Evaluation of a Novel Synthetic Peptide Derived from Cytolytic Mycotoxin Candidalysin
Abstract
1. Introduction
2. Results
2.1. Rational Design of the Ca-MAP1 Peptide
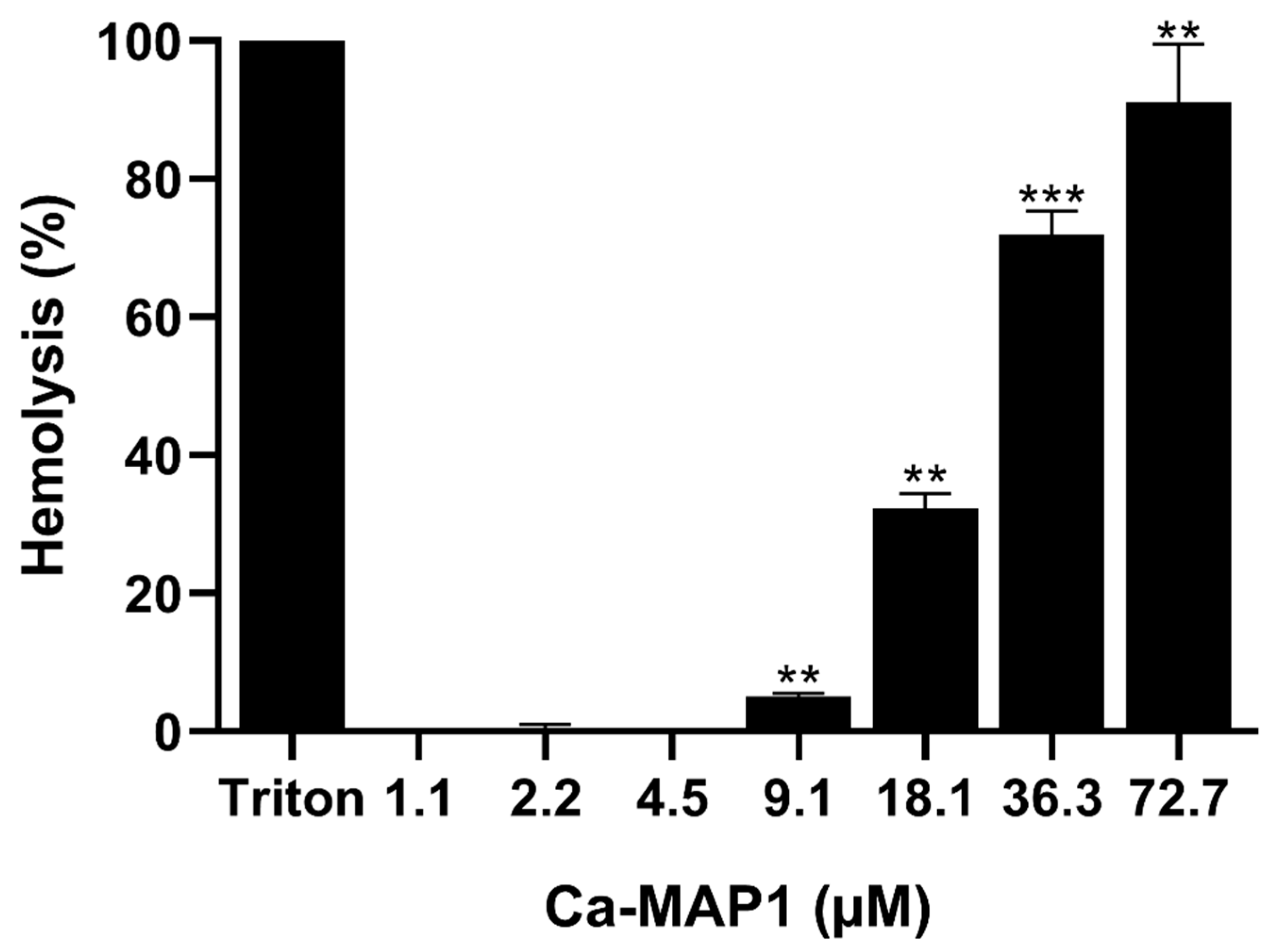
2.2. Hemolytic Activity
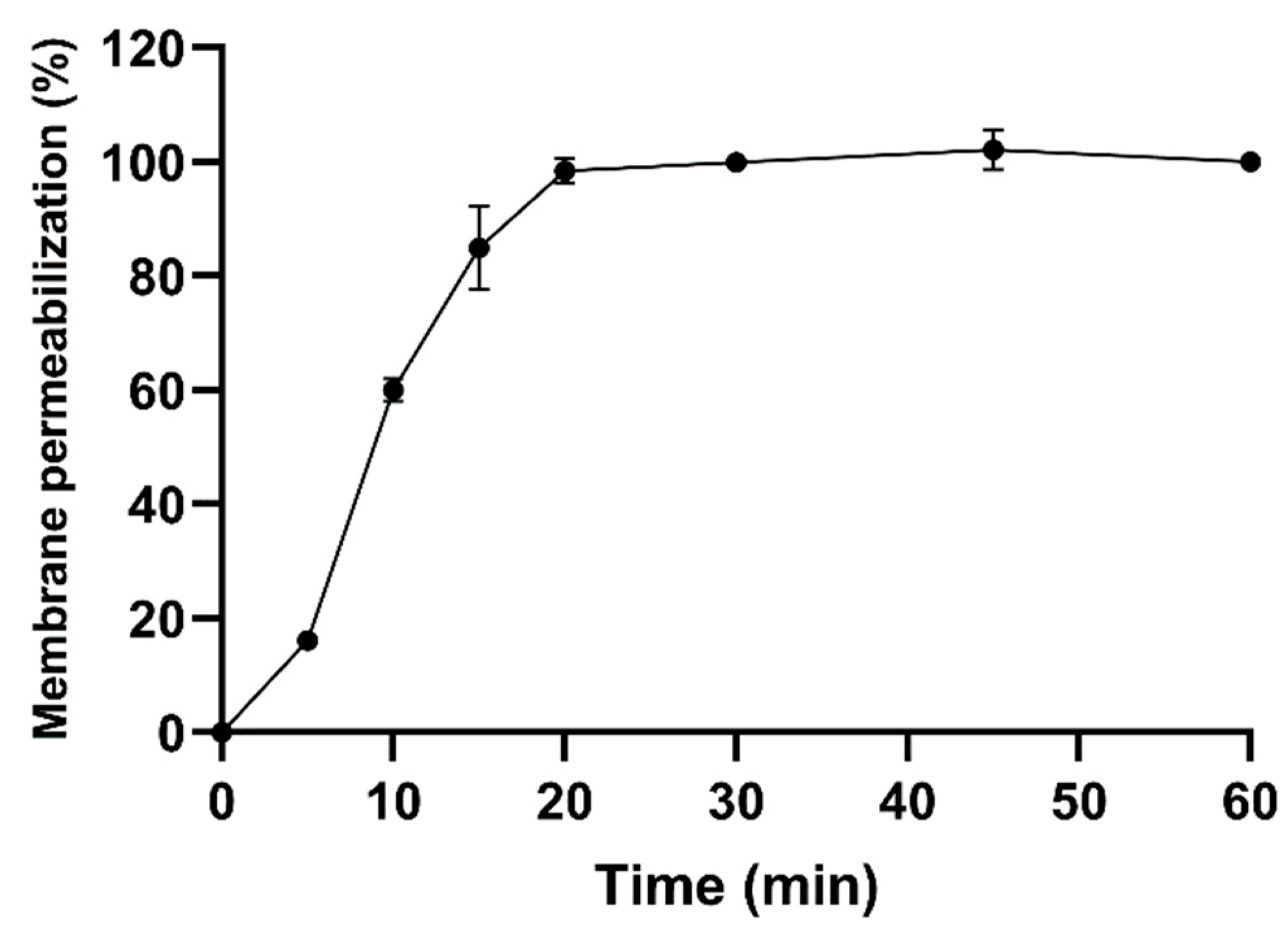
2.3. Antibacterial Activity
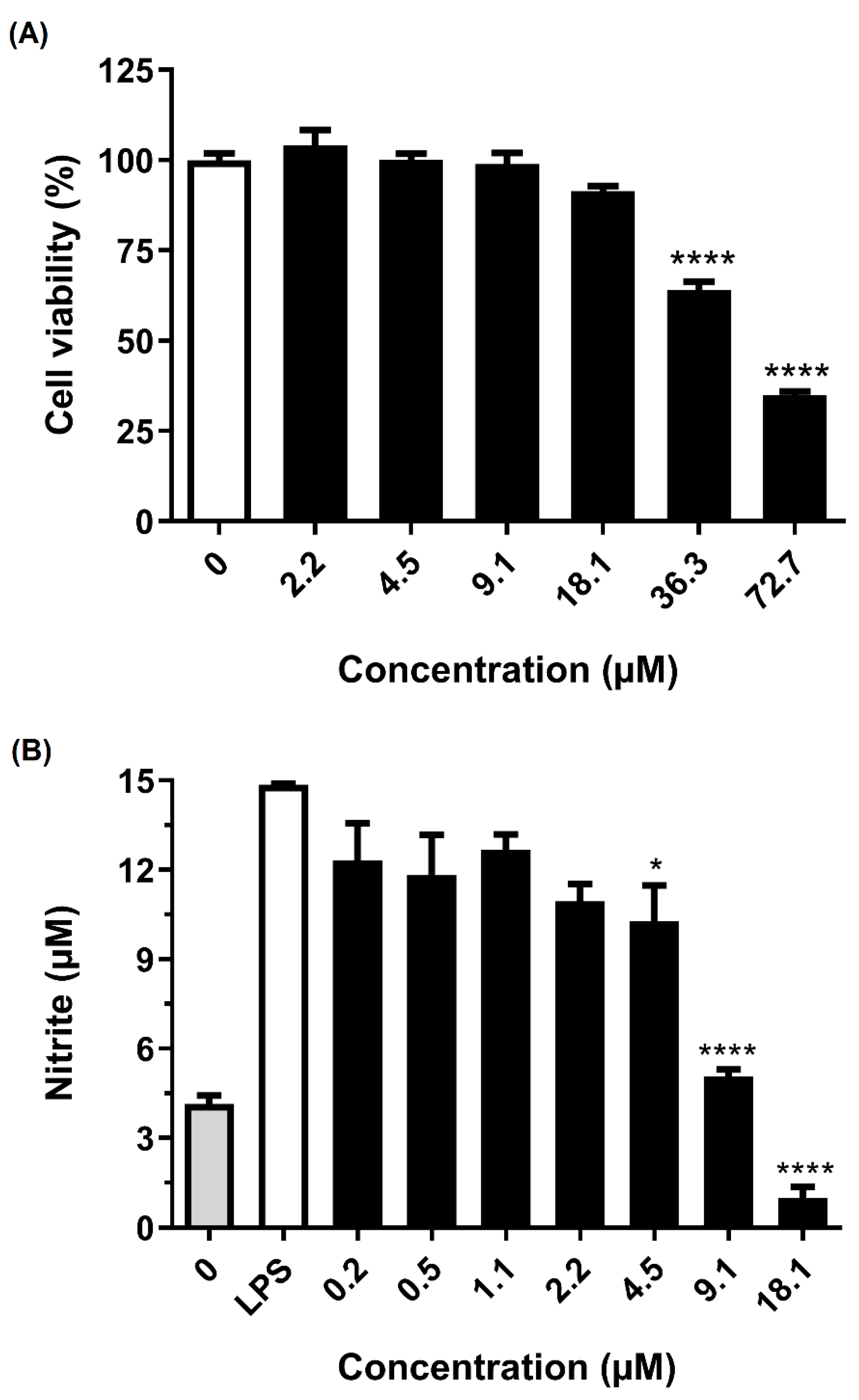
2.4. Anti-Neuroinflammatory Activity
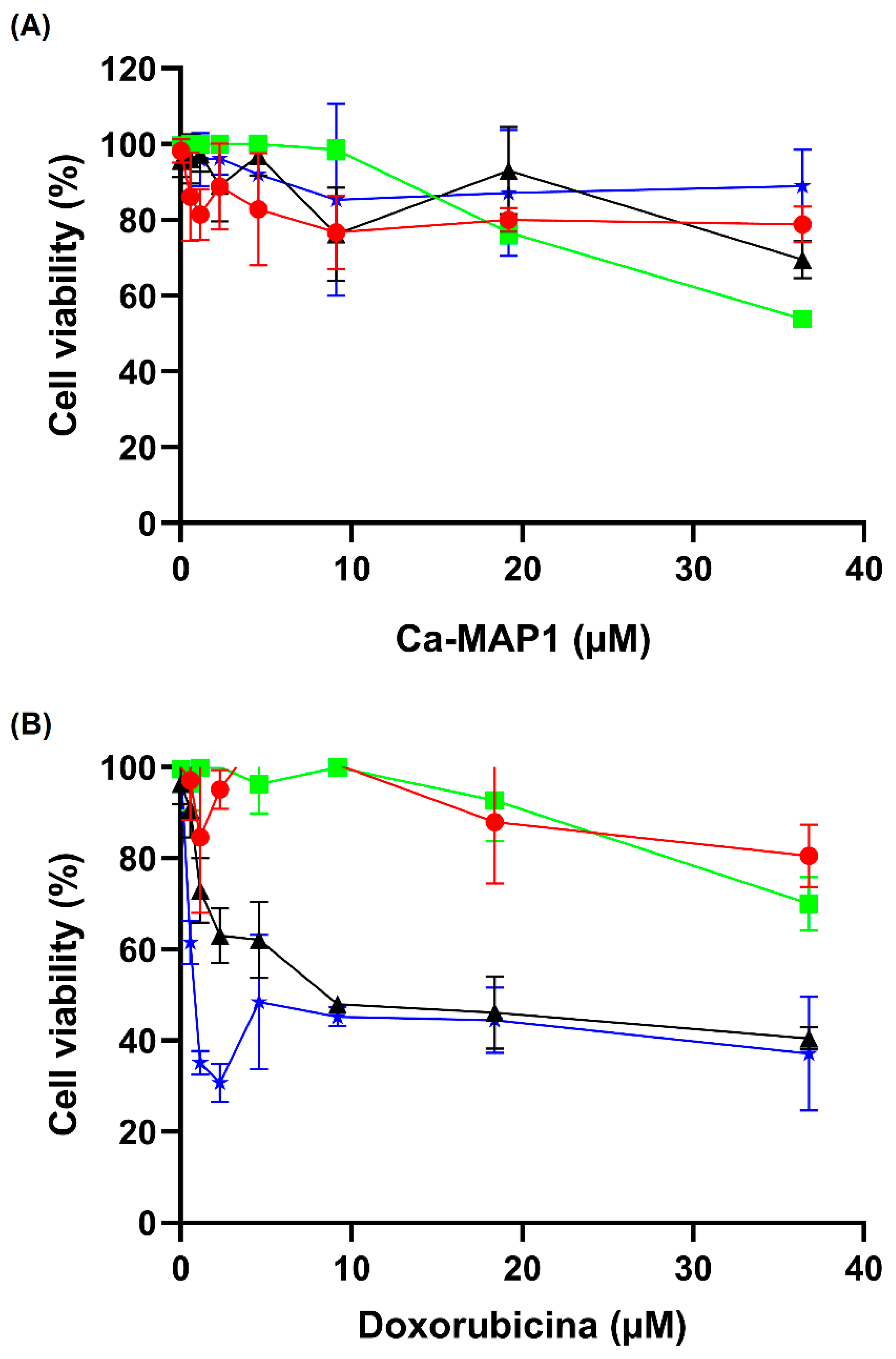
2.5. Anticancer Activity
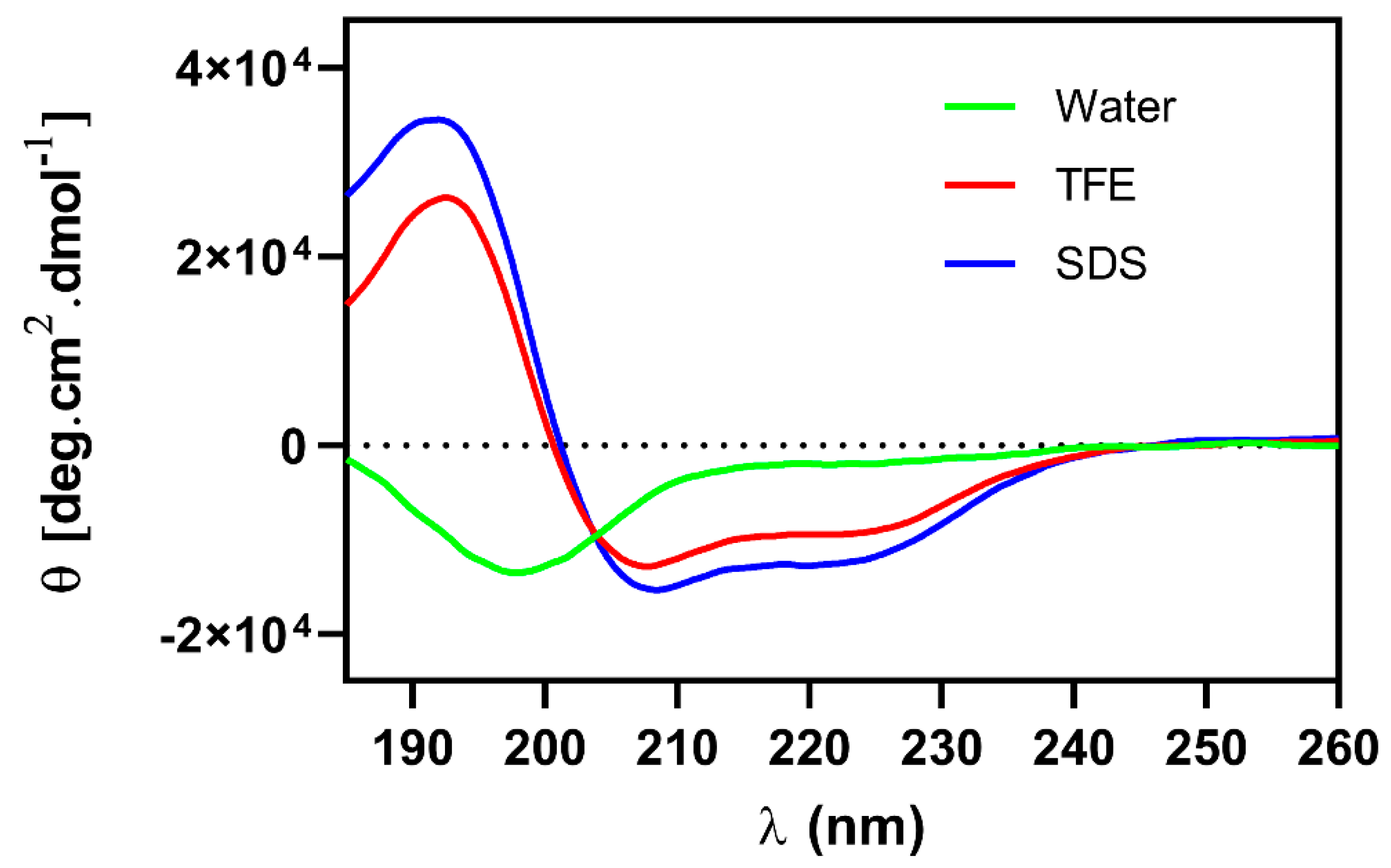
2.6. Circular Dichroism
2.7. Molecular Modeling
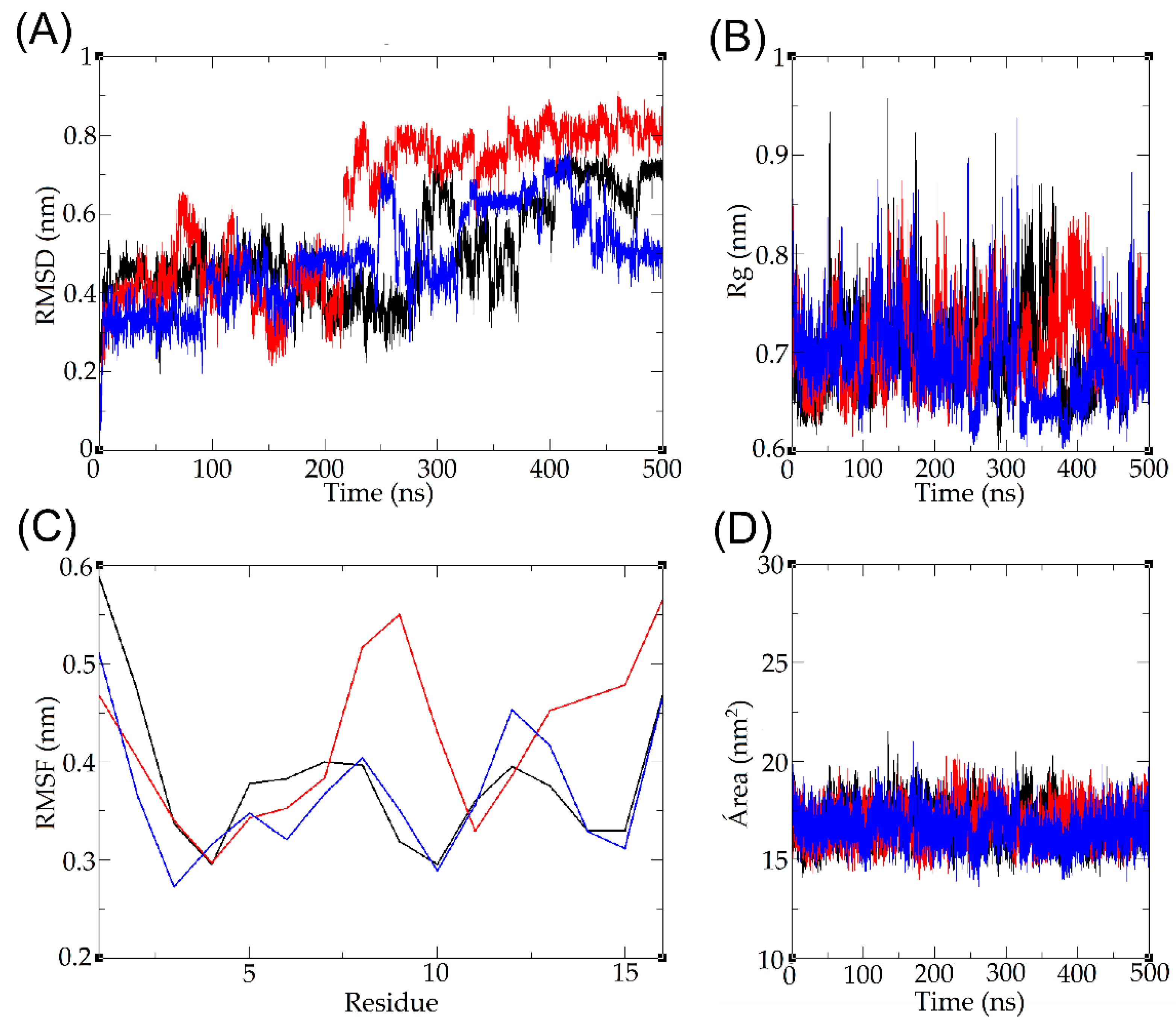
2.8. Molecular Dynamics
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Rational Design
5.2. Peptide Synthesis
5.3. Hemolytic Assay
5.4. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Assays
5.5. E. coli Membrane Permeabilization
5.6. Cell Cultures
5.7. Cell Viability Test Using MTT Methodology
5.8. Inhibition of Microglial Activation by LPS
5.9. Circular Dichroism (CD)
5.10. Comparative Modeling and Validation
5.11. Molecular Dynamics in Water
5.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boleti, A.P.D.A.; Flores, T.M.D.O.; Moreno, S.E.; dos Anjos, L.; Mortari, M.R.; Migliolo, L. Neuroinflammation: An Overview of Neurodegenerative and Metabolic Diseases and of Biotechnological Studies. Neurochem. Int. 2020, 136, 104714. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.Y.F.; Rawji, K.S.; Ghorbani, S.; Xue, M.; Yong, V.W. The Benefits of Neuroinflammation for the Repair of the Injured Central Nervous System. Cell. Mol. Immunol. 2019, 16, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Xiang, D.; Dang, R.; Xu, P.; Wang, J.; Han, W.; Fu, Y.; Yao, D.; Cao, L.; Jiang, P. Neuroprotective Effects of Dl-3-n-Butylphthalide against Doxorubicin-Induced Neuroinflammation, Oxidative Stress, Endoplasmic Reticulum Stress, and Behavioral Changes. Oxid. Med. Cell. Longev. 2018, 2018, 9125601. [Google Scholar] [CrossRef] [PubMed]
- Boleti, A.P.D.A.; Frihling, B.E.F.; Silva, P.S.E.; Cardoso, P.H.D.O.; de Moraes, L.F.R.N.; Rodrigues, T.A.A.; Biembengute, M.E.F.; Koolen, H.H.F.; Migliolo, L. Biochemical Aspects and Therapeutic Mechanisms of Cannabidiol in Epilepsy. Neurosci. Biobehav. Rev. 2022, 132, 1214–1228. [Google Scholar] [CrossRef]
- Batista, C.R.; Gomes, G.F.; Candelario-Jalil, E.; Fiebich, B.L.; de Oliveira, A.C. Lipopolysaccharide-Induced Neuroinflammation as a Bridge to Understand Neurodegeneration. Int. J. Mol. Sci. 2019, 20, 2293. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Pirri, G.; Rinaldi, A.C. Antimicrobial Peptides: The LPS Connection. In Antimicrobial Peptides; Springer: Berlin/Heidelberg, Germany, 2010; pp. 137–154. [Google Scholar]
- Di Lorenzo, F.; De Castro, C.; Silipo, A.; Molinaro, A. Lipopolysaccharide Structures of Gram-Negative Populations in the Gut Microbiota and Effects on Host Interactions. FEMS Microbiol. Rev. 2019, 43, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Mansour, S.C.; Hancock, R.E.W. Antimicrobial Peptides: An Introduction. Methods Mol. Biol. 2017, 1548, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Fosgerau, K.; Hoffmann, T. Peptide Therapeutics: Current Status and Future Directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef]
- Kombrink, A.; Tayyrov, A.; Essig, A.; Stöckli, M.; Micheller, S.; Hintze, J.; van Heuvel, Y.; Dürig, N.; Lin, C.; Kallio, P.T.; et al. Induction of Antibacterial Proteins and Peptides in the Coprophilous Mushroom Coprinopsis Cinerea in Response to Bacteria. ISME J. 2019, 13, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial Peptides (AMPs): Ancient Compounds That Represent Novel Weapons in the Fight against Bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Künzler, M. How Fungi Defend Themselves against Microbial Competitors and Animal Predators. PLoS Pathog. 2018, 14, e1007184. [Google Scholar] [CrossRef] [PubMed]
- Blagojevic, M.; Camilli, G.; Maxson, M.; Hube, B.; Moyes, D.L.; Richardson, J.P.; Naglik, J.R. Candidalysin Triggers Epithelial Cellular Stresses That Induce Necrotic Death. Cell. Microbiol. 2021, 23, e13371. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Wickramasinghe, D.N.; Nikou, S.-A.; Hube, B.; Richardson, J.P.; Naglik, J.R. Candidalysin Is a Potent Trigger of Alarmin and Antimicrobial Peptide Release in Epithelial Cells. Cells 2020, 9, 699. [Google Scholar] [CrossRef] [PubMed]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin Is a Fungal Peptide Toxin Critical for Mucosal Infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Sothiselvam, S.; Lu, T.K.; de la Fuente-Nunez, C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431, 3547–3567. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A Web Server to Screen Sequences with Specific Alpha-Helical Properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Almeida, C.V.; de Oliveira, C.F.R.; dos Santos, E.L.; dos Santos, H.F.; Júnior, E.C.; Marchetto, R.; da Cruz, L.A.; Ferreira, A.M.T.; Gomes, V.M.; Taveira, G.B.; et al. Differential Interactions of the Antimicrobial Peptide, RQ18, with Phospholipids and Cholesterol Modulate Its Selectivity for Microorganism Membranes. Biochim. Biophys. Acta-Gen. Subj. 2021, 1865, 129937. [Google Scholar] [CrossRef]
- Mohanram, H.; Bhattacharjya, S. Salt-Resistant Short Antimicrobial Peptides. Biopolymers 2016, 106, 345–356. [Google Scholar] [CrossRef]
- Green, L.C.; Wagner, D.A.; Glogowski, J.; Skipper, P.L.; Wishnok, J.S.; Tannenbaum, S.R. Analysis of Nitrate, Nitrite, and [15N]Nitrate in Biological Fluids. Anal. Biochem. 1982, 126, 131–138. [Google Scholar] [CrossRef]
- Silva, O.N.; Porto, W.F.; Migliolo, L.; Mandal, S.M.; Gomes, D.G.; Holanda, H.H.S.; Silva, R.S.P.; Dias, S.C.; Costa, M.P.; Costa, C.R.; et al. Cn-AMP1: A New Promiscuous Peptide with Potential for Microbial Infections Treatment. Biopolymers 2012, 98, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Migliolo, L.; Felício, M.R.; Cardoso, M.H.; Silva, O.N.; Xavier, M.-A.E.; Nolasco, D.O.; de Oliveira, A.S.; Roca-Subira, I.; Vila Estape, J.; Teixeira, L.D.; et al. Structural and Functional Evaluation of the Palindromic Alanine-Rich Antimicrobial Peptide Pa-MAP2. Biochim. Biophys. Acta 2016, 1858 Pt A, 1488–1498. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using Circular Dichroism Spectra to Estimate Protein Secondary Structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.L.S.; Miles, A.J.; Whitmore, L.; Wallace, B.A. Distinct Circular Dichroism Spectroscopic Signatures of Polyproline II and Unordered Secondary Structures: Applications in Secondary Structure Analyses. Protein Sci. 2014, 23, 1765–1772. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35 (Suppl. 2), 407–410. [Google Scholar] [CrossRef]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall III, W.B.; Snoeyink, J.; Richardson, J.S. MolProbity: All-Atom Contacts and Structure Validation for Proteins and Nucleic Acids. Nucleic Acids Res. 2007, 35 (Suppl. 2), W375–W383. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and Better Reference Data for Improved All-atom Structure Validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Yan, J.; Bhadra, P.; Li, A.; Sethiya, P.; Qin, L.; Tai, H.K.; Wong, K.H.; Siu, S.W.I. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids 2020, 20, 882–894. [Google Scholar] [CrossRef]
- Kim, H.; Jang, J.H.; Kim, S.C.; Cho, J.H. De Novo Generation of Short Antimicrobial Peptides with Enhanced Stability and Cell Specificity. J. Antimicrob. Chemother. 2014, 69, 121–132. [Google Scholar] [CrossRef]
- Ramesh, S.; Govender, T.; Kruger, H.G.; de la Torre, B.G.; Albericio, F. Short AntiMicrobial Peptides (SAMPs) as a Class of Extraordinary Promising Therapeutic Agents. J. Pept. Sci. 2016, 22, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Mant, C.T.; Vasil, M.; Hodges, R.S. Role of Positively Charged Residues on the Polar and Non-Polar Faces of Amphipathic α-Helical Antimicrobial Peptides on Specificity and Selectivity for Gram-Negative Pathogens. Chem. Biol. Drug Des. 2018, 91, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Béven, L.; Castano, S.; Dufourcq, J.; Wieslander, A.; Wróblewski, H. The Antibiotic Activity of Cationic Linear Amphipathic Peptides: Lessons from the Action of Leucine/Lysine Copolymers on Bacteria of the Class Mollicutes. Eur. J. Biochem. 2003, 270, 2207–2217. [Google Scholar] [CrossRef]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of Peptide Hydrophobicity in the Mechanism of Action of Alpha-Helical Antimicrobial Peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; Pedron, C.N.; Higashikuni, Y.; Kramer, R.M.; Cardoso, M.H.; Oshiro, K.G.N.; Franco, O.L.; Silva Junior, P.I.; Silva, F.D.; Oliveira Junior, V.X.; et al. Structure-Function-Guided Exploration of the Antimicrobial Peptide Polybia-CP Identifies Activity Determinants and Generates Synthetic Therapeutic Candidates. Commun. Biol. 2018, 1, 221. [Google Scholar] [CrossRef]
- Takechi-Haraya, Y.; Ohgita, T.; Kotani, M.; Kono, H.; Saito, C.; Tamagaki-Asahina, H.; Nishitsuji, K.; Uchimura, K.; Sato, T.; Kawano, R.; et al. Effect of Hydrophobic Moment on Membrane Interaction and Cell Penetration of Apolipoprotein E-Derived Arginine-Rich Amphipathic α-Helical Peptides. Sci. Rep. 2022, 12, 4959. [Google Scholar] [CrossRef]
- Porto, W.F.; Ferreira, K.C.V.; Ribeiro, S.M.; Franco, O.L. Sense the Moment: A Highly Sensitive Antimicrobial Activity Predictor Based on Hydrophobic Moment. Biochim. Biophys. Acta-Gen. Subj. 2022, 1866, 130070. [Google Scholar] [CrossRef]
- Höfs, S. Identification of Candidalysin: A Candida Albicans Peptide Toxin Involved in Epithelial Damage; Friedrich-Schiller-Universität: Jena, Germany, 2016. [Google Scholar]
- Oddo, A.; Hansen, P.R. Hemolytic Activity of Antimicrobial Peptides. In Antimicrobial Peptides; Springer: Berlin/Heidelberg, Germany, 2017; pp. 427–435. [Google Scholar]
- Shakil, M.M.; Hossain, M.A.J.; Karal, M.A.S.; Islam, M.Z.; Chowdhury, M.S.; Hasan; Haque, M.A.; Alam, M.M. Comparative Time-Kill Assessment of Cationic Antimicrobial Peptide and Fluoroquinolone against Gram-Negative Bacteria. EC Pharmacol. Toxicol. 2020, 8, 117–124. [Google Scholar]
- Mortimer, F.C.; Mason, D.J.; Gant, V.A. Flow Cytometric Monitoring of Antibiotic-Induced Injury in Escherichia Coli Using Cell-Impermeant Fluorescent Probes. Antimicrob. Agents Chemother. 2000, 44, 676–681. [Google Scholar] [CrossRef]
- Wang, W.Y.; Tan, M.S.; Yu, J.T.; Tan, L. Role of Pro-Inflammatory Cytokines Released from Microglia in Alzheimer’s Disease. Ann. Transl. Med. 2015, 3, 136. [Google Scholar] [CrossRef]
- Wang, X.; Rousset, C.I.; Hagberg, H.; Mallard, C. Lipopolysaccharide-Induced Inflammation and Perinatal Brain Injury. Semin. Fetal Neonatal Med. 2006, 11, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Shabab, T.; Khanabdali, R.; Moghadamtousi, S.Z.; Kadir, H.A.; Mohan, G. Neuroinflammation Pathways: A General Review. Int. J. Neurosci. 2017, 127, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Huang, Z.; Meng, H.; Shi, X.; Xie, J. Immunomodulation Effect of Polysaccharides from Liquid Fermentation of Monascus Purpureus 40269 via Membrane TLR-4 to Activate the MAPK and NF-ΚB Signaling Pathways. Int. J. Biol. Macromol. 2022, 201, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.-C.; Jeong, Y.H.; Li, W.; Go, Y. Angelicae Gigantis Radix Regulates LPS-Induced Neuroinflammation in BV2 Microglia by Inhibiting NF-ΚB and MAPK Activity and Inducing Nrf-2 Activity. Molecules 2019, 24, 3755. [Google Scholar] [CrossRef] [PubMed]
- Zandbergen, N.; de Rooij, B.H.; Vos, M.C.; Pijnenborg, J.M.A.; Boll, D.; Kruitwagen, R.F.P.M.; van de Poll-Franse, L.V.; Ezendam, N.P.M. Changes in Health-Related Quality of Life among Gynecologic Cancer Survivors during the Two Years after Initial Treatment: A Longitudinal Analysis. Acta Oncol. 2019, 58, 790–800. [Google Scholar] [CrossRef]
- Xu, Z.; Luo, F.; Wang, Y.; Zou, B.; Man, Y.; Liu, J.; Li, H.; Arshad, B.; Li, H.; Li, S. Cognitive Impairments in Breast Cancer Survivors Treated with Chemotherapy: A Study Based on Event-Related Potentials. Cancer Chemother. Pharmacol. 2020, 85, 61–67. [Google Scholar] [CrossRef]
- Yamada, T.H.; Denburg, N.L.; Beglinger, L.J.; Schultz, S.K. Neuropsychological Outcomes of Older Breast Cancer Survivors: Cognitive Features Ten or More Years after Chemotherapy. J. Neuropsychiatry Clin. Neurosci. 2010, 22, 48–54. [Google Scholar] [CrossRef]
- Wang, L.; Apple, A.C.; Schroeder, M.P.; Ryals, A.J.; Voss, J.L.; Gitelman, D.; Sweet, J.J.; Butt, Z.A.; Cella, D.; Wagner, L.I. Reduced Prefrontal Activation during Working and Long-term Memory Tasks and Impaired Patient-reported Cognition among Cancer Survivors Postchemotherapy Compared with Healthy Controls. Cancer 2016, 122, 258–268. [Google Scholar] [CrossRef]
- Groves, T.R.; Farris, R.; Anderson, J.E.; Alexander, T.C.; Kiffer, F.; Carter, G.; Wang, J.; Boerma, M.; Allen, A.R. 5-Fluorouracil Chemotherapy Upregulates Cytokines and Alters Hippocampal Dendritic Complexity in Aged Mice. Behav. Brain Res. 2017, 316, 215–224. [Google Scholar] [CrossRef]
- Lee, B.E.; Choi, B.Y.; Hong, D.K.; Kim, J.H.; Lee, S.H.; Kho, A.R.; Kim, H.; Choi, H.C.; Suh, S.W. The Cancer Chemotherapeutic Agent Paclitaxel (Taxol) Reduces Hippocampal Neurogenesis via down-Regulation of Vesicular Zinc. Sci. Rep. 2017, 7, 11667. [Google Scholar] [CrossRef]
- Jiang, Z.-G.; Winocur, G.; Wojtowicz, J.M.; Shevtsova, O.; Fuller, S.; Ghanbari, H.A. PAN-811 Prevents Chemotherapy-Induced Cognitive Impairment and Preserves Neurogenesis in the Hippocampus of Adult Rats. PLoS ONE 2018, 13, e0191866. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Aya, L.F.; Gonzalez-Angulo, A.M. Adjuvant Systemic Therapies in Breast Cancer. Surg. Clin. 2013, 93, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, J.M.; Alfredo, T.M.; Antunes, K.Á.; Cunha, J.D.S.M.D.; Costa, E.M.A.; Lima, E.S.; Silva, D.B.; Carollo, C.A.; Schmitz, W.O.; Boleti, A.P.D.A. Guazuma Ulmifolia Lam. Decreases Oxidative Stress in Blood Cells and Prevents Doxorubicin-Induced Cardiotoxicity. Oxid. Med. Cell. Longev. 2018, 2018, 2935051. [Google Scholar] [CrossRef]
- Olmedo, M.E.; Forster, M.; Moreno, V.; López-Criado, M.P.; Braña, I.; Flynn, M.; Doger, B.; de Miguel, M.; López-Vilariño, J.A.; Núñez, R. Efficacy and Safety of Lurbinectedin and Doxorubicin in Relapsed Small Cell Lung Cancer. Results from an Expansion Cohort of a Phase I Study. Investig. New Drugs 2021, 39, 1275–1283. [Google Scholar] [CrossRef]
- McGowan, J.V.; Chung, R.; Maulik, A.; Piotrowska, I.; Walker, J.M.; Yellon, D.M. Anthracycline Chemotherapy and Cardiotoxicity. Cardiovasc. Drugs Ther. 2017, 31, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, C.I.F.D.S.; André, S.A.; Correia, J.M.F.; Matos, C.L.A.M.D.; Nogueira, F.J.D. Carcinoma Mucoepidermóide Do Pulmão—Estado Da Arte a Propósito de Um Caso Clínico Mucoepidermoid Carcinoma of the Lung—State of Art with Regard to a Case Report. Rev. Soc. Bras. Clin. Med. 2016, 14, 41–44. [Google Scholar]
- Kashkin, K.N.; Musatkina, E.A.; Komelkov, A.V.; Favorskaya, I.A.; Trushkin, E.V.; Shleptsova, V.A.; Sakharov, D.A.; Vinogradova, T.V.; Kopantzev, E.P.; Zinovyeva, M.V. Expression Profiling and Putative Mechanisms of Resistance to Doxorubicin of Human Lung Cancer Cells. In Doklady. Biochemistry and Biophysics; Springer Nature BV: London, UK, 2010; Volume 430, p. 20. [Google Scholar]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to Study Proteins by Circular Dichroism. Biochim. Biophys. Acta-Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Kaminský, J.; Kubelka, J.; Bour, P. Theoretical Modeling of Peptide α-Helical Circular Dichroism in Aqueous Solution. J. Phys. Chem. A 2011, 115, 1734–1742. [Google Scholar] [CrossRef]
- Khezri, A.; Karimi, A.; Yazdian, F.; Jokar, M.; Mofradnia, S.R.; Rashedi, H.; Tavakoli, Z. Molecular Dynamic of Curcumin/Chitosan Interaction Using a Computational Molecular Approach: Emphasis on Biofilm Reduction. Int. J. Biol. Macromol. 2018, 114, 972–978. [Google Scholar] [CrossRef]
- Guo, X.; Ma, C.; Du, Q.; Wei, R.; Wang, L.; Zhou, M.; Chen, T.; Shaw, C. Two Peptides, TsAP-1 and TsAP-2, from the Venom of the Brazilian Yellow Scorpion, Tityus Serrulatus: Evaluation of Their Antimicrobial and Anticancer Activities. Biochimie 2013, 95, 1784–1794. [Google Scholar] [CrossRef]
- Frihling, B.E.F.; Boleti, A.P.D.A.; de Oliveira, C.F.R.; Sanches, S.C.; Cardoso, P.H.d.O.; Verbisck, N.; Macedo, M.L.R.; Rita, P.H.S.; Carvalho, C.M.E.; Migliolo, L. Purification, Characterization and Evaluation of the Antitumoral Activity of a Phospholipase A2 from the Snake Bothrops Moojeni. Pharmaceuticals 2022, 15, 724. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.S.; Oh, J.; Lim, J.S.; Kim, H.J.; Kim, J.-S. Anti-Inflammatory Effect of Wasp Venom in BV-2 Microglial Cells in Comparison with Bee Venom. Insects 2021, 12, 297. [Google Scholar] [CrossRef] [PubMed]
- Migliolo, L.; Silva, O.N.; Silva, P.A.; Costa, M.P.; Costa, C.R.; Nolasco, D.O.; Barbosa, J.A.R.G.; Silva, M.R.R.; Bemquerer, M.P.; Lima, L.M.P.; et al. Structural and Functional Characterization of a Multifunctional Alanine-Rich Peptide Analogue from Pleuronectes Americanus. PLoS ONE 2012, 7, e47047. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Lu, X.; Deng, Z.; Xiao, S.; Yuan, B.; Yang, K. How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane. Molecules 2019, 24, 1775. [Google Scholar] [CrossRef]
- Kim, M.K.; Kang, H.K.; Ko, S.J.; Hong, M.J.; Bang, J.K.; Seo, C.H.; Park, Y. Mechanisms Driving the Antibacterial and Antibiofilm Properties of Hp1404 and Its Analogue Peptides against Multidrug-Resistant Pseudomonas Aeruginosa. Sci. Rep. 2018, 8, 1763. [Google Scholar] [CrossRef]
- Souza E Silva, P.; Ferreira, M.A.; de Moraes, L.F.R.; de Barros, E.; Preza, S.L.E.; Cardoso, M.H.; Franco, O.L.; Migliolo, L. Synthetic Peptides Bioinspired in Temporin-PTa with Antibacterial and Antibiofilm Activity. Chem. Biol. Drug Des. 2022, 100, 51–63. [Google Scholar] [CrossRef]
- Mishra, B.; Wang, G. Ab Initio Design of Potent Anti-MRSA Peptides Based on Database Filtering Technology. J. Am. Chem. Soc. 2012, 134, 12426–12429. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid Phase Synthesis. Science 1986, 232, 341–348. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.C.; Kim, M.H.; Lim, H.T.; Park, Y.; Hahm, K.S. Antimicrobial Activity Studies on a Trypsin–Chymotrypsin Protease Inhibitor Obtained from Potato. Biochem. Biophys. Res. Commun. 2005, 330, 921–927. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., III.; De Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure Validation by Cα Geometry: ϕ, ψ and Cβ Deviation. Proteins Struct. Funct. Bioinform. 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, E.; Hess, B.; Van Der Spoel, D. GROMACS 3.0: A Package for Molecular Simulation and Trajectory Analysis. J. Mol. Model. 2001, 7, 306–317. [Google Scholar] [CrossRef]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B. GROMACS User Manual Version 5.0.4; Royal Institute of Technology and Uppsala University: Uppsala, Sweden, 2014. [Google Scholar]
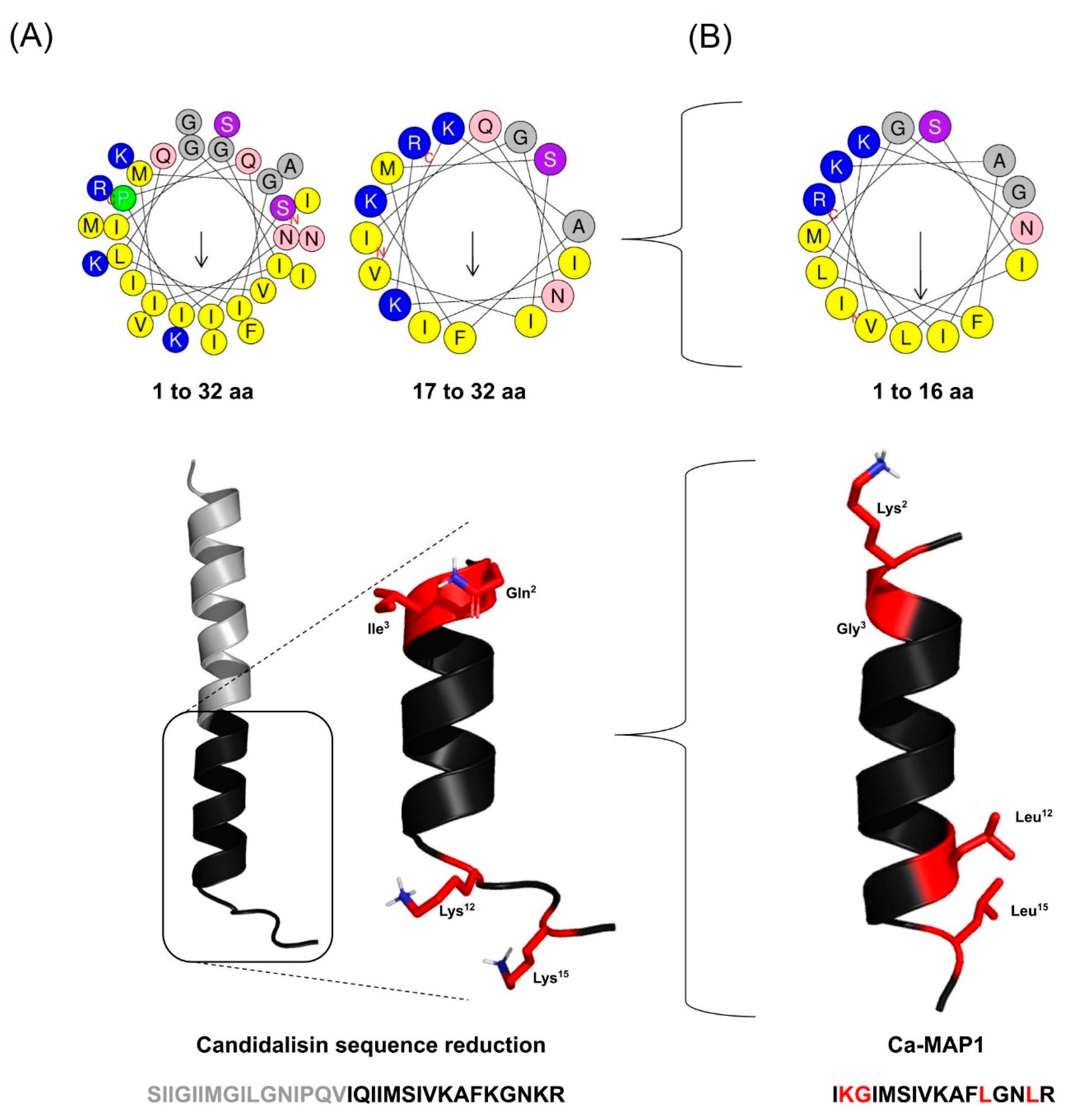
| Name | Alignment | N° of Residues |
|---|---|---|
| Candidalysin | SIIGIIMGILGNIPQVIQIIMSIVKAFKGNKR | 32 |
| Ca-MAP1 | ----------------IKGIMSIVKAFLGNLR | 16 |
| Name | <Z> | Apolar (%) | <H> | <µH> | Mass (Da) |
|---|---|---|---|---|---|
| Candidalysin | +4 | 56.25 | 0.679 | 0.408 | 3464.05 |
| Ca-MAP1 | +3 | 56.25 | 0.607 | 0.631 | 1759.05 |
| Bacteria | Ca-MAP1 (µM) | Ciprofloxacin (µM) | ||
|---|---|---|---|---|
| MIC | MBC | MIC | MBC | |
| Gram-negative | ||||
| Acinetobacter baumannii | 18.1 | >72.7 | 96.5 | >386.3 |
| Escherichia coli | 9.1 | 9.1 | 96.5 | >386.3 |
| Escherichia coli (KPC + 001812446) | 9.1 | 9.1 | 386.3 | 96.5 |
| Klebsiella pneumoniae (ATCC + 13883) | 18.1 | 18.1 | 6 | 193.1 |
| Pseudomonas aeruginosa | >72.7 | >72.7 | 6 | 6 |
| Gram-positive | ||||
| Staphylococcus aureus | 36.3 | 36.3 | 6 | 6 |
| Name | C-Score | RMSD | TM-Score | Z-Score | Rama-Z |
|---|---|---|---|---|---|
| Ca-MAP1 | 0.05 | 0.6 ± 0.6Å | 0.72 ± 0.11 | −0.35 | −1.85 ± 1.76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoso, P.H.d.O.; Boleti, A.P.d.A.; Silva, P.S.e.; Mukoyama, L.T.H.; Guindo, A.S.; Moraes, L.F.R.N.d.; Oliveira, C.F.R.d.; Macedo, M.L.R.; Carvalho, C.M.E.; de Castro, A.P.; et al. Evaluation of a Novel Synthetic Peptide Derived from Cytolytic Mycotoxin Candidalysin. Toxins 2022, 14, 696. https://doi.org/10.3390/toxins14100696
Cardoso PHdO, Boleti APdA, Silva PSe, Mukoyama LTH, Guindo AS, Moraes LFRNd, Oliveira CFRd, Macedo MLR, Carvalho CME, de Castro AP, et al. Evaluation of a Novel Synthetic Peptide Derived from Cytolytic Mycotoxin Candidalysin. Toxins. 2022; 14(10):696. https://doi.org/10.3390/toxins14100696
Chicago/Turabian StyleCardoso, Pedro Henrique de Oliveira, Ana Paula de Araújo Boleti, Patrícia Souza e Silva, Lincoln Takashi Hota Mukoyama, Alexya Sandim Guindo, Luiz Filipe Ramalho Nunes de Moraes, Caio Fernando Ramalho de Oliveira, Maria Ligia Rodrigues Macedo, Cristiano Marcelo Espínola Carvalho, Alinne Pereira de Castro, and et al. 2022. "Evaluation of a Novel Synthetic Peptide Derived from Cytolytic Mycotoxin Candidalysin" Toxins 14, no. 10: 696. https://doi.org/10.3390/toxins14100696
APA StyleCardoso, P. H. d. O., Boleti, A. P. d. A., Silva, P. S. e., Mukoyama, L. T. H., Guindo, A. S., Moraes, L. F. R. N. d., Oliveira, C. F. R. d., Macedo, M. L. R., Carvalho, C. M. E., de Castro, A. P., & Migliolo, L. (2022). Evaluation of a Novel Synthetic Peptide Derived from Cytolytic Mycotoxin Candidalysin. Toxins, 14(10), 696. https://doi.org/10.3390/toxins14100696