Antimicrobial, Insecticidal and Cytotoxic Activity of Linear Venom Peptides from the Pseudoscorpion Chelifer cancroides
Abstract
:1. Introduction
2. Results
2.1. Checacin1 Is Highly Active against Bacteria and Fungi
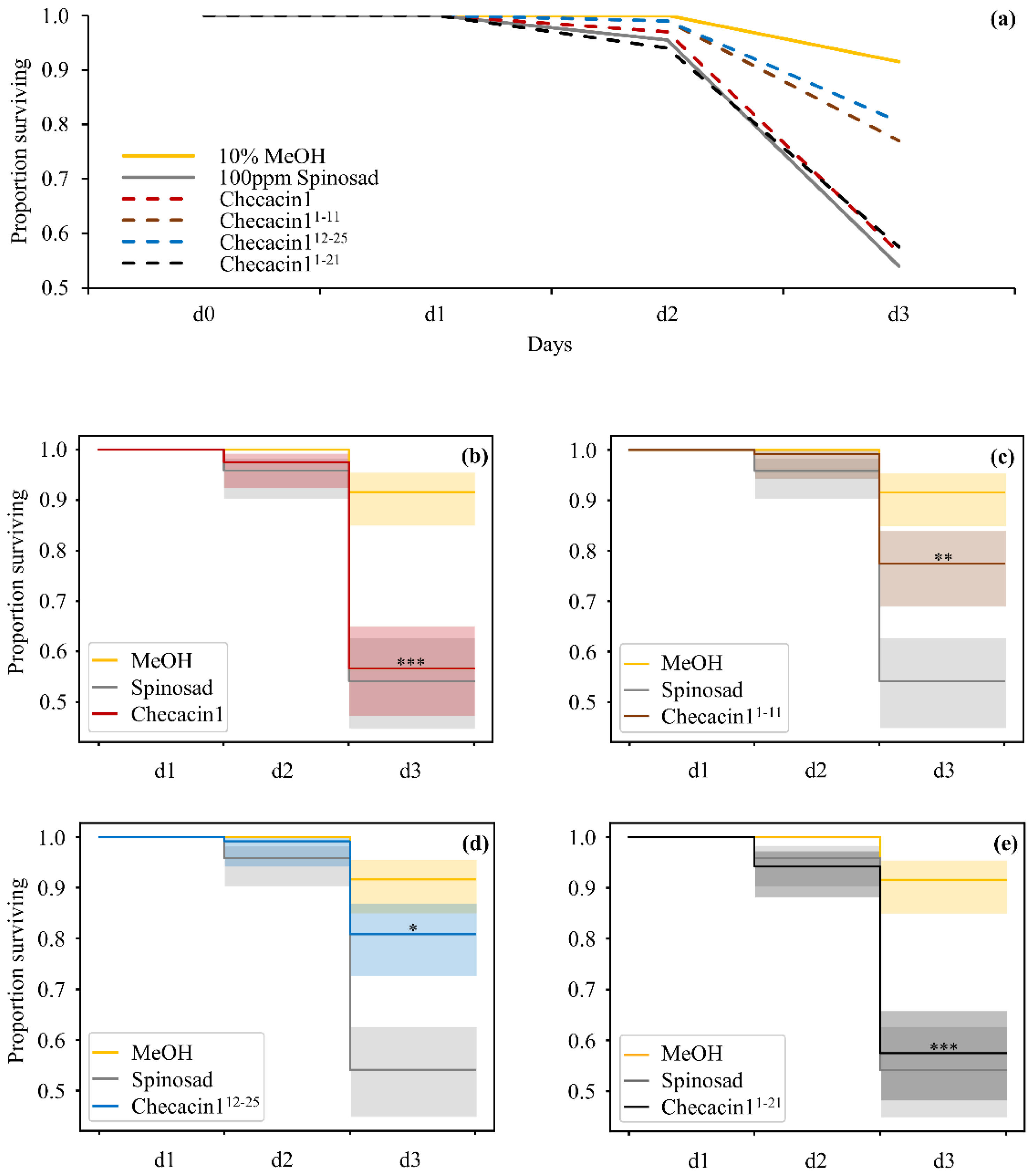
2.2. Orally Administered Checacin1 Is Active against Pea Aphids (Acyrthosiphon pisum)
2.3. Cytotoxic Activity of Checacin1
3. Discussion
4. Conclusions
5. Material and Methods
5.1. Peptide Synthesis
5.2. Feeding Assay on Pea Aphids (A. pisum)
5.3. Antimicrobial Activity Assays
5.4. Cytotoxicity Assays
5.4.1. Cell Culture
5.4.2. Cell Viability Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Bahar, A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion Venom Components as Potential Candidates for Drug Development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef]
- de Barros, E.; Gonçalves, R.M.; Cardoso, M.H.; Santos, N.C.; Franco, O.L.; Cândido, E.S. Snake Venom Cathelicidins as Natural Antimicrobial Peptides. Front. Pharmacol. 2019, 10, 1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebou, A.; Koua, D.; Addablah, A.; Kakou-Ngazoa, S.; Dutertre, S. Combined Proteotranscriptomic-Based Strategy to Discover Novel Antimicrobial Peptides from Cone Snails. Biomedicines 2021, 9, 344. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Harrison, P.L.; Abdel-Rahman, M.A.; Miller, K.; Strong, P.N. Antimicrobial Peptides from Scorpion Venoms. Toxicon 2014, 88, 115–137. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.M.; Reis, P.V.; Pimenta, A.M.C. Antimicrobial Peptides in Spider Venoms. In Spider Venoms; Gopalakrishnakone, P., Corzo, G.A., de Lima, M.E., Diego-García, E., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 361–377. ISBN 978-94-007-6388-3. [Google Scholar]
- Lüddecke, T.; Herzig, V.; Reumont, B.M.; Vilcinskas, A. The Biology and Evolution of Spider Venoms. Biol. Rev. 2021, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Wahed, A.; Yosri, N.; Sakr, H.H.; Du, M.; Algethami, A.F.M.; Zhao, C.; Abdelazeem, A.H.; Tahir, H.E.; Masry, S.H.D.; Abdel-Daim, M.M.; et al. Wasp Venom Biochemical Components and Their Potential in Biological Applications and Nanotechnological Interventions. Toxins 2021, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, K.; Zhu, W.; Ye, X.; Ding, L.; Jiang, H.; Li, F.; Chen, Z.; Luo, X. Two New Cationic α-Helical Peptides Identified from the Venom Gland of Liocheles australasiae Possess Antimicrobial Activity against Methicillin-Resistant Staphylococci. Toxicon 2021, 196, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Sani, M.-A.; Silva-Sanchez, J.; Jiménez-Vargas, J.M.; Reyna-Flores, F.; Winkel, K.D.; Wright, C.E.; Possani, L.D.; Separovic, F. Membrane Interactions and Biological Activity of Antimicrobial Peptides from Australian Scorpion. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 2140–2148. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Yang, F.; Li, F.; Li, Z.; Lang, Y.; Shen, B.; Wu, Y.; Li, W.; Harrison, P.L.; Strong, P.N.; et al. Therapeutic Potential of a Scorpion Venom-Derived Antimicrobial Peptide and Its Homologs Against Antibiotic-Resistant Gram-Positive Bacteria. Front. Microbiol. 2018, 9, 1159. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramirez, K.; Skaljac, M.; Grotmann, J.; Kirfel, P.; Vilcinskas, A. Orally Delivered Scorpion Antimicrobial Peptides Exhibit Activity against Pea Aphid (Acyrthosiphon pisum) and Its Bacterial Symbionts. Toxins 2017, 9, 261. [Google Scholar] [CrossRef]
- Van Emden, H.F.; Harrington, R. (Eds.) Aphids as Crop Pests, 2nd ed.; CABI: Wallingford/Oxfordshire, UK; Boston, MA, USA, 2017; ISBN 978-1-78064-709-8. [Google Scholar]
- Harvey, M.S. Pseudoscorpions of the World, Version 3.0. Available online: http://www.museum.wa.gov.au/catalogues/pseudoscorpions (accessed on 29 November 2021).
- Ontano, A.Z.; Gainett, G.; Aharon, S.; Ballesteros, J.A.; Benavides, L.R.; Corbett, K.F.; Gavish-Regev, E.; Harvey, M.S.; Monsma, S.; Santibáñez-López, C.E.; et al. Taxonomic Sampling and Rare Genomic Changes Overcome Long-Branch Attraction in the Phylogenetic Placement of Pseudoscorpions. Mol. Biol. Evol. 2021, 38, 2446–2467. [Google Scholar] [CrossRef] [PubMed]
- Lüddecke, T.; Vilcinskas, A.; Lemke, S. Phylogeny-Guided Selection of Priority Groups for Venom Bioprospecting: Harvesting Toxin Sequences in Tarantulas as a Case Study. Toxins 2019, 11, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santibáñez-López, C.; Ontano, A.; Harvey, M.; Sharma, P. Transcriptomic Analysis of Pseudoscorpion Venom Reveals a Unique Cocktail Dominated by Enzymes and Protease Inhibitors. Toxins 2018, 10, 207. [Google Scholar] [CrossRef] [Green Version]
- Lebenzon, J.E.; Toxopeus, J.; Anthony, S.E.; Sinclair, B.J. De Novo Assembly and Characterisation of the Transcriptome of the Beringian Pseudoscorpion. Can. Entomol. 2021, 153, 301–313. [Google Scholar] [CrossRef]
- Smith, J.; Undheim, E. True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra subspinipes. Toxins 2018, 10, 96. [Google Scholar] [CrossRef] [Green Version]
- Krämer, J.; Pohl, H.; Predel, R. Venom Collection and Analysis in the Pseudoscorpion Chelifer cancroides (Pseudoscorpiones: Cheliferidae). Toxicon 2019, 162, 15–23. [Google Scholar] [CrossRef]
- Krämer, J.; Peigneur, S.; Tytgat, J.; Jenner, R.A.; van Toor, R.; Predel, R. A Pseudoscorpion’s Promising Pinch: The Venom of Chelifer cancroides Contains a Rich Source of Novel Compounds. Toxicon 2021, S0041010121002233. [Google Scholar] [CrossRef] [PubMed]
- Diego-García, E.; Caliskan, F.; Tytgat, J. The Mediterranean Scorpion Mesobuthus gibbosus (Scorpiones, Buthidae): Transcriptome Analysis and Organization of the Genome Encoding Chlorotoxin-like Peptides. BMC Genom. 2014, 15, 295. [Google Scholar] [CrossRef] [Green Version]
- Hancock, R.E.W.; Falla, T.; Brown, M. Cationic Bactericidal Peptides. In Advances in Microbial Physiology; Elsevier: Amsterdam, The Netherlands, 1995; Volume 37, pp. 135–175. ISBN 978-0-12-027737-7. [Google Scholar]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [Green Version]
- Shafee, T.M.A.; Lay, F.T.; Phan, T.K.; Anderson, M.A.; Hulett, M.D. Convergent Evolution of Defensin Sequence, Structure and Function. Cell. Mol. Life Sci. 2017, 74, 663–682. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Langenegger, N.; Heller, M.; Koua, D.; Nentwig, W. The Dual Prey-Inactivation Strategy of Spiders—In-Depth Venomic Analysis of Cupiennius salei. Toxins 2019, 11, 167. [Google Scholar] [CrossRef] [Green Version]
- Lüddecke, T.; Schulz, S.; Steinfartz, S.; Vences, M. A Salamander’s Toxic Arsenal: Review of Skin Poison Diversity and Function in True Salamanders, Genus Salamandra. Sci. Nat. 2018, 105, 56. [Google Scholar] [CrossRef] [PubMed]
- Bacalum, M.; Radu, M. Cationic Antimicrobial Peptides Cytotoxicity on Mammalian Cells: An Analysis Using Therapeutic Index Integrative Concept. Int. J. Pept. Res. Ther. 2015, 21, 47–55. [Google Scholar] [CrossRef]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Rational Design of α-Helical Antimicrobial Peptides with Enhanced Activities and Specificity/Therapeutic Index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [Green Version]
- Gremski, L.H.; da Silveira, R.B.; Chaim, O.M.; Probst, C.M.; Ferrer, V.P.; Nowatzki, J.; Weinschutz, H.C.; Madeira, H.M.; Gremski, W.; Nader, H.B.; et al. A Novel Expression Profile of the Loxosceles intermedia Spider Venomous Gland Revealed by Transcriptome Analysis. Mol. Biosyst. 2010, 6, 2403. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, B.; Kuhn-Nentwig, L.; Tromp, J.; Kampfer, U.; Schaller, J.; Schurch, S.; Nentwig, W. CSTX-13, a Highly Synergistically Acting Two-Chain Neurotoxic Enhancer in the Venom of the Spider Cupiennius salei (Ctenidae). Proc. Natl. Acad. Sci. USA 2004, 101, 11251–11256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heep, J.; Skaljac, M.; Grotmann, J.; Kessel, T.; Seip, M.; Schmidtberg, H.; Vilcinskas, A. Identification and Functional Characterization of a Novel Insecticidal Decapeptide from the Myrmicine Ant Manica rubida. Toxins 2019, 11, 562. [Google Scholar] [CrossRef] [Green Version]
- Heep, J.; Klaus, A.; Kessel, T.; Seip, M.; Vilcinskas, A.; Skaljac, M. Proteomic Analysis of the Venom from the Ruby Ant Myrmica rubra and the Isolation of a Novel Insecticidal Decapeptide. Insects 2019, 10, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akey, D.H.; Beck, S.D. Continuous Rearing of the Pea Aphid, Acyrthosiphon pisum,1 on a Holidic Diet2. Ann. Entomol. Soc. Am. 1971, 64, 353–356. [Google Scholar] [CrossRef]
- Sadeghi, A.; Van Damme, E.J.M.; Smagghe, G. Evaluation of the Susceptibility of the Pea Aphid, Acyrthosiphon pisum, to a Selection of Novel Biorational Insecticides Using an Artificial Diet. J. Insect Sci. 2009, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, G.; Simmons, N.L. Identification of two Strains of Cultured Canine Renal Epithelial Cells (MDCK Cells) which Display Entirely Different Physiological Properties. Q. J. Exp. Physiol. 1981, 66, 61–72. [Google Scholar] [CrossRef] [PubMed]




| Compounds | MIC (µM) | ||||||
|---|---|---|---|---|---|---|---|
| Ec | Pa | Ms | Sa | Af | Ca | ||
| ATCC 35218 | ATCC27853 | ATCC607 | ATCC33592 | ATCC9170 | FH2173 | ||
| TEM-1 | TEM-1 | MRSA | |||||
| CAMHII | CAMH-C | CAMHII | CAMHII | CAMHII | CAMHII | CAMHII | |
| Checacin1 | 1.6–0.8 | 1.6 | 12.5 | 25 | 1.6 | 50 | 6.25 |
| Checacin11−11 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| Checacin112−25 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| Checacin11−21 | >50 | >50 | >50 | >50 | 12.5 | >50 | >50 |
| Rifampicin | 4.9 | 19.4 | 38.9 | 38.9–19.4 | >77.8 | NA | NA |
| Tetracycline | 4.5 | 36–18 | >144.1 | 0.14 | >72 | NA | NA |
| Gentamycin */Isoniazid ’’ | 2.1–1 | 1–0.5 | 1 | 29.2–14.6 ’’ | 0.5–0.26 | NA | NA |
| Tebuconazole | NA | NA | NA | NA | NA | 0.19 | 0.19–0.09 |
| Amphotericin B | NA | NA | NA | NA | NA | 1.1 | 8.6–4.3 |
| Nystatin | NA | NA | NA | NA | NA | 1.1 | 8.6 |
| readout | MTT | MTT | MTT | BTG | MTT | BTG | BTG |
| Component/Company ID | Sequence | Purity |
|---|---|---|
| Checacin1/D-4040 | FFGAIAKLAMKFLPAIYKQIQKKRK * | 96.75% |
| Checacin11−11/D-4041 | FFGAIAKLAMK | 97.84% |
| Checacin112−25/D-4042 | FLPAIYKQIQKKRK * | 98.07% |
| Checacin11−21/D-4043 | FFGAIAKLAMKFLPAIYKQIQ | 96.87% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krämer, J.; Lüddecke, T.; Marner, M.; Maiworm, E.; Eichberg, J.; Hardes, K.; Schäberle, T.F.; Vilcinskas, A.; Predel, R. Antimicrobial, Insecticidal and Cytotoxic Activity of Linear Venom Peptides from the Pseudoscorpion Chelifer cancroides. Toxins 2022, 14, 58. https://doi.org/10.3390/toxins14010058
Krämer J, Lüddecke T, Marner M, Maiworm E, Eichberg J, Hardes K, Schäberle TF, Vilcinskas A, Predel R. Antimicrobial, Insecticidal and Cytotoxic Activity of Linear Venom Peptides from the Pseudoscorpion Chelifer cancroides. Toxins. 2022; 14(1):58. https://doi.org/10.3390/toxins14010058
Chicago/Turabian StyleKrämer, Jonas, Tim Lüddecke, Michael Marner, Elena Maiworm, Johanna Eichberg, Kornelia Hardes, Till F. Schäberle, Andreas Vilcinskas, and Reinhard Predel. 2022. "Antimicrobial, Insecticidal and Cytotoxic Activity of Linear Venom Peptides from the Pseudoscorpion Chelifer cancroides" Toxins 14, no. 1: 58. https://doi.org/10.3390/toxins14010058
APA StyleKrämer, J., Lüddecke, T., Marner, M., Maiworm, E., Eichberg, J., Hardes, K., Schäberle, T. F., Vilcinskas, A., & Predel, R. (2022). Antimicrobial, Insecticidal and Cytotoxic Activity of Linear Venom Peptides from the Pseudoscorpion Chelifer cancroides. Toxins, 14(1), 58. https://doi.org/10.3390/toxins14010058