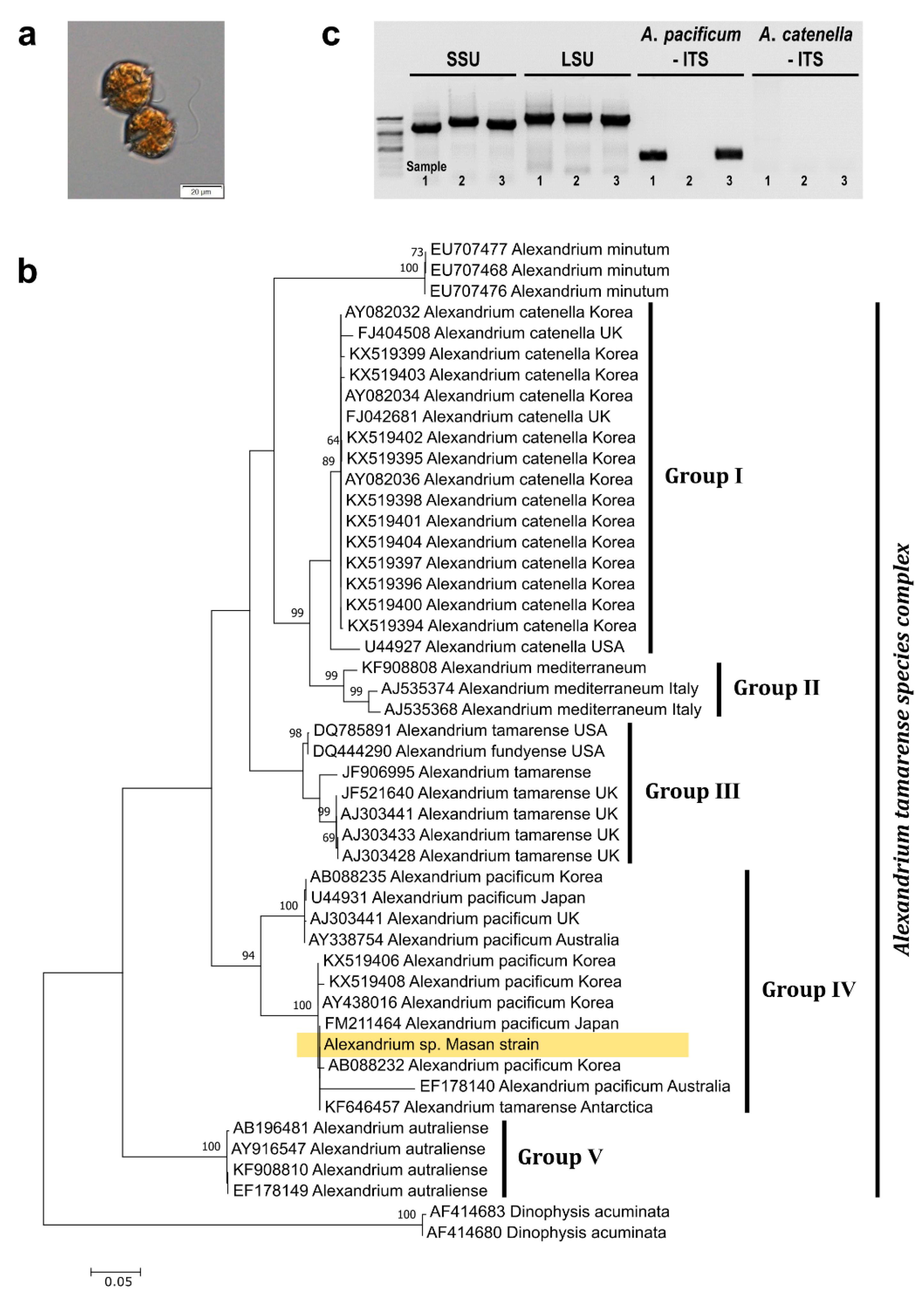
2.1. Identification of the Masan Strain of Alexandrium pacificum
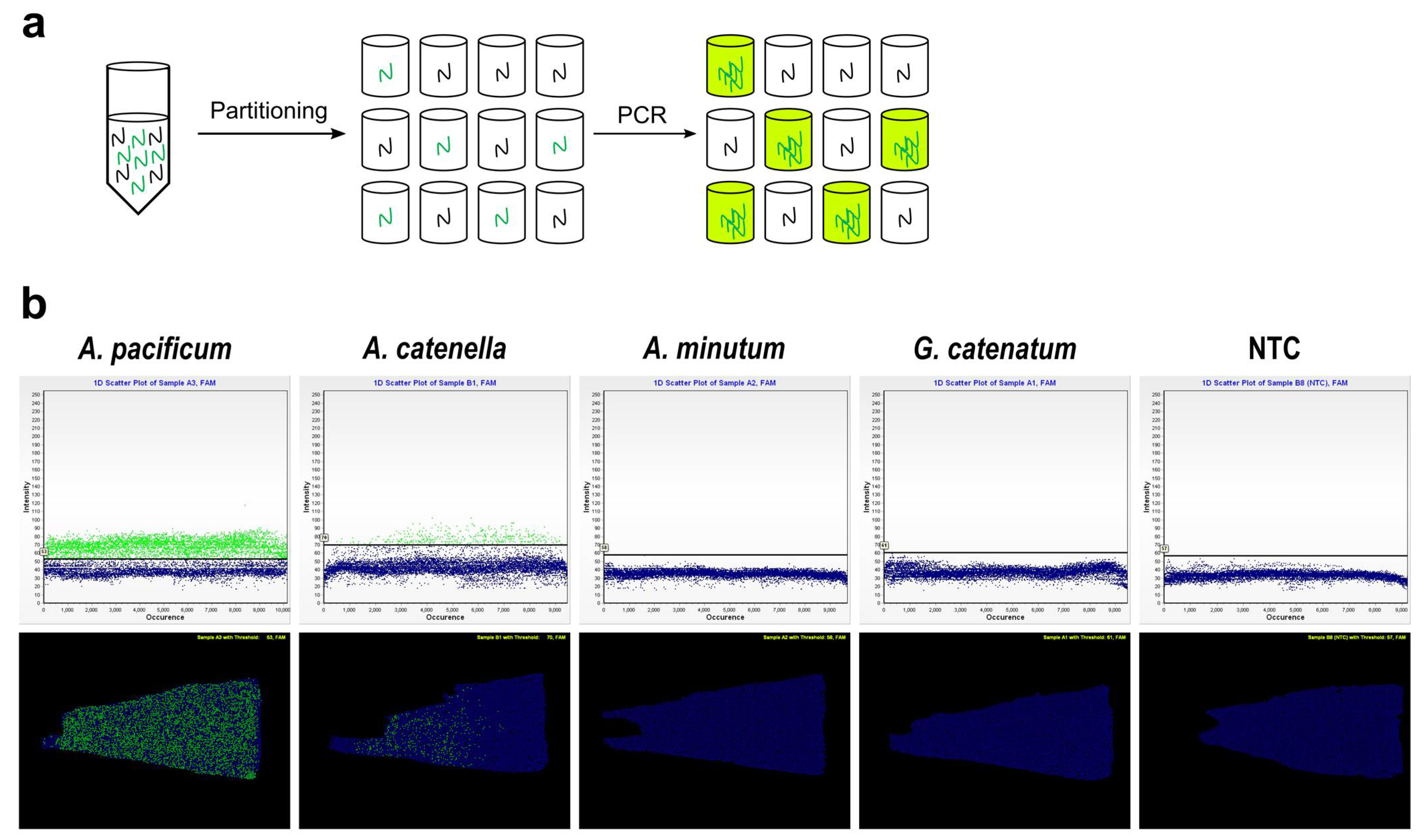
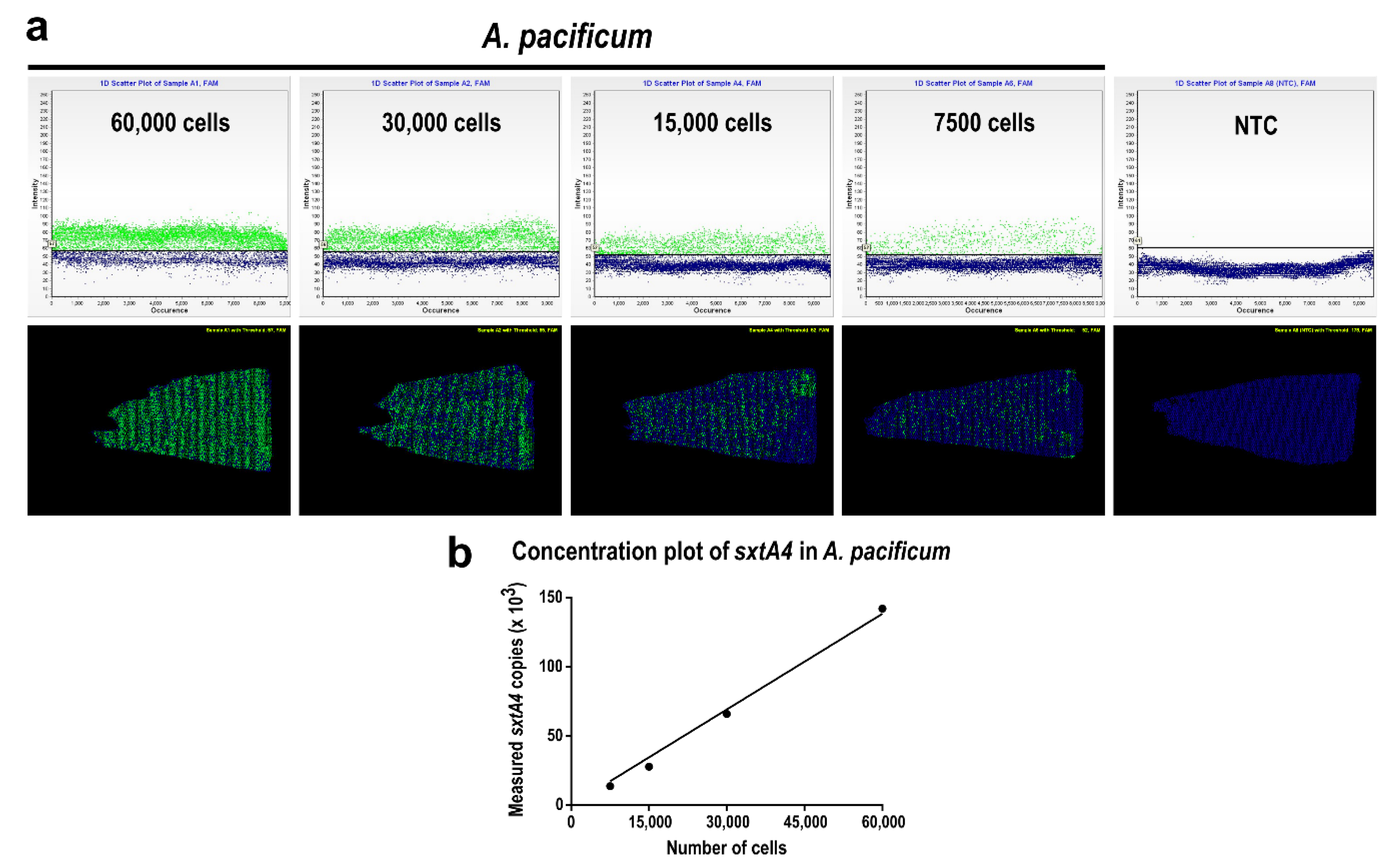
2.2. Quantifying Alexandrium pacificum Cells via Chip-Based dPCR Targeting sxtA4
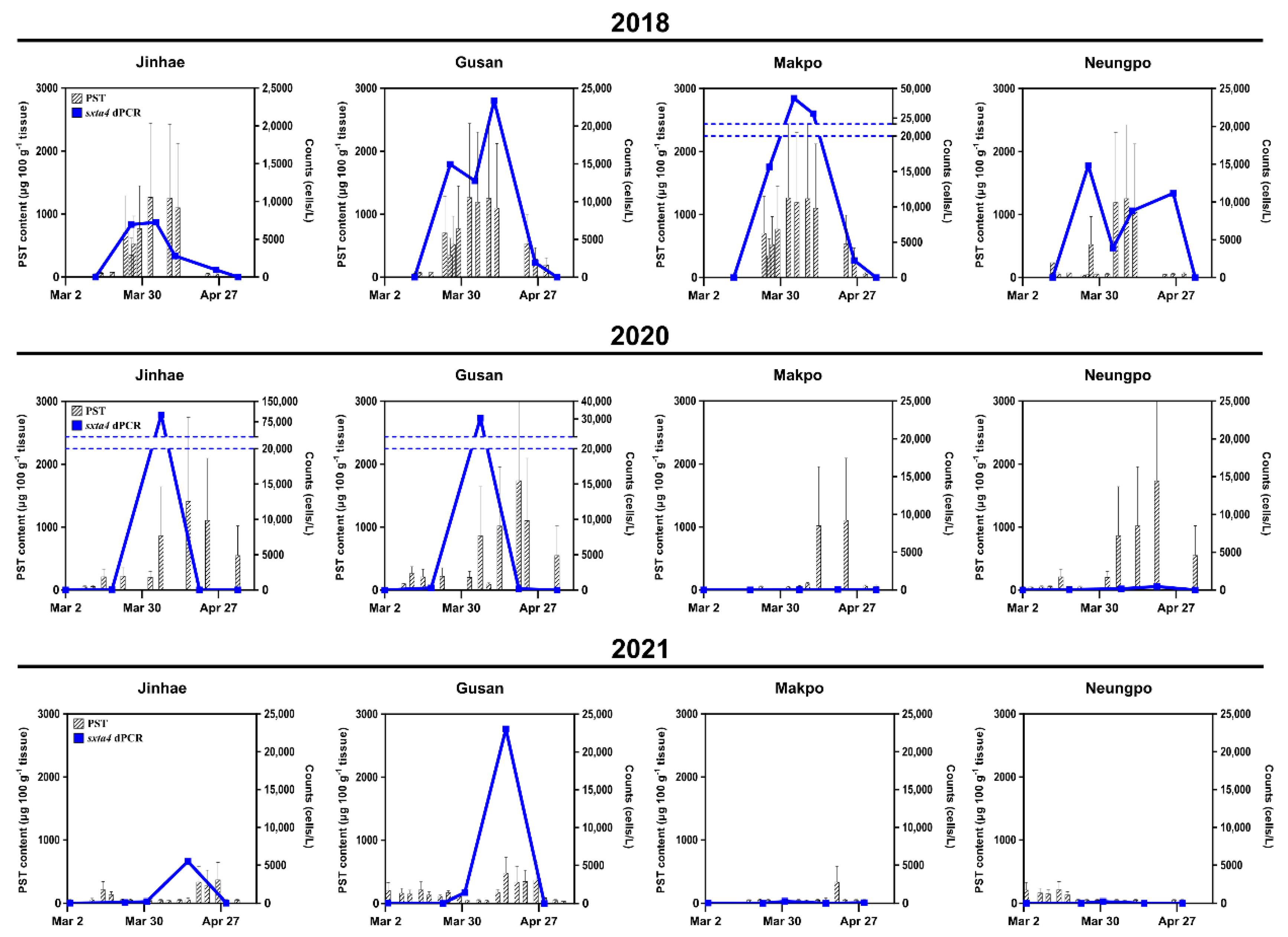
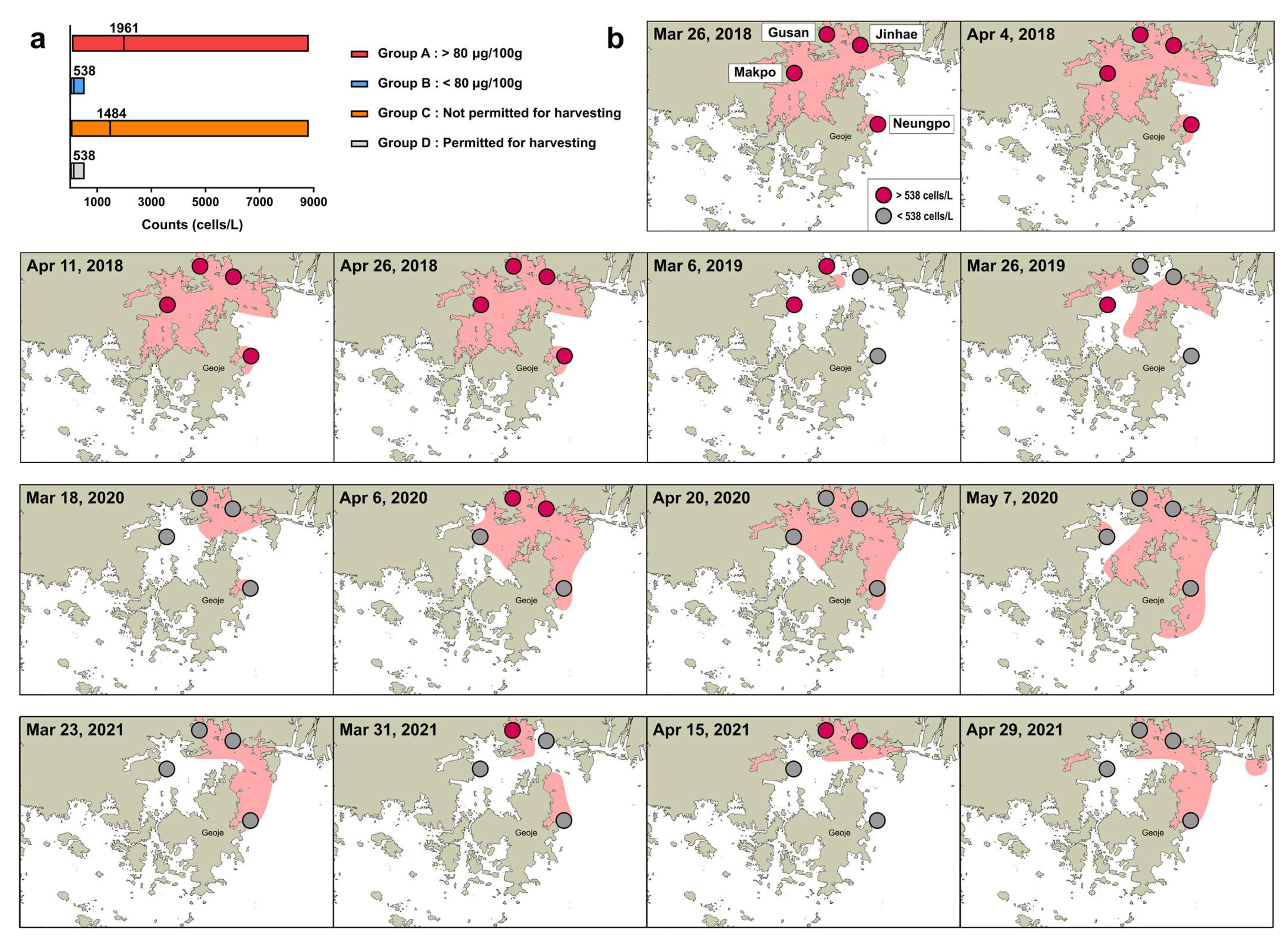
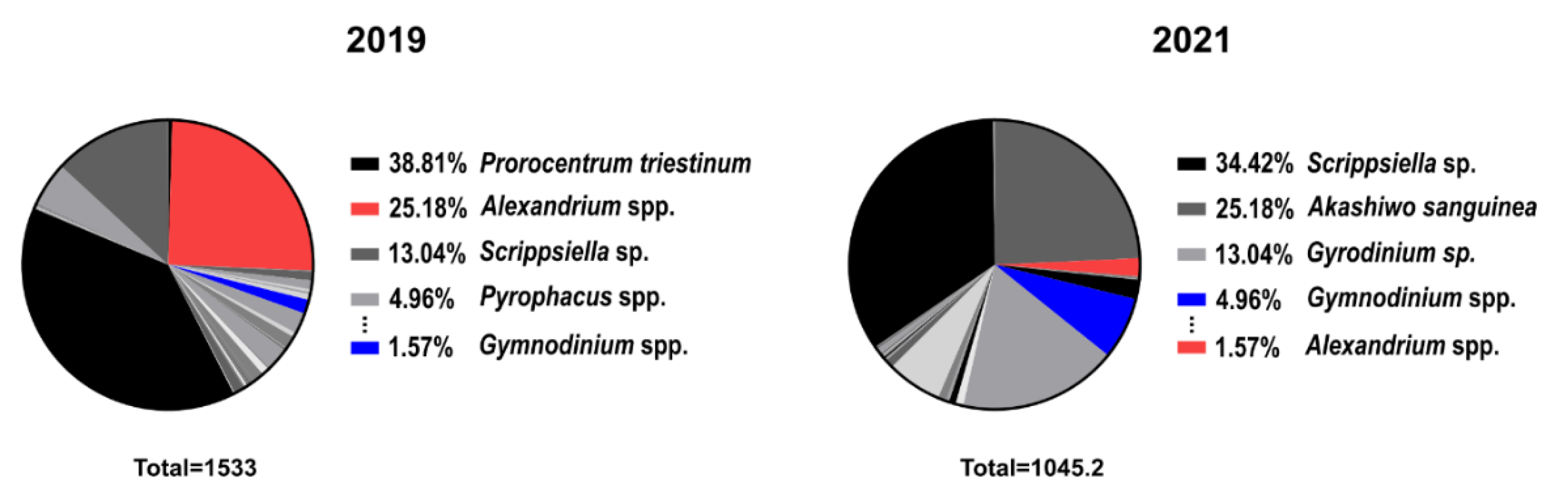
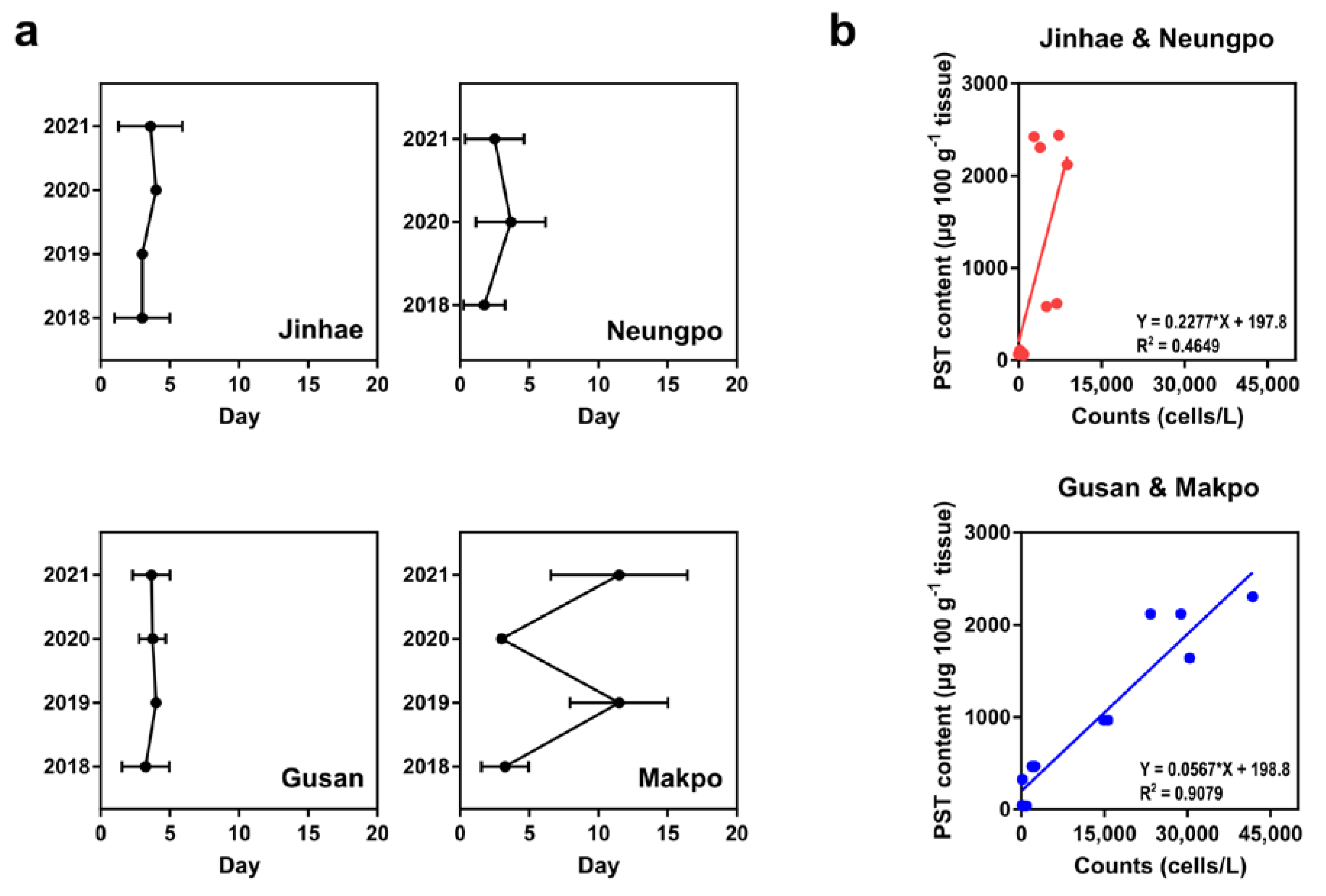
2.3. Comparison of Cell Abundance and PST Content in Field Samples
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Strain Setup
5.2. DNA Extraction and PCR
5.3. Species Identification
5.4. Quantification of the sxtA4 Gene by Chip-Based dPCR and qPCR
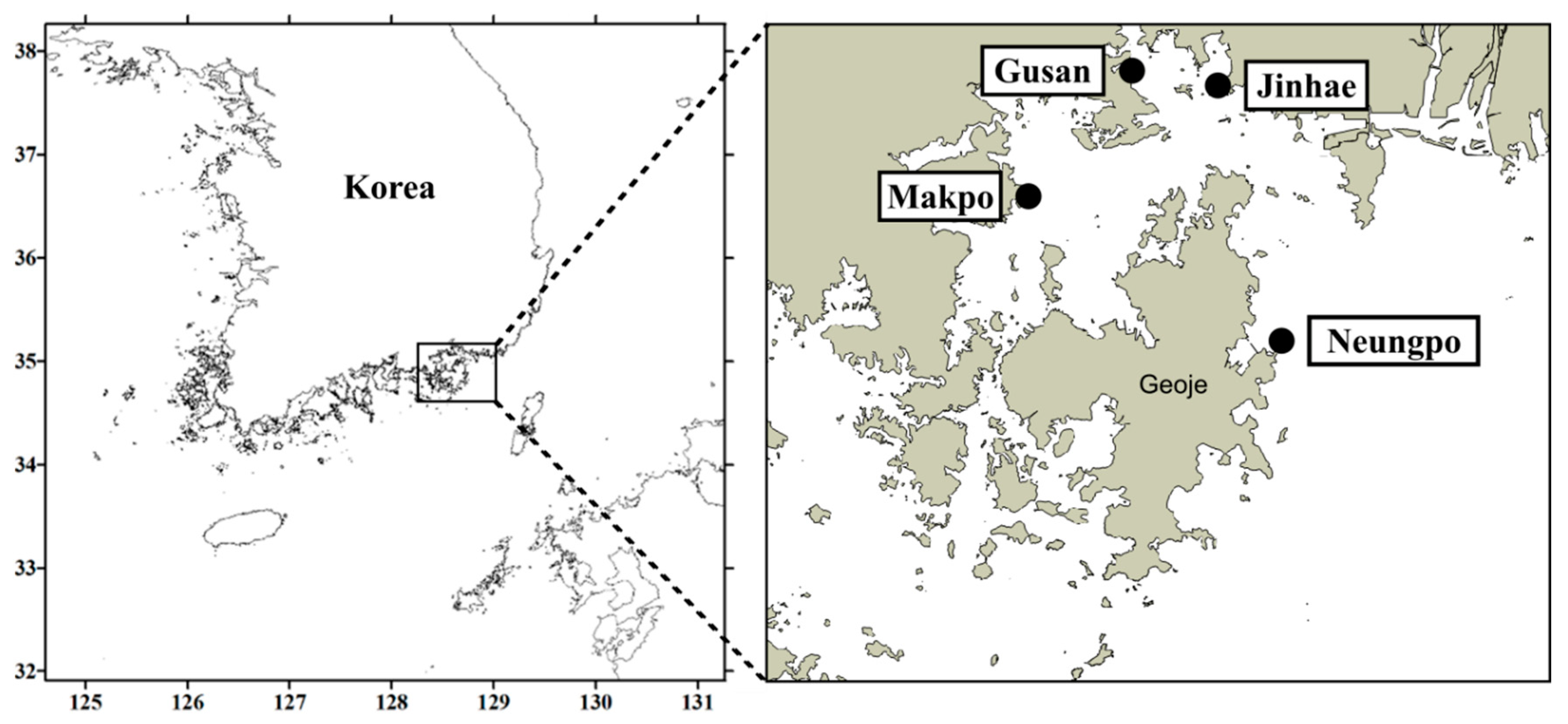
5.5. Field Sampling
5.6. Comparative Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, D.M.; Alpermann, T.J.; Cembella, A.D.; Collos, Y.; Masseret, E.; Montresor, M. The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 2012, 14, 10–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grattan, L.M.; Holobaugh, S.; Morris, J.G., Jr. Harmful algal blooms and public health. Harmful Algae 2016, 57, 2–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morabito, S.; Silvestro, S.; Faggio, C. How the marine biotoxins affect human health. Nat. Prod. Res. 2018, 32, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Oshima, Y.; Yasumoto, T. Structures of two paralytic shellfish toxins, gonyautoxins V and VI, isolated from a tropical dinoflagellate, Pyrodinium bahamense var. compressa. Agric. Biol. Chem. 1982, 46, 1861–1864. [Google Scholar] [CrossRef] [Green Version]
- Oshima, Y.; Hasegawa, M.; Yasumoto, T.; Hallegraeff, G.; Blackburn, S. Dinoflagellate Gymnodinium catenatum as the source of paralytic shellfish toxins in Tasmanian shellfish. Toxicon 1987, 25, 1105–1111. [Google Scholar] [CrossRef]
- Mackenzie, L.; White, D.; Oshima, Y.; Kapa, J. The resting cyst and toxicity of Alexandrium ostenfeldii (Dinophyceae) in New Zealand. Phycologia 1996, 35, 148–155. [Google Scholar] [CrossRef]
- Bolch, C.J.; de Salas, M.F. A review of the molecular evidence for ballast water introduction of the toxic dinoflagellates Gymnodinium catenatum and the Alexandrium “tamarensis complex” to Australasia. Harmful Algae 2007, 6, 465–485. [Google Scholar] [CrossRef]
- Chang, D.-S.; Shin, I.-S.; Pyeun, J.-H.; Park, Y.-H. A Study on Paralytic Shellfish Poison of Sea Mussel, Mytilus edulis-Food Poisoning Accident in Gamchun Bay, Pusan, Korea, 1986. Korean J. Fish. Aquat. Sci. 1987, 20, 293–299. [Google Scholar]
- Han, M.-S.; Jeon, J.-K.; Kim, Y.-O. Occurrence of dinoflagellate Alexandrium tamarense, a causative organism of paralytic shellfish poisoning in Chinhae Bay, Korea. J. Plankton Res. 1992, 14, 1581–1592. [Google Scholar] [CrossRef]
- Lee, J.-S.; Shin, I.-S.; Kim, Y.-M.; CHANG, D.-S. Paralytic shellfish toxins in the mussel, Mytilus edulis, caused the shellfish poisoning accident at Geoje, Korea, in 1996. Korean J. Fish. Aquat. Sci. 1997, 30, 158–160. [Google Scholar]
- Kim, Y.-O.; Park, M.-H.; Han, M.-S. Role of cyst germination in the bloom initiation of Alexandrium tamarense (Dinophyceae) in Masan Bay, Korea. Aquat. Microb. Ecol. 2002, 29, 279–286. [Google Scholar] [CrossRef]
- Shin, H.H.; Li, Z.; Kim, H.J.; Park, B.S.; Lee, J.; Shin, A.-Y.; Park, T.-G.; Lee, K.-W.; Han, K.H.; Youn, J.Y. Alexandrium catenella (Group I) and A. pacificum (Group IV) cyst germination, distribution, and toxicity in Jinhae-Masan Bay, Korea. Harmful Algae 2021, 110, 102122. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.J.; Kwon, S.J.; Jung, Y.J.; Son, K.T.; Ha, K.S.; Mok, J.S.; Kim, J.H. Comparison of analytical methods for the detection of paralytic shellfish toxins (PSTs). Korean J. Fish. Aquat. Sci. 2017, 50, 669–674. [Google Scholar]
- Lee, H.-G.; Kim, H.M.; Min, J.; Park, C.; Jeong, H.J.; Lee, K.; Kim, K.Y. Quantification of the paralytic shellfish poisoning dinoflagellate Alexandrium species using a digital PCR. Harmful Algae 2020, 92, 101726. [Google Scholar] [CrossRef] [PubMed]
- Jellett, J.F.; Marks, L.J.; Stewart, J.E.; Dorey, M.L.; Watson-Wright, W.; Lawrence, J.F. Paralytic shellfish poison (saxitoxin family) bioassays: Automated endpoint determination and standardization of the in vitro tissue culture bioassay, and comparison with the standard mouse bioassay. Toxicon 1992, 30, 1143–1156. [Google Scholar] [CrossRef]
- Doucette, G.J.; Logan, M.M.; Ramsdell, J.S.; Van Dolah, F.M. Development and preliminary validation of a microtiter plate-based receptor binding assay for paralytic shellfish poisoning toxins. Toxicon 1997, 35, 625–636. [Google Scholar] [CrossRef]
- Fonfría, E.S.; Vilariño, N.; Campbell, K.; Elliott, C.; Haughey, S.A.; Ben-Gigirey, B.; Vieites, J.M.; Kawatsu, K.; Botana, L.M. Paralytic shellfish poisoning detection by surface plasmon resonance-based biosensors in shellfish matrixes. Anal. Chem. 2007, 79, 6303–6311. [Google Scholar] [CrossRef]
- Humpage, A.; Magalhaes, V.; Froscio, S. Comparison of analytical tools and biological assays for detection of paralytic shellfish poisoning toxins. Anal. Bioanal. Chem. 2010, 397, 1655–1671. [Google Scholar] [CrossRef]
- Murray, S.A.; Ruvindy, R.; Kohli, G.S.; Anderson, D.M.; Brosnahan, M.L. Evaluation of sxtA and rDNA qPCR assays through monitoring of an inshore bloom of Alexandrium catenella Group 1. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Scholin, C.A.; Herzog, M.; Sogin, M.; Anderson, D.M. Identification of Group-and Strain-specific Genetic Markers for Globally Distributed Alexandrium (dinophyceae). ii. Sequence Analysis of a Fragment of the Lsu rrna Gene 1. J. Phycol. 1994, 30, 999–1011. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Itakura, S.; Imai, I.; Ishida, Y. A rapid and precise technique for enumeration of resting cysts of Alexandrium spp. (Dinophyceae) in natural sediments. Phycologia 1995, 34, 207–214. [Google Scholar] [CrossRef]
- Fabro, E.; Almandoz, G.O.; Ferrario, M.; John, U.; Tillmann, U.; Toebe, K.; Krock, B.; Cembella, A. Morphological, molecular, and toxin analysis of field populations of Alexandrium genus from the Argentine Sea. J. Phycol. 2017, 53, 1206–1222. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.A.; Wiese, M.; Stüken, A.; Brett, S.; Kellmann, R.; Hallegraeff, G.; Neilan, B.A. sxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl. Environ. Microbiol. 2011, 77, 7050–7057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Yu, R.-C.; Murray, S.A.; Chen, J.-H.; Kang, Z.-J.; Zhang, Q.-C.; Kong, F.-Z.; Zhou, M.-J. High specificity of a quantitative PCR assay targeting a saxitoxin gene for monitoring toxic algae associated with paralytic shellfish toxins in the Yellow Sea. Appl. Environ. Microbiol. 2015, 81, 6973–6981. [Google Scholar] [CrossRef] [Green Version]
- Penna, A.; Perini, F.; Dell’Aversano, C.; Capellacci, S.; Tartaglione, L.; Giacobbe, M.G.; Casabianca, S.; Fraga, S.; Ciminiello, P.; Scardi, M. The sxt gene and paralytic shellfish poisoning toxins as markers for the monitoring of toxic Alexandrium species blooms. Environ. Sci. Technol. 2015, 49, 14230–14238. [Google Scholar] [CrossRef] [Green Version]
- Mendoza-Flores, A.; Leyva-Valencia, I.; Band-Schmidt, C.J.; Galindo-Sánchez, C.E.; Bustillos-Guzmán, J.J. Identification of the gene sxtA (Domains sxtA1 and sxtA4) in Mexican strains of Gymnodinium catenatum (Dinophyceae) and their evolution. Front. Mar. Sci. 2018, 5, 289. [Google Scholar] [CrossRef]
- Galluzzi, L.; Penna, A.; Bertozzini, E.; Vila, M.; Garcés, E.; Magnani, M. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 2004, 70, 1199–1206. [Google Scholar] [CrossRef] [Green Version]
- Hosoi-Tanabe, S.; Sako, Y. Species-Specific Detection and Quantification of Toxic Marine Dinoflagellates Alexandrium tamarense and A. catenella byReal-Time PCR Assay. Mar. Biotechnol. 2005, 7, 506–514. [Google Scholar] [CrossRef]
- Erdner, D.L.; Percy, L.; Keafer, B.; Lewis, J.; Anderson, D.M. A quantitative real-time PCR assay for the identification and enumeration of Alexandrium cysts in marine sediments. Deep Sea Res. Part II: Top. Stud. Oceanogr. 2010, 57, 279–287. [Google Scholar] [CrossRef] [Green Version]
- Garneau, M.-È.; Schnetzer, A.; Countway, P.D.; Jones, A.C.; Seubert, E.L.; Caron, D.A. Examination of the seasonal dynamics of the toxic dinoflagellate Alexandrium catenella at Redondo Beach, California, by quantitative PCR. Appl. Environ. Microbiol. 2011, 77, 7669–7680. [Google Scholar] [CrossRef] [Green Version]
- Vandersea, M.W.; Kibler, S.R.; Van Sant, S.B.; Tester, P.A.; Sullivan, K.; Eckert, G.; Cammarata, C.; Reece, K.; Scott, G.; Place, A. qPCR assays for Alexandrium fundyense and A. ostenfeldii (Dinophyceae) identified from Alaskan waters and a review of species-specific Alexandrium molecular assays. Phycologia 2017, 56, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Roose-Amsaleg, C.; Garnier-Sillam, E.; Harry, M. Extraction and purification of microbial DNA from soil and sediment samples. Appl. Soil Ecol. 2001, 18, 47–60. [Google Scholar] [CrossRef]
- Flekna, G.; Schneeweiss, W.; Smulders, F.J.; Wagner, M.; Hein, I. Real-time PCR method with statistical analysis to compare the potential of DNA isolation methods to remove PCR inhibitors from samples for diagnostic PCR. Mol. Cell. Probes 2007, 21, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Ellison, S.L.; Emslie, K.R.; Kassir, Z. A standard additions method reduces inhibitor-induced bias in quantitative real-time PCR. Anal. Bioanal. Chem. 2011, 401, 3221–3227. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Yarimizu, K.; Sildever, S.; Hamamoto, Y.; Tazawa, S.; Oikawa, H.; Yamaguchi, H.; Basti, L.; Mardones, J.I.; Paredes-Mella, J.; Nagai, S. Development of an absolute quantification method for ribosomal RNA gene copy numbers per eukaryotic single cell by digital PCR. Harmful Algae 2021, 103, 102008. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-G.; Kim, H.M.; Min, J.; Kim, K.; Park, M.G.; Jeong, H.J.; Kim, K.Y. An advanced tool, droplet digital PCR (ddPCR), for absolute quantification of the red-tide dinoflagellate, Cochlodinium polykrikoides Margalef (Dinophyceae). Algae 2017, 32, 189–197. [Google Scholar] [CrossRef] [Green Version]
- Stüken, A.; Orr, R.J.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE 2011, 6, e20096. [Google Scholar] [CrossRef] [Green Version]
- Murray, S.A.; Diwan, R.; Orr, R.J.; Kohli, G.S.; John, U. Gene duplication, loss and selection in the evolution of saxitoxin biosynthesis in alveolates. Mol. Phylogenetics Evol. 2015, 92, 165–180. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.H.; Li, Z.; Kim, E.S.; Park, J.-W.; Lim, W.A. Which species, Alexandrium catenella (Group I) or A. pacificum (Group IV), is really responsible for past paralytic shellfish poisoning outbreaks in Jinhae-Masan Bay, Korea? Harmful Algae 2017, 68, 31–39. [Google Scholar] [CrossRef]
- KIM, H.G.; MATSUOKA, K.; LEE, S.G.; AN, K.H. The occurrence of a dinoflagellate Gymnodinium catenatum from Chinhae Bay, Korea. Korean J. Fish. Aquat. Sci. 1996, 29, 837–842. [Google Scholar]
- Park, T.G.; Kim, C.H.; Oshima, Y. Paralytic shellfish toxin profiles of different geographic populations of Gymnodinium catenatum (Dinophyceae) in Korean coastal waters. Phycol. Res. 2004, 52, 300–305. [Google Scholar] [CrossRef]
- Baek, S.H.; Choi, J.M.; Lee, M.; Park, B.S.; Zhang, Y.; Arakawa, O.; Takatani, T.; Jeon, J.-K.; Kim, Y.O. Change in paralytic shellfish toxins in the mussel Mytilus galloprovincialis depending on dynamics of harmful Alexandrium catenella (Group I) in the Geoje coast (South Korea) during bloom season. Toxins 2020, 12, 442. [Google Scholar] [CrossRef]
- Dundas, I.; Johannessen, O.; Berge, G.; Heimdal, B. Toxic algal bloom in Scandinavian waters, May-June 1988. Oceanography 1989, 2, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Litaker, R.W.; Vandersea, M.W.; Kibler, S.R.; Reece, K.S.; Stokes, N.A.; Steidinger, K.A.; Millie, D.F.; Bendis, B.J.; Pigg, R.J.; Tester, P.A. Identification of Pfiesteria Piscicida (Dinophyceae) and Pfiesteria-like Organisms Using Internal Transcribed Spacer-Specific Pcr Assays 1. J. Phycol. 2003, 39, 754–761. [Google Scholar] [CrossRef]
- Weekers, P.; Gast, R.J.; Fuerst, P.A.; Byers, T.J. Sequence variations in small-subunit ribosomal RNAs of Hartmannella vermiformis and their phylogenetic implications. Mol. Biol. Evol. 1994, 11, 684–690. [Google Scholar]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.; Herrmann, J.; Armishaw, P.; Corbisier, P.; Emslie, K.R. Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal. Bioanal. Chem. 2009, 394, 457–467. [Google Scholar] [CrossRef]
- Dong, L.; Meng, Y.; Sui, Z.; Wang, J.; Wu, L.; Fu, B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Low, H.; Chan, S.-J.; Soo, G.-H.; Ling, B.; Tan, E.-L. Clarity™ digital PCR system: A novel platform for absolute quantification of nucleic acids. Anal. Bioanal. Chem. 2017, 409, 1869–1875. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Primer Region | Sequence (5′–3′) | Reference |
|---|---|---|---|
| EukA | Forward, SSU | AACCTGGTTGATCCTGCCAG | [45] |
| G18R | Reverse, SSU | GCATCACAGACCTGTTATTG | [46] |
| 570F | Forward, SSU | GTAATTCCAGCTCCAATAGC | [47] |
| EukB | Reverse, SSU | TGATCCTTCTGCAGGTTCACCTAC | [45] |
| ITSF2 | Forward, ITS | TACGTCCCTGCCCTTTGTAC | [46] |
| LSUB | Reverse, LSU | ACGAACGATTTGCACGTCAG | [46] |
| LSU500R | Reverse, LSU | CCCTCATGCTACTTGTTTGC | [46] |
| LSU500F | Forward, LSU | GCAAACAAGTACCATGAGGG | [46] |
| ITSFR2 | Reverse, ITS | TCCCTGTTCATTCGCCATTAC | [46] |
| Sxt007 | Forward, sxtA4 | ATGCTCAACATGGGAGTCATCC | [38] |
| Sxt008 | Reverse, sxtA4 | GGGTCCAGTAGATGTTGACGATG | [38] |
| SxtA4MAPF | Forward, sxtA4 of the Masan strain of A. pacificum | GCTGGACTACGCGGAGAA | this study |
| SxtA4MAPR | Reverse, sxtA4 of the Masan strain of A. pacificum | GGCGAATTGAACGCCTTGCT | this study |
| SxtA4MAPP | Probe, sxtA4 of the Masan strain of A. pacificum | AACATCATCTACGCCGGGCAGC | this study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hyung, J.-H.; Hwang, J.; Moon, S.-J.; Kim, E.-J.; Kim, D.-W.; Park, J. Development of a Method for Detecting Alexandrium pacificum Based on the Quantification of sxtA4 by Chip-Based Digital PCR. Toxins 2022, 14, 111. https://doi.org/10.3390/toxins14020111
Hyung J-H, Hwang J, Moon S-J, Kim E-J, Kim D-W, Park J. Development of a Method for Detecting Alexandrium pacificum Based on the Quantification of sxtA4 by Chip-Based Digital PCR. Toxins. 2022; 14(2):111. https://doi.org/10.3390/toxins14020111
Chicago/Turabian StyleHyung, Jun-Ho, Jinik Hwang, Seung-Joo Moon, Eun-Joo Kim, Dong-Wook Kim, and Jaeyeon Park. 2022. "Development of a Method for Detecting Alexandrium pacificum Based on the Quantification of sxtA4 by Chip-Based Digital PCR" Toxins 14, no. 2: 111. https://doi.org/10.3390/toxins14020111
APA StyleHyung, J.-H., Hwang, J., Moon, S.-J., Kim, E.-J., Kim, D.-W., & Park, J. (2022). Development of a Method for Detecting Alexandrium pacificum Based on the Quantification of sxtA4 by Chip-Based Digital PCR. Toxins, 14(2), 111. https://doi.org/10.3390/toxins14020111





