Proteomic Identification and Quantification of Snake Venom Biomarkers in Venom and Plasma Extracellular Vesicles
Abstract
:1. Introduction
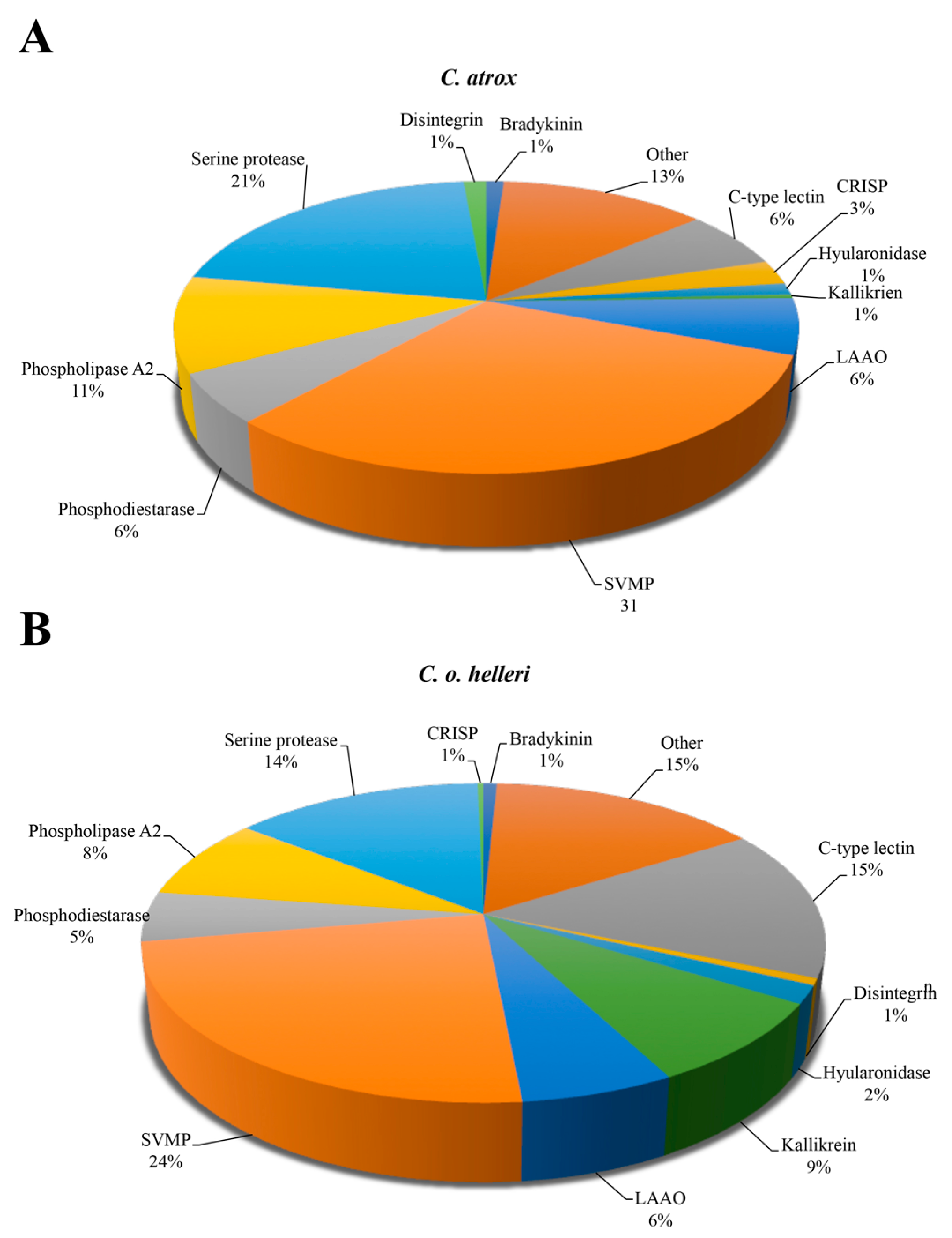
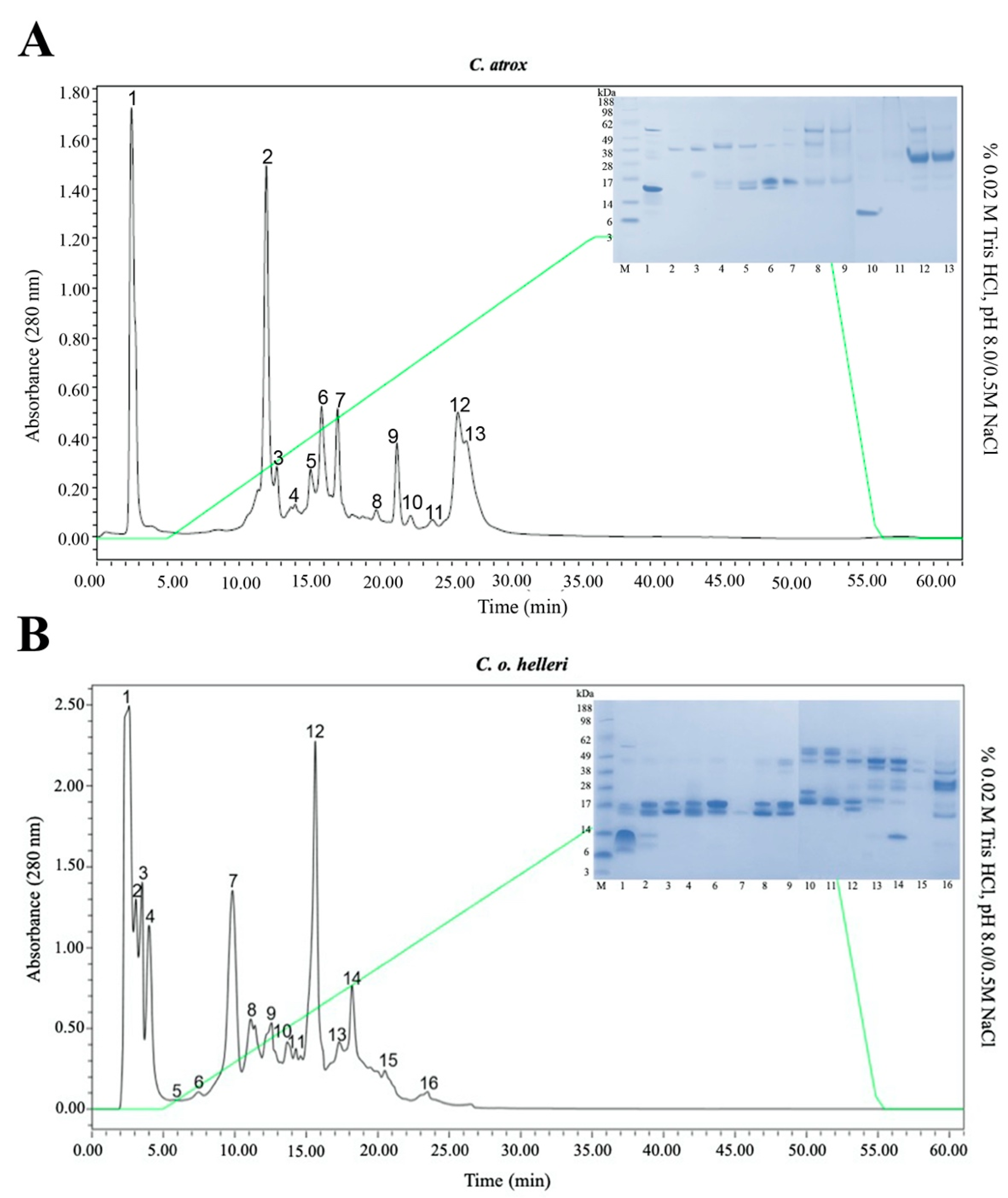
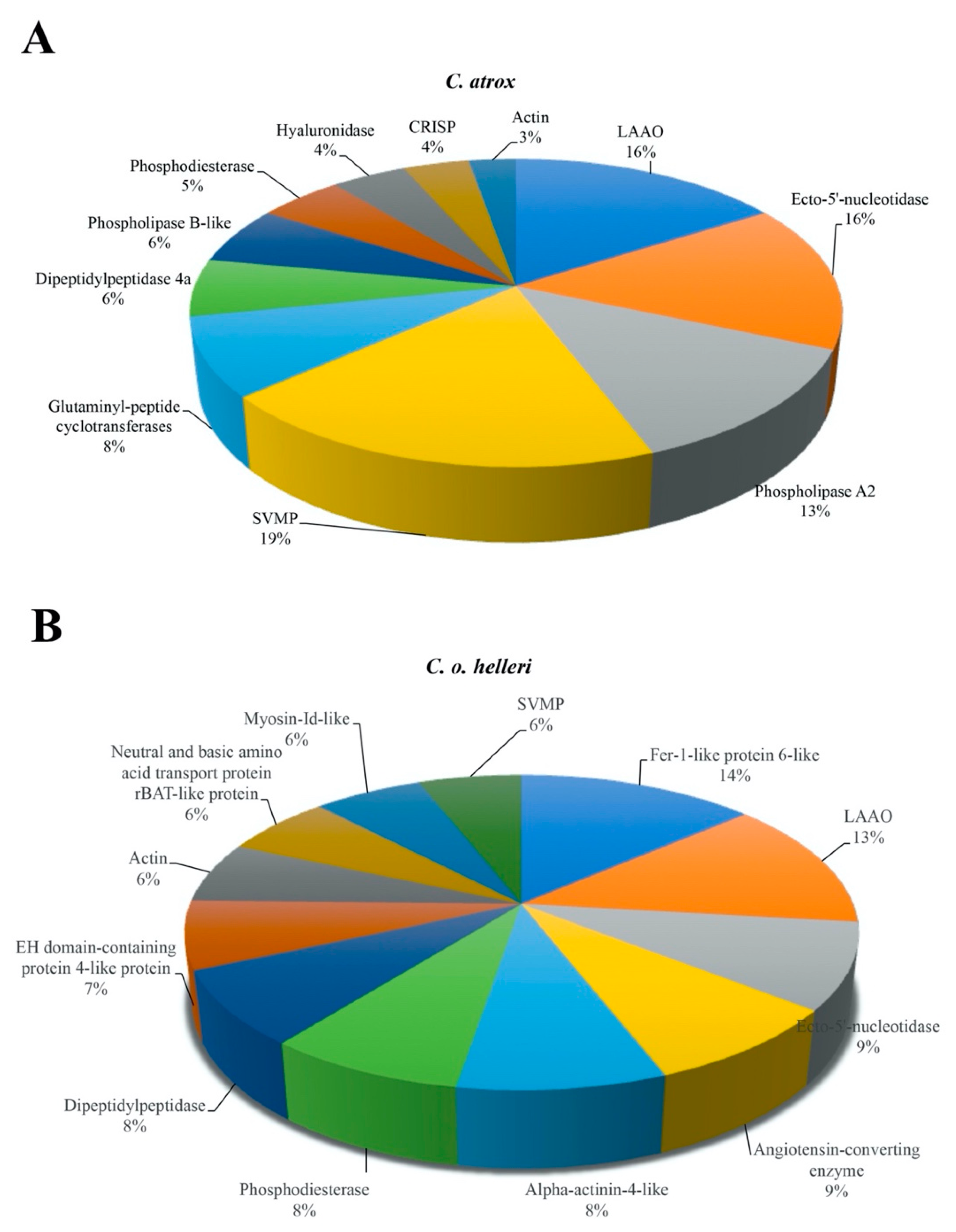
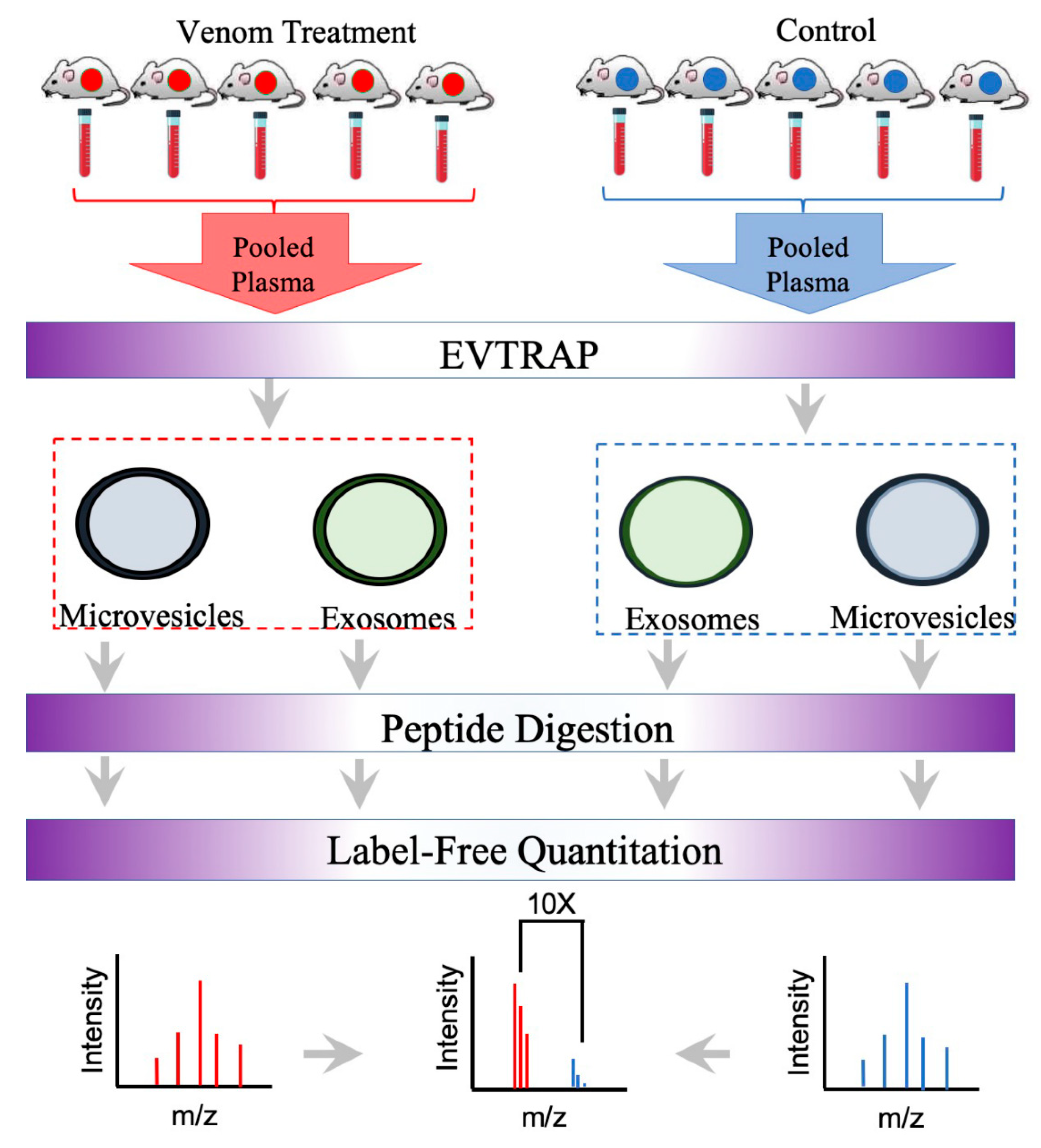
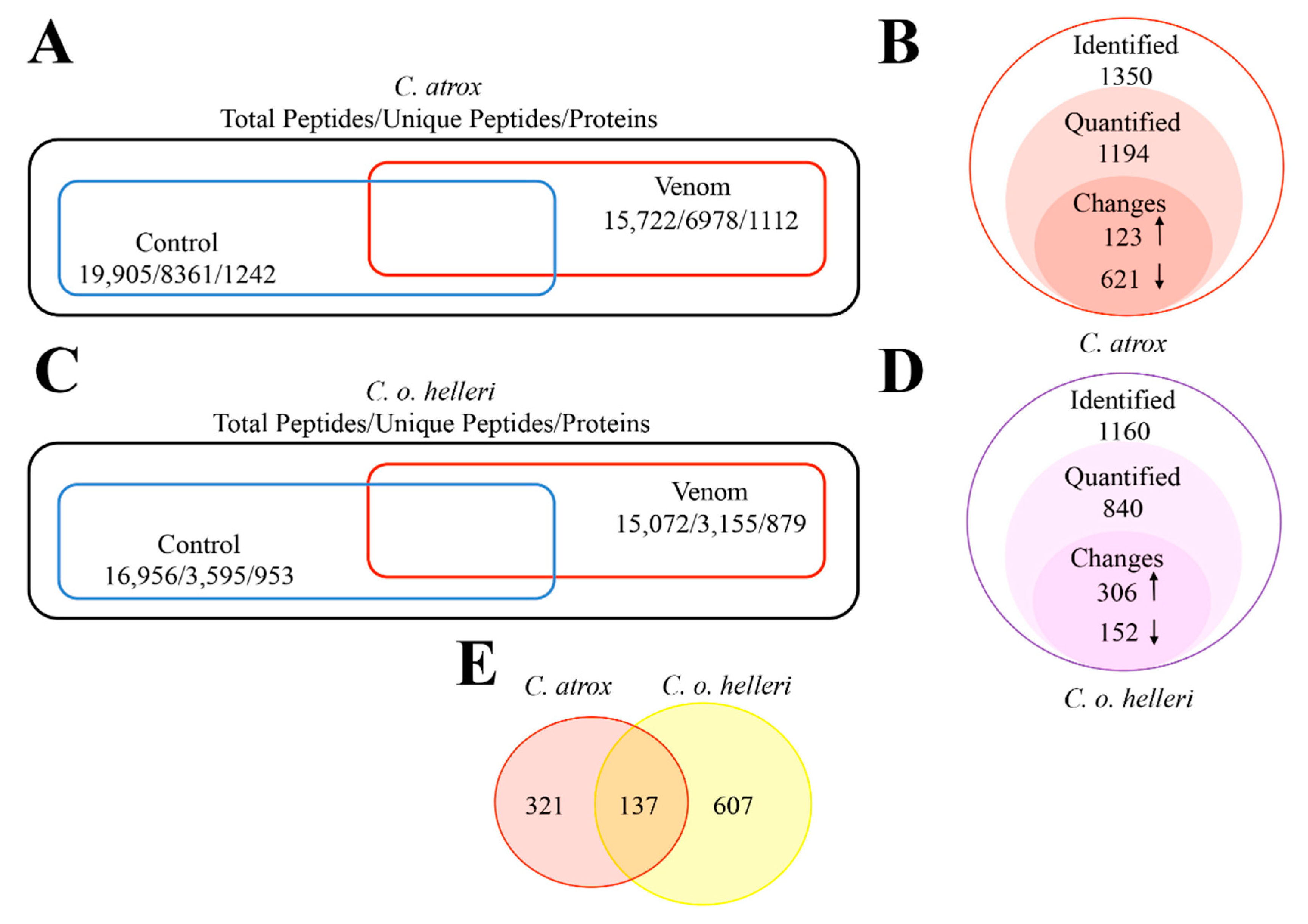
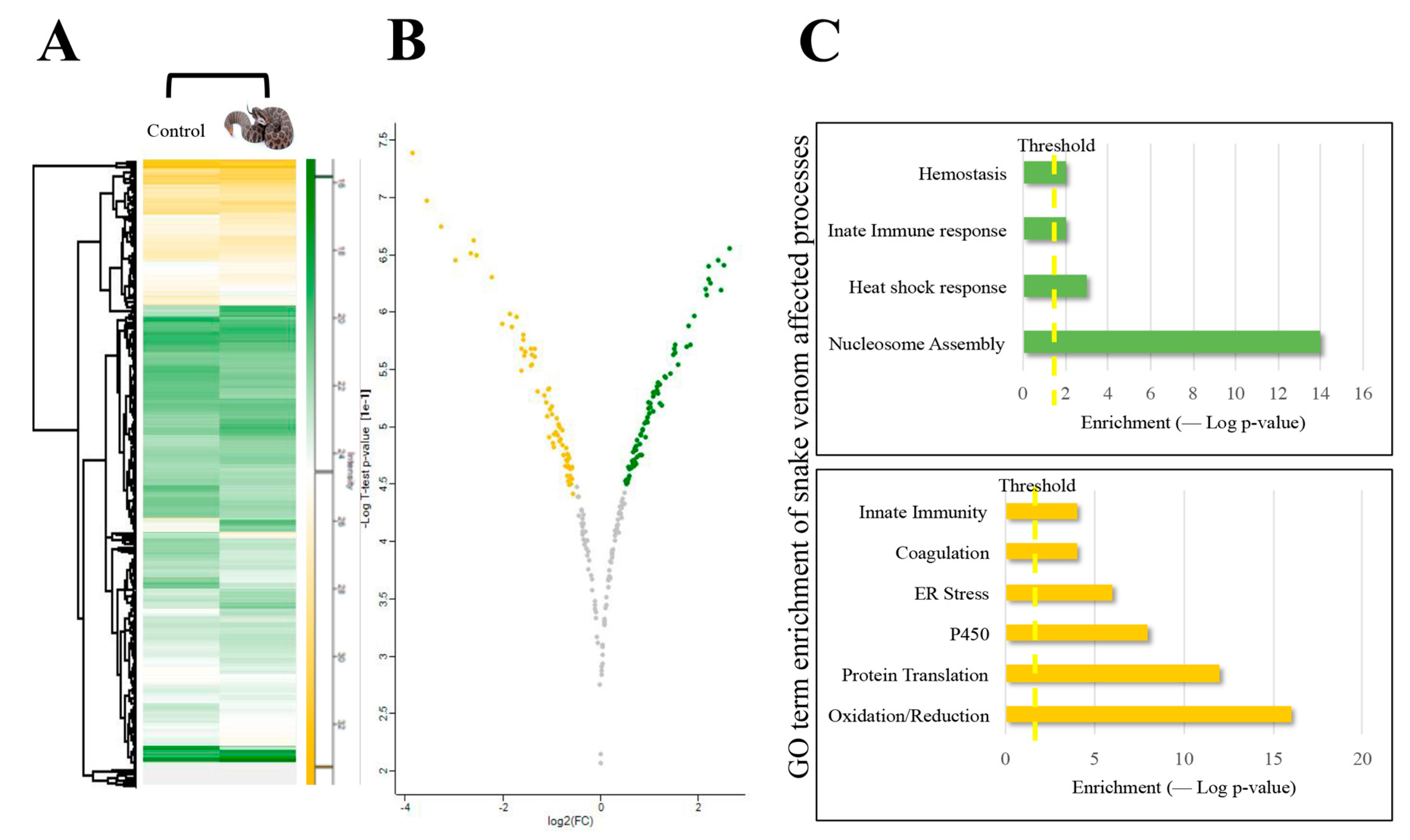
2. Results and Discussion
3. Conclusions
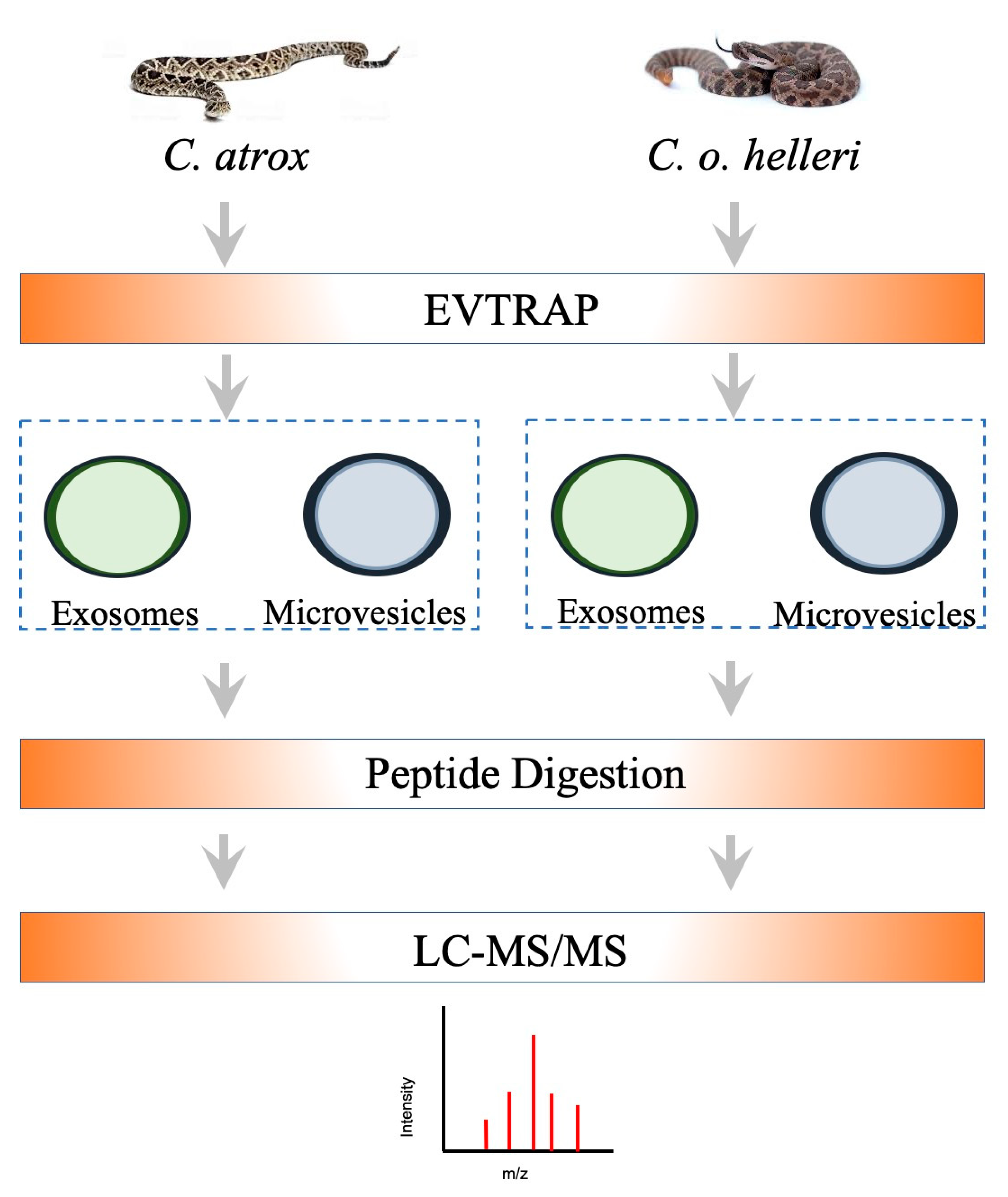
4. Materials and Methods
4.1. Venom Collection
4.2. Snake Venom and Mouse Plasma Extracellular Vesicles Enrichment
4.3. Anion Exchange DEAE Chromatography
4.4. Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
4.5. Subcutaneous Mouse Injection
4.6. Ethical Procedures
4.7. Snake Venom and svEV Proteomic Analysis
4.8. LC–MS–MS
4.9. Data Acquisition, Quantification, and Bioinformatics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perez, J.C.; Sanchez, E.E. Natural protease inhibitors to hemorrhagins in snake venoms and their potential use in medicine. Toxicon 1999, 37, 703–728. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A Review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, D.J.; Faiz, M.A.; Abela-Ridder, B.; Ainsworth, S.; Bulfone, T.C.; Nickerson, A.D.; Habib, A.G.; Junghanss, T.; Fan, H.W.; Turner, M.; et al. Strategy for a globally coordinated response to a priority neglected tropical disease: Snakebite envenoming. PLoS Negl. Trop. Dis. 2019, 13, e0007059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, J.M. Global availability of antivenoms: The relevance of public manufacturing laboratories. Toxins 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, S.A.; Boyer, L.V.; Benson, B.E.; Rogers, J.J. AAPCC database characterization of native U.S. venomous snake exposures, 2001–2005. Clin. Toxicol (Phila) 2009, 47, 327–335. [Google Scholar] [CrossRef]
- Ernst, C.H.; Ernst, E.M. Venomous Reptiles of the United States, Canada, and Northern Mexico; John Hopkins University Press: Baltimore, MD, USA, 2012. [Google Scholar]
- Klauber, L.M. Their Habits, Life Histories, and Influence on Mankind; University of California Press: Berkeley, CA, USA, 1956. [Google Scholar]
- Campbell, J.; Lamar, W.W. Venomous Reptiles of the Western Hemisphere; Cornell University Press: Ithaca, NY, USA, 2004. [Google Scholar]
- Lavonas, E.J.; Khatri, V.; Daugherty, C.; Bucher-Bartelson, B.; King, T.; Dart, R.C. Medically significant late bleeding after treated crotaline envenomation: A systematic review. Ann. Emerg. Med. 2014, 63, 71–78. [Google Scholar] [CrossRef]
- Boyer, L.V.; Seifert, S.A.; Clark, R.F.; McNally, J.T.; Williams, S.R.; Nordt, S.P.; Walter, F.G.; Dart, R.C. Recurrent and persistent coagulopathy following pit viper envenomation. Arch. Intern. Med. 1999, 159, 706–710. [Google Scholar] [CrossRef] [Green Version]
- Dart, R.C.; McNally, J. Efficacy, safety, and use of snake antivenoms in the United States. Ann. Emerg. Med. 2001, 37, 181–188. [Google Scholar] [CrossRef]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef] [Green Version]
- Calvete, J.J.; Juarez, P.; Sanz, L. Snake venomics. Strategy and applications. J. Mass Spectrom. 2007, 42, 1405–1414. [Google Scholar] [CrossRef]
- Moura-da-Silva, A.M.; Almeida, M.T.; Portes-Junior, J.A.; Nicolau, C.A.; Gomes-Neto, F.; Valente, R.H. Processing of Snake Venom Metalloproteinases: Generation of Toxin Diversity and Enzyme Inactivation. Toxins 2016, 8, 183. [Google Scholar] [CrossRef]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.M. Exosomes biological significance: A concise review. Blood Cells Mol. Dis. 2006, 36, 315–321. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranyai, T.; Herczeg, K.; Onodi, Z.; Voszka, I.; Modos, K.; Marton, N.; Nagy, G.; Mager, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciregia, F.; Urbani, A.; Palmisano, G. Extracellular vesicles in brain tumors and neurodegenerative diseases. Front. Mol. Neurosci. 2017, 10, 276. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.; Marban, E. Exosomes: Fundamental biology and roles in cardiovascular physiology. Annu. Rev. Physiol. 2016, 78, 67–83. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Warshawsky, H.; Haddad, A.; Goncalves, R.P.; Valeri, V.; De Lucca, F.L. Fine structure of the venom gland epithelium of the South American rattlesnake and radioautographic studies of protein formation by the secretory cells. Am. J. Anat. 1973, 138, 79–119. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Murayama, N.; Fujita, Y.; Yanoshita, R. Characterization and cDNA cloning of aminopeptidase A from the venom of Gloydius blomhoffi brevicaudus. Toxicon 2007, 49, 1172–1181. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, S.M.; Fernandes, W.; Sant’Anna, S.S.; Yamanouye, N. Microvesicles in the venom of Crotalus durissus terrificus (Serpentes, Viperidae). Toxicon 2007, 49, 106–110. [Google Scholar] [CrossRef]
- Souza-Imberg, A.; Carneiro, S.M.; Giannotti, K.C.; Sant’Anna, S.S.; Yamanouye, N. Origin and characterization of small membranous vesicles present in the venom of Crotalus durissus terrificus. Toxicon 2017, 136, 27–33. [Google Scholar] [CrossRef]
- Carregari, V.C.; Rosa-Fernandes, L.; Baldasso, P.; Bydlowski, S.P.; Marangoni, S.; Larsen, M.R.; Palmisano, G. Snake Venom Extracellular vesicles (SVEVs) reveal wide molecular and functional proteome diversity. Sci Rep. 2018, 8, 12067. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, L.; Iliuk, A.; Tao, W.A. Highly efficient phosphoproteome capture and analysis from urinary extracellular vesicles. J. Proteome Res. 2018, 17, 3308–3316. [Google Scholar] [CrossRef] [PubMed]
- Iliuk, A.; Wu, X.; Li, L.; Sun, J.; Hadisurya, M.; Boris, R.S.; Tao, W.A. Plasma-derived extracellular vesicle phosphoproteomics through chemical affinity purification. J. Proteome Res. 2020, 19, 2563–2574. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Sanz, L.; Angulo, Y.; Lomonte, B.; Gutierrez, J.M. Venoms, venomics, antivenomics. FEBS Lett. 2009, 583, 1736–1743. [Google Scholar] [CrossRef] [Green Version]
- Ayres, L.R.; Recio Ados, R.; Burin, S.M.; Pereira, J.C.; Martins, A.C.; Sampaio, S.V.; de Castro, F.A.; Pereira-Crott, L.S. Bothrops snake venoms and their isolated toxins, an L-amino acid oxidase and a serine protease, modulate human complement system pathways. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burin, S.M.; Menaldo, D.L.; Sampaio, S.V.; Frantz, F.G.; Castro, F.A. An overview of the immune modulating effects of enzymatic toxins from snake venoms. Int. J. Biol. Macromol. 2018, 109, 664–671. [Google Scholar] [CrossRef]
- Sales, P.B.; Santoro, M.L. Nucleotidase and DNase activities in Brazilian snake venoms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008, 147, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Arun, R.; Arafat, A.S.; D’Souza, C.J.; Sivaramakrishnan, V.; Dhananjaya, B.L. Vanillin analog–vanillyl mandelic acid, a novel specific inhibitor of snake venom 5′-nucleotidase. Arch. Pharm. (Weinheim) 2014, 347, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, T.; Araki, S.; Mori, H.; Takeda, S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007, 581, 2416–2422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, H.F.; Mellows, B.A.; Mitchell, R.; Sfyri, P.; Layfield, H.J.; Salamah, M.; Vaiyapuri, R.; Collins-Hooper, H.; Bicknell, A.B.; Matsakas, A.; et al. Mechanisms underpinning the permanent muscle damage induced by snake venom metalloprotease. PLoS Negl. Trop. Dis. 2019, 13, e0007041. [Google Scholar] [CrossRef] [Green Version]
- Ullah, A.; Ullah, K.; Ali, H.; Betzel, C.; Ur Rehman, S. The sequence and a three-dimensional structural analysis reveal substrate specificity among snake venom phosphodiesterases. Toxins 2019, 11, 625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aird, S.D. Snake venom dipeptidyl peptidase IV: Taxonomic distribution and quantitative variation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 150, 222–228. [Google Scholar] [CrossRef]
- Peterfi, O.; Boda, F.; Szabo, Z.; Ferencz, E.; Baba, L. Hypotensive Snake Venom Components–A Mini-Review. Molecules 2019, 24, 2778. [Google Scholar] [CrossRef] [Green Version]
- Dreyer, F.; Baur, A. Biogenesis and Functions of Exosomes and Extracellular Vesicles. Methods Mol. Biol. 2016, 1448, 201–216. [Google Scholar] [CrossRef]
- Tang, H.Y.; Cai, M. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell Biol. 1996, 16, 4897–4914. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Zhang, L.; Guo, A.; Liu, L.C.; Yu, F.; Diao, N.; Xu, C.; Wang, D. Overexpression of FER1L4 promotes the apoptosis and suppresses epithelial-mesenchymal transition and stemness markers via activating PI3K/AKT signaling pathway in osteosarcoma cells. Pathol. Res. Pract. 2019, 215, 152412. [Google Scholar] [CrossRef] [PubMed]
- Waiddyanatha, S.; Silva, A.; Siribaddana, S.; Isbister, G.K. Long-term Effects of Snake Envenoming. Toxins 2019, 11, 193. [Google Scholar] [CrossRef] [Green Version]
- Enjeti, A.K.; Lincz, L.F.; Seldon, M.; Isbister, G.K. Circulating microvesicles in snakebite patients with microangiopathy. Res. Pract. Thromb. Haemost. 2019, 3, 121–125. [Google Scholar] [CrossRef]
- Matsui, T.; Fujimura, Y.; Titani, K. Snake venom proteases affecting hemostasis and thrombosis. Biochim. Biophys. Acta 2000, 1477, 146–156. [Google Scholar] [CrossRef]
- de Carvalho, A.E.Z.; Giannotti, K.; Junior, E.L.; Matsubara, M.; Santos, M.C.D.; Fortes-Dias, C.L.; Teixeira, C. Crotalus durissus ruruima Snake Venom and a Phospholipase A2 Isolated from This Venom Elicit Macrophages to Form Lipid Droplets and Synthesize Inflammatory Lipid Mediators. J. Immunol. Res. 2019, 2019, 2745286. [Google Scholar] [CrossRef] [Green Version]
- Ghneim, H.K.; Al-Sheikh, Y.A.; Aboul-Soud, M.A. The effect of Walterinnesia aegyptia venom proteins on TCA cycle activity and mitochondrial NAD(+)-redox state in cultured human fibroblasts. Biomed. Res. Int. 2015, 2015, 738147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza, T.P.; Abreu, D.S.; Carepo, M.S.P.; Silva, M.A.S.; Zampieri, D.; Eberlin, M.N.; Paulo, T.F.; Sousa, E.H.S.; Longhinotti, E.; Diogenes, I.C.N. Effect of Crotalus basiliscus snake venom on the redox reaction of myoglobin. J. Biol. Inorg. Chem. 2019, 24, 171–178. [Google Scholar] [CrossRef]
- Masuda, T.; Tomita, M.; Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008, 7, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]



| Accession No. | Protein | Fold Change |
|---|---|---|
| Q05421 | Cytochrome P450 2E1 | >100 |
| O88451 | Retinol dehydrogenase 7 | >100 |
| Q4VAA2 | Protein CDV3 | >100 |
| Q9CQW3 | Vitamin K-dependent protein Z | >100 |
| Q61316 | Heat shock 70 kDa protein 4 | >100 |
| Q99KC8 | von Willebrand factor A domain-containing protein 5A | >100 |
| P50172 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | >100 |
| O88587 | Catechol O-methyltransferase | >100 |
| Q64152 | Transcription factor BTF3 | >100 |
| A0A0R4IZY2 | Cytochrome P450 2D26 | >100 |
| Q20BD0 | Heterogeneous nuclear ribonucleoprotein A/B | >100 |
| Q9R0H2 | Endomucin | >100 |
| P06880 | Somatotropin | >100 |
| P97429 | Annexin A4 | >100 |
| P56654 | Cytochrome P450 2C37 | >100 |
| P70296 | Phosphatidylethanolamine-binding protein 1 | >100 |
| P56656 | Cytochrome P450 2C39 | >100 |
| Q8K0C5 | Zymogen granule membrane protein 16 | >100 |
| H3BKH6 | S-formylglutathione hydrolase | >100 |
| Accession No. | Protein | Fold Change |
|---|---|---|
| Q9QYG0 | Protein NDRG2 | >100 |
| B7ZW98 | Ank1 protein | >100 |
| Q9Z239 | Phospholemman | >100 |
| Q8BYH8 | Chromodomain-helicase-DNA-binding protein 9 | >100 |
| P22437 | Prostaglandin G/H synthase 1 | >100 |
| Q9CZE3 | Ras-related protein Rab-32 | >100 |
| Q8BP47 | Asparagine--tRNA ligase, cytoplasmic | >100 |
| A0A0A6YVW3 | Protein Ighv1-23 (Fragment) | >100 |
| Q80XI4 | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | >100 |
| Q91YE6 | Importin-9 | >100 |
| D3YZZ5 | Protein Tmed7 | >100 |
| Q9CR86 | Calcium-regulated heat stable protein 1 | >100 |
| O70435 | Proteasome subunit alpha type-3 | >100 |
| P61953 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-11 | >100 |
| P01748 | Ig heavy chain V region 23 | >100 |
| Q9Z1P6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 7 | >100 |
| Q3TVI8 | Pre-B-cell leukemia transcription factor-interacting protein 1 | >100 |
| P56393 | Cytochrome c oxidase subunit 7B, mitochondrial | >100 |
| D3Z2F8 | Actin-related protein 2/3 complex subunit 3 | >100 |
| O08795 | Glucosidase 2 subunit beta | >100 |
| A6PWS5 | Gelsolin (Fragment) | >100 |
| P62334 | 26S protease regulatory subunit 10B | >100 |
| P55821 | Stathmin-2 | >100 |
| Accession No. | Protein | Fold Change |
|---|---|---|
| A8DUK4 | Beta-globin | >100 |
| A0A0R4J0I9 | Low-density lipoprotein receptor-related protein 1 | >100 |
| P01644 | Ig kappa chain V–V region | >100 |
| G3UXX3 | Sepiapterin reductase | >100 |
| A0A1W2P7F1 | Complement component 1, s subcomponent 2 | >100 |
| B2RT14 | UDP-glucuronosyltransferase | >100 |
| Q01279 | Epidermal growth factor receptor | >100 |
| Q8R0Y6 | Cytosolic 10-formyltetrahydrofolate dehydrogenase | >100 |
| P55258 | Ras-related protein Rab-8A | >100 |
| Q9D1D4 | Transmembrane emp24 domain-containing protein 10 | >100 |
| Q9QXF8 | Glycine N-methyltransferase | >100 |
| Q3TNA1 | Xylulose kinase | >100 |
| Q91YI0 | Argininosuccinate lyase | >100 |
| Q9R257 | Heme-binding protein 1 | >100 |
| D3YYS6 | Monoglyceride lipase | >100 |
| P47738 | Aldehyde dehydrogenase, mitochondrial | >100 |
| Q91X52 | L-xylulose reductase | >100 |
| A0A0G2JDE1 | Immunoglobulin-heavy variable V8-12 (Fragment) | >100 |
| A0A1L1SSA8 | Transmembrane protein 205 (Fragment) | >100 |
| Accession No. | Protein | Fold Change |
|---|---|---|
| Q64514 | Tripeptidyl-peptidase 2 | >100 |
| A0A140LHR4 | Serpin H1 (Fragment) | >100 |
| Q1XH17 | Tripartite motif-containing protein 72 | >100 |
| P11404 | Fatty acid-binding protein, heart | >100 |
| Q8K274 | Ketosamine-3-kinase | >100 |
| E9Q4M2 | Hormone-sensitive lipase | >100 |
| Q9JHK5 | Pleckstrin | >100 |
| H3BKL6 | Melanoma inhibitory activity protein 2 | >100 |
| Q64314 | Hematopoietic progenitor cell antigen CD34 | >100 |
| Q920L1 | Acyl-CoA (8-3)-desaturase | >100 |
| Q9CVB6 | Actin-related protein 2/3 complex subunit 2 | >100 |
| P08207 | Protein S100-A10 | >100 |
| Q3B7Z2 | Oxysterol-binding protein 1 | >100 |
| Q99MA9 | Homeobox protein Nkx-6.1 | >100 |
| Q7TMY4 | THO complex subunit 7 homolog | >100 |
| F7BJH9 | Predicted gene 21970 (Fragment) | >100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willard, N.K.; Salazar, E.; Oyervides, F.A.; Wiebe, C.S.; Ocheltree, J.S.; Cortez, M.; Perez, R.P.; Markowitz, H.; Iliuk, A.; Sanchez, E.E.; et al. Proteomic Identification and Quantification of Snake Venom Biomarkers in Venom and Plasma Extracellular Vesicles. Toxins 2021, 13, 654. https://doi.org/10.3390/toxins13090654
Willard NK, Salazar E, Oyervides FA, Wiebe CS, Ocheltree JS, Cortez M, Perez RP, Markowitz H, Iliuk A, Sanchez EE, et al. Proteomic Identification and Quantification of Snake Venom Biomarkers in Venom and Plasma Extracellular Vesicles. Toxins. 2021; 13(9):654. https://doi.org/10.3390/toxins13090654
Chicago/Turabian StyleWillard, Nicholas Kevin, Emelyn Salazar, Fabiola Alejandra Oyervides, Cierra Siobhrie Wiebe, Jack Sutton Ocheltree, Mario Cortez, Ricardo Pedro Perez, Harry Markowitz, Anton Iliuk, Elda Eliza Sanchez, and et al. 2021. "Proteomic Identification and Quantification of Snake Venom Biomarkers in Venom and Plasma Extracellular Vesicles" Toxins 13, no. 9: 654. https://doi.org/10.3390/toxins13090654
APA StyleWillard, N. K., Salazar, E., Oyervides, F. A., Wiebe, C. S., Ocheltree, J. S., Cortez, M., Perez, R. P., Markowitz, H., Iliuk, A., Sanchez, E. E., Suntravat, M., & Galan, J. A. (2021). Proteomic Identification and Quantification of Snake Venom Biomarkers in Venom and Plasma Extracellular Vesicles. Toxins, 13(9), 654. https://doi.org/10.3390/toxins13090654






