Finding Species-Specific Extracellular Surface-Facing Proteomes in Toxic Dinoflagellates
Abstract
:1. Introduction
2. Results
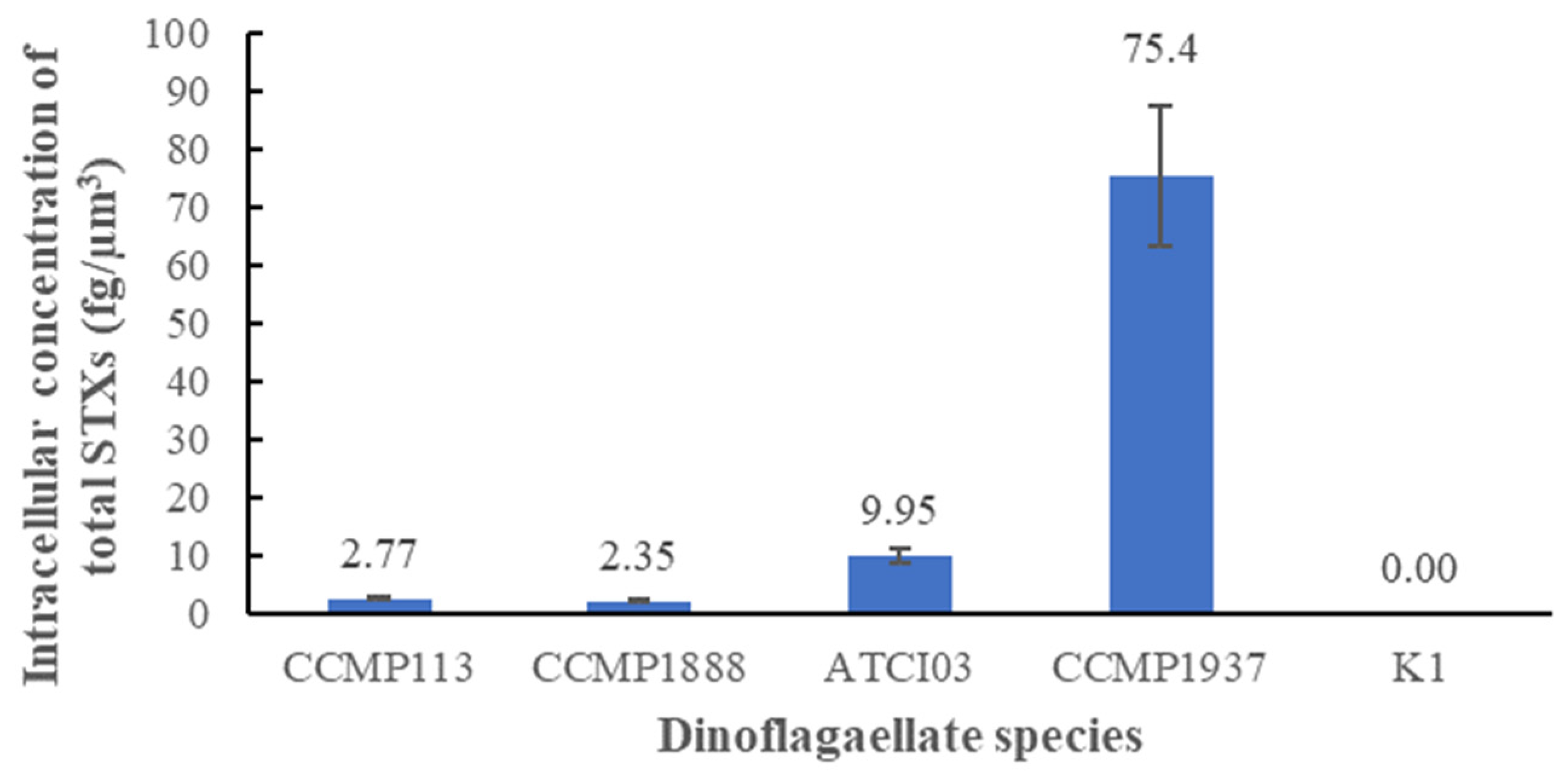
2.1. Molecular Identification and Quantification of STXs for the Selected Dinoflagellate Species
2.2. Transcriptomic Libraries and Orthogroup Inference
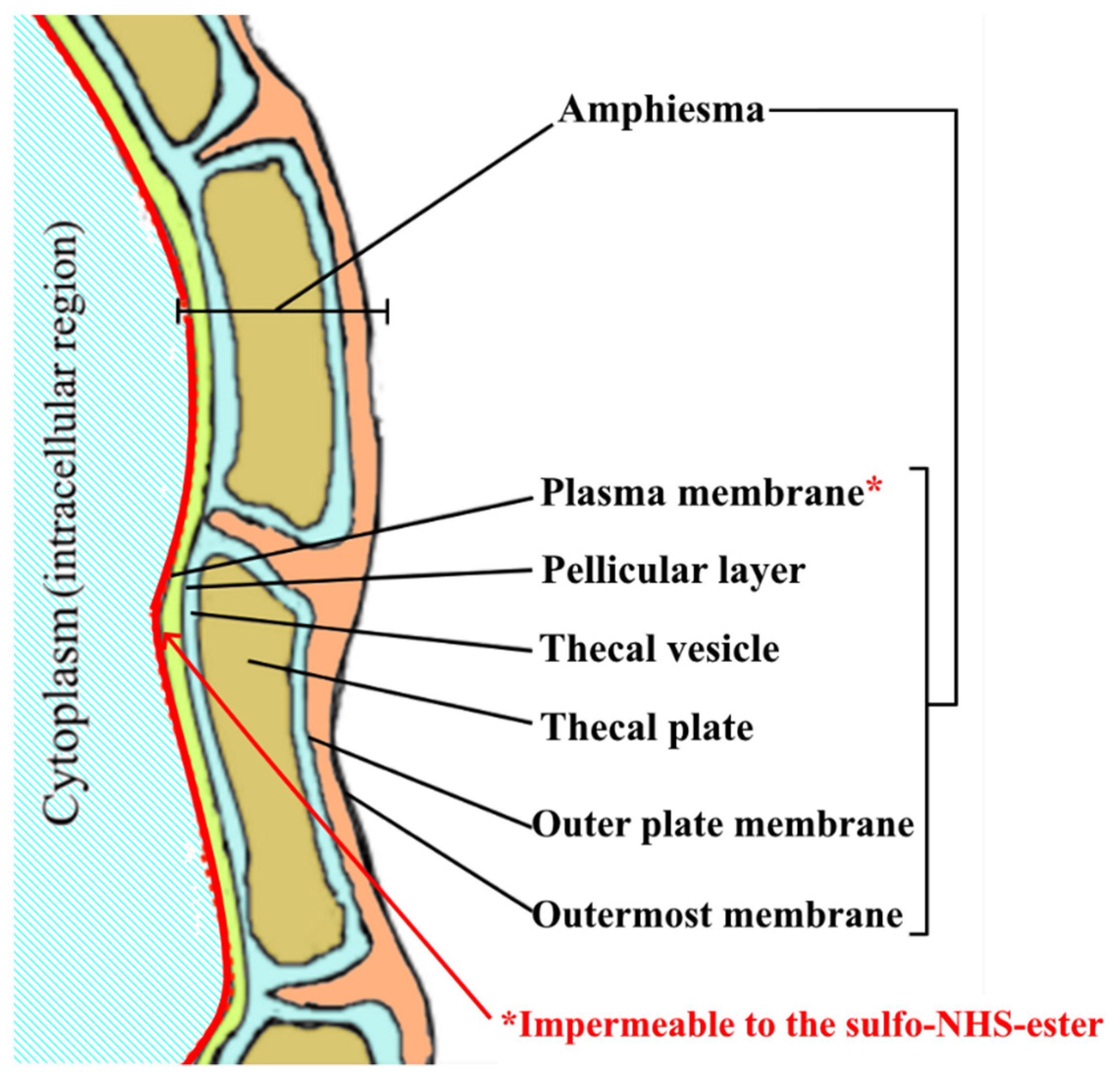
2.3. Efficiency of Our Novel Method in Labeling Amphiesmal Protein
2.4. Proteomic Orthogroup Analysis for the Surface Amphiesmal Protein Extracts
2.4.1. Species-Specific Protein Orthologs
2.4.2. Stxs-Producing-Species-Specific Orthologs
3. Discussion
3.1. Extracellular-Surface Facing Transporters
3.2. On-Surface STX-Producing-Species-Specific Proteins
3.3. On-Surface Specific Proteins
3.4. Advancements Made in Labeling and Extraction Techniques
4. Materials and Methods
4.1. Culture Conditions
4.2. Molecular Identification
4.3. Extraction and Quantification of Saxitoxins
4.4. Whole Lysate Extractions
4.5. Cell Surface Labeling
4.6. Surface Amphiesmal Protein Extractions
4.7. Sample Preparation for Mass Spectrometry
4.8. RNA Extraction and Construction of Transcriptomic Databases for Proteomic Search
4.9. Nano-LC Coupled ESI-Orbitrap-MS/MS and Proteomic Search
4.10. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patockaa, J.; Stredab, L. Brief review of natural nonprotein neurotoxins. ASA Newslett. 2002, 89, 16–24. [Google Scholar]
- Deeds, J.; Landsberg, J.; Etheridge, S.; Pitcher, G.; Longan, S. Non-Traditional Vectors for Paralytic Shellfish Poisoning. Mar. Drugs 2008, 6, 308–348. [Google Scholar] [CrossRef]
- Kellmann, R.; Mihali, T.K.; Jeon, Y.J.; Pickford, R.; Pomati, F.; Neilan, B.A. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, S.A.; Wiese, M.; Stuken, A.; Brett, S.; Kellmann, R.; Hallegraeff, G.; Neilan, B.A. sxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl. Environ. Microbiol. 2011, 77, 7050–7057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, F.W.-F.; Ho, K.C.; Lo, S.C.-L. Rapid identification of dinoflagellates using protein profiling with matrix-assisted laser desorption/ionization mass spectrometry. Harmful Algae 2008, 7, 551–559. [Google Scholar] [CrossRef]
- Lee, F.W.-F.; Ho, K.-C.; Mak, Y.-L.; Lo, S.C.-L. Authentication of the proteins expression profiles (PEPs) identification methodology in a bloom of Karenia digitata, the most damaging harmful algal bloom causative agent in the history of Hong Kong. Harmful Algae 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Cordwell, S.J.; Wilkins, M.R.; Cerpa-Poljak, A.; Gooley, A.A.; Duncan, M.; Williams, K.L.; Humphery-Smith, I. Cross-species identification of proteins separated by two-dimensional gel electrophoresis using matrix-assisted laser desorption ionisation/time-of-flight mass spectrometry and amino acid composition. Electrophoresis 1995, 16, 438–443. [Google Scholar] [CrossRef]
- Wasinger, V.C.; Cordwell, S.J.; Cerpa-Poljak, A.; Yan, J.X.; Gooley, A.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphery-Smith, I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef]
- Cordwell, S.J.; Basseal, D.J.; Humphery-Smith, I. Proteome analysis of Spiroplasma melliferum (A56) and protein characterisation across species boundaries. Electrophoresis 1997, 18, 1335–1346. [Google Scholar] [CrossRef]
- Lee, F.W.-F.; Ho, K.C.; Mak, D.Y.-L.; Liang, J.; Chen, C.; Gao, Y.; Lo, S.C.-L. Cellular Protein/Peptide Expression Profiles (PEPs): An alternative approach for easy identification of cyanobacterial species. J. Integr. OMICS 2013, 3, 174–184. [Google Scholar]
- Hou, Y.; Lin, S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: Gene content estimation for dinoflagellate genomes. PLoS ONE 2009, 4, e6978. [Google Scholar] [CrossRef] [Green Version]
- Höhfeld, I.; Melkonian, M. Amphiesmal Ultrastructure of Dinoflagellates: A Reevaluation of Pellicle Formation. J. Phycol. 1992, 28, 82–89. [Google Scholar] [CrossRef]
- Gualtieri, P. Algae: Anatomy, biochemistry, and biotechnology; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Kwok, A.; Wong, J. Cellulose Synthesis Is Coupled to Cell Cycle Progression at G1 in the Dinoflagellate Crypthecodinium cohnii. Plant Physiol. 2003, 131, 1681–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vera, K.; Olga, M.; Mariia, B.; Sergei, S. Trophic strategies in dinoflagellates: How nutrients pass through the amphiesma. Protistology 2018, 12, 3–11. [Google Scholar]
- Wang, D.-Z.; Zhang, S.-F.; Zhang, Y.; Lin, L. Paralytic shellfish toxin biosynthesis in cyanobacteria and dinoflagellates: A molecular overview. J. Proteom. 2016, 135, 132–140. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.-F.; Lin, L.; Wang, D.-Z. Comparative Transcriptome Analysis of a Toxin-Producing Dinoflagellate Alexandrium catenella and Its Non-Toxic Mutant. Mar. Drugs 2014, 12, 5698–5718. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Barua, A.; Ruvindy, R.; Savela, H.; Ajani, P.A.; Murray, S.A. The Genetic Basis of Toxin Biosynthesis in Dinoflagellates. Microorganisms 2019, 7, 222. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.-Z.; Dong, H.-P.; Li, C.; Xie, Z.-X.; Lin, L.; Hong, H.-S. Identification and Characterization of Cell Wall Proteins of a Toxic Dinoflagellate Alexandrium catenella Using 2-D DIGE and MALDI TOF-TOF Mass Spectrometry. Evid. Based Complement. Altern. Med. 2011, 2011, 984080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Feinstein, T.N.; Zhang, H.; Carpenter, E.J. Development of an immunofluorescence technique for detecting Pfiesteria piscicida. Harmful Algae 2003, 2, 223–231. [Google Scholar] [CrossRef]
- Chan, L.L.; Li, X.; Sit, W.-H.; Lam, P.K.-S.; Leung, K.M.-Y. Development of theca specific antisera for the profiling of cell surface proteins in the marine toxic dinoflagellate genus Alexandrium Halim. Harmful Algae 2012, 16, 58–62. [Google Scholar] [CrossRef]
- Bozarth, C.S.; Mullet, J.E.; Boyer, J.S. Cell Wall Proteins at Low Water Potentials. Plant Physiol. 1987, 85, 261. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.; Mitchell, G.P.; Gilroy, J.S.; Gerrish, C.; Bolwell, G.P.; Slabas, A.R. Differential extraction and protein sequencing reveals major differences in patterns of primary cell wall proteins from plants. J. Biol. Chem. 1997, 272, 15841–15848. [Google Scholar] [CrossRef] [Green Version]
- Morrill, L.; Loeblich, A.R. Cell division and reformation of the amphiesma in the pelliculate dinoflagellate, Heterocapsa niei. J. Mar. Biol. Assoc. UK 1984, 64, 939–953. [Google Scholar] [CrossRef]
- Pozdnyakov, I.; Skarlato, S. Dinoflagellate amphiesma at different stages of the life cycle. Protistology 2012, 7, 108–115. [Google Scholar]
- Olga, M. Cellular mechanisms of dinoflagellate cyst development and ecdysis-many questions to answer. Protistology 2019, 13, 47–56. [Google Scholar]
- Morrill, L.C.; Loeblich, A.R. Ultrastructure of the Dinoflagellate Amphiesma. In International Review of Cytology; Bourne, G.H., Danielli, J.F., Jeon, K.W., Eds.; Academic Press: Cambridge, MA, USA, 1983; Volume 82, pp. 151–180. [Google Scholar]
- Chan, W.S.; Kwok, A.C.M.; Wong, J.T.Y. Knockdown of dinoflagellate cellulose synthase CesA1 resulted in malformed intracellular cellulosic thecal plates and severely impeded cyst-to-swarmer transition. Front. Microbiol. 2019, 10, 546. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A.; Beardall, J. Energizing the plasmalemma of marine photosynthetic organisms: The role of primary active transport. J. Mar. Biol. Assoc. UK 2020, 100, 333–346. [Google Scholar] [CrossRef]
- Zhang, S.; Sui, Z.; Chang, L.; Kang, K.; Ma, J.; Kong, F.; Zhou, W.; Wang, J.; Guo, L.; Geng, H.; et al. Transcriptome de novo assembly sequencing and analysis of the toxic dinoflagellate Alexandrium catenella using the Illumina platform. Gene 2014, 537, 285–293. [Google Scholar] [CrossRef]
- Meng, F.-Q.; Song, J.-T.; Zhou, J.; Cai, Z.-H. Transcriptomic Profile and Sexual Reproduction-Relevant Genes of Alexandrium minutum in Response to Nutritional Deficiency. Front. Microbiol. 2019, 10, 2629. [Google Scholar] [CrossRef] [Green Version]
- Orr, R.J.S.; Stüken, A.; Murray, S.A.; Jakobsen, K.S. Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates. Mar. Drugs 2013, 11, 2814–2828. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Guo, R.; Lim, W.-A.; Allen, A.E.; Ki, J.-S. Comparative transcriptomics of toxin synthesis genes between the non-toxin producing dinoflagellate Cochlodinium polykrikoides and toxigenic Alexandrium pacificum. Harmful Algae 2020, 93, 101777. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.-F.; Lin, L.; Wang, D.-Z. Whole Transcriptomic Analysis Provides Insights into Molecular Mechanisms for Toxin Biosynthesis in a Toxic Dinoflagellate Alexandrium catenella (ACHK-T). Toxins 2017, 9, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.-F.; Zhang, Y.; Lin, L.; Wang, D.-Z. iTRAQ-Based Quantitative Proteomic Analysis of a Toxigenic Dinoflagellate Alexandrium catenella and Its Non-toxigenic Mutant Exposed to a Cell Cycle Inhibitor Colchicine. Front. Microbiol. 2018, 9, 650. [Google Scholar] [CrossRef] [PubMed]
- Veith, T.; Buchel, C. The monomeric photosystem I-complex of the diatom Phaeodactylum tricornutum binds specific fucoxanthin chlorophyll proteins (FCPs) as light-harvesting complexes. Biochim. Biophys. Acta 2007, 1767, 1428–1435. [Google Scholar] [CrossRef] [Green Version]
- Nagao, R.; Yokono, M.; Teshigahara, A.; Akimoto, S.; Tomo, T. Light-Harvesting Ability of the Fucoxanthin Chlorophyll a/c-Binding Protein Associated with Photosystem II from the Diatom Chaetoceros gracilis As Revealed by Picosecond Time-Resolved Fluorescence Spectroscopy. J. Phys. Chem. B 2014, 118, 5093–5100. [Google Scholar] [CrossRef]
- Iglesias-Prieto, R.; Govind, N.S.; Trench, R.K. Isolation and characterization of three membranebound chlorophyll-protein complexes from four dinoflagellate species. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 1993, 340, 381–392. [Google Scholar]
- Boczar, B.A.; Prezelin, B.B. Organization and Comparison of Chlorophyll-Protein Complexes from Two Fucoxanthin-Containing Algae:Nitzschia closterium(Bacillariophyceae) andIsochrysis galbana (Prymnesiophyceae)1. Plant Cell Physiol. 1989, 30, 1047–1056. [Google Scholar]
- Waller, R.F.; Kořený, L. Plastid complexity in dinoflagellates: A picture of gains, losses, replacements and revisions. Adv. Bot. Res. 2017, 84, 105–143. [Google Scholar]
- Janouškovec, J.; Gavelis, G.S.; Burki, F.; Dinh, D.; Bachvaroff, T.R.; Gornik, S.G.; Bright, K.J.; Imanian, B.; Strom, S.L.; Delwiche, C.F. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. USA 2017, 114, E171–E180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackett, J.D.; Anderson, D.M.; Erdner, D.L.; Bhattacharya, D. Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 2004, 91, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Tse, S.P.-K.; Lee, F.W.-F.; Mak, D.Y.-L.; Kong, H.-K.; Chan, K.K.-Y.; Lo, P.-Y.; Lo, S.C.-L. Production of Paralytic Shellfish Toxins (PSTs) in Toxic Alexandrium catenella is Intertwined with Photosynthesis and Energy Production. Toxins 2020, 12, 477. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.-P.; Cembella, A.D. Effects of salinity, light and inorganic nitrogen on growth and toxigenicity of the marine dinoflagellate Alexandrium tamarense from northeastern Canada. J. Plankton Res. 1999, 21, 939–955. [Google Scholar] [CrossRef]
- Carneiro, R.L.; dos Santos, M.E.V.; Pacheco, A.B.F.; de Oliveira e Azevedo, S.M.F. Effects of light intensity and light quality on growth and circadian rhythm of saxitoxins production in Cylindrospermopsis raciborskii (Cyanobacteria). J. Plankton Res. 2009, 31, 481–488. [Google Scholar] [CrossRef]
- Southern, J.A.; Parker, J.R.; Woods, D.R. Novel structure, properties and inactivation of glutamine synthetase cloned from Bacteroides fragilis. Microbiology 1987, 133, 2437–2446. [Google Scholar] [CrossRef] [Green Version]
- Unno, H.; Uchida, T.; Sugawara, H.; Kurisu, G.; Sugiyama, T.; Yamaya, T.; Sakakibara, H.; Hase, T.; Kusunoki, M. Atomic Structure of Plant Glutamine Synthetase: A Key Enzyme For Plant Productivity. J. Biol. Chem. 2006, 281, 29287–29296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inokuchi, R.; Kuma, K.I.; Miyata, T.; Okada, M. Nitrogen-assimilating enzymes in land plants and algae: Phylogenic and physiological perspectives. Physiol. Plant. 2002, 116, 1–11. [Google Scholar] [CrossRef]
- Cruz, C.; Bio, A.F.M.; Domínguez-Valdivia, M.D.; Aparicio-Tejo, P.M.; Lamsfus, C.; Martins-Loução, M.A. How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 2006, 223, 1068–1080. [Google Scholar] [CrossRef] [PubMed]
- Konig, A.; Pearson, H.; Silva, S.A. Ammonia toxicity to algal growth in waste stabilization ponds. Water Sci. Technol. 1987, 19, 115–122. [Google Scholar] [CrossRef]
- Abeliovich, A.; Azov, Y. Toxicity of ammonia to algae in sewage oxidation ponds. Appl. Environ. Microbiol. 1976, 31, 801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Chen, P.; Addy, M.; Zhang, R.; Deng, X.; Ma, Y.; Cheng, Y.; Hussain, F.; Chen, C.; Liu, Y.; et al. Carbon-dependent alleviation of ammonia toxicity for algae cultivation and associated mechanisms exploration. Bioresour. Technol. 2018, 249, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Azov, Y.; Goldman, J.C. Free Ammonia Inhibition of Algal Photosynthesis in Intensive Cultures. Appl. Environ. Microbiol. 1982, 43, 735. [Google Scholar] [CrossRef] [Green Version]
- Collos, Y.; Harrison, P.J. Acclimation and toxicity of high ammonium concentrations to unicellular algae. Mar. Pollut. Bull. 2014, 80, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Källqvist, T.; Svenson, A. Assessment of ammonia toxicity in tests with the microalga, Nephroselmis pyriformis, Chlorophyta. Water Res. 2003, 37, 477–484. [Google Scholar] [CrossRef]
- Gálvez, S.; Lancien, M.; Hodges, M. Are isocitrate dehydrogenases and 2-oxoglutarate involved in the regulation of glutamate synthesis? Trends Plant Sci. 1999, 4, 484–490. [Google Scholar] [CrossRef]
- Taroncher-Oldenburg, G.; Kulis, D.M.; Anderson, D.M. Coupling of saxitoxin biosynthesis to the G1 phase of the cell cycle in the dinoflagellate Alexandrin fundyense: Temperature and nutrient effects. Nat. Toxins 1999, 7, 207–219. [Google Scholar] [CrossRef]
- Lemieux, C.; Otis, C.; Turmel, M. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature 2000, 403, 649–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, A.; Minden, A.; Martinetto, H.; Claret, F.X.; Lange-Carter, C.; Mercurio, F.; Johnson, G.L.; Karin, M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science 1995, 268, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Bellei, E.; Bergamini, S.; Monari, E.; Fantoni, L.I.; Cuoghi, A.; Ozben, T.; Tomasi, A. High-abundance proteins depletion for serum proteomic analysis: Concomitant removal of non-targeted proteins. Amino Acids 2011, 40, 145–156. [Google Scholar] [CrossRef]
- Andersen, R.; Berges, J.; Harrison, P.; Watanabe, M.M. Appendix A—Recipes for freshwater and seawater media. Algal Cult. Tech. 2005, 1, 429–539. [Google Scholar]
- Watanabe, R.; Matsushima, R.; Harada, T.; Oikawa, H.; Murata, M.; Suzuki, T. Quantitative determination of paralytic shellfish toxins in cultured toxic algae by LC-MS/MS. Food Addit. Contam. Part A 2013, 30, 1351–1357. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| BLAST Search Results | |||||
|---|---|---|---|---|---|
| Species | Primer * | Accession No. of Best Match | % Ident. | E-Value | Source Organism |
| CCMP113 | Pair1 | FJ823523.1 | 99 | 0 | Alexandrium minutum |
| Pair2 | JF521634.1 | 99 | 0 | Alexandrium minutum | |
| CCMP1888 | Pair1 | JF906999.1 | 99 | 0 | Alexandrium lusitanicum |
| ATCI03 | Pair2 | JF906992.1 | 99 | 0 | Alexandrium tamarense |
| CCMP1937 | Pair1 | DQ779989.2 | 99 | 0 | Gymnodinium catenatum |
| K1 | Pair1 | KU314866.1 | 99 | 0 | Karenia mikimotoi |
| Pair2 | LC055227.1 | 99 | 0 | Karenia mikimotoi | |
| Statistical Categories | Quantity |
|---|---|
| Genes inputted | |
| No. of genes | 695,887 |
| No. of genes in orthogroups | 386,478 |
| No. of unassigned genes | 309,409 |
| No. of genes in species-specific orthogroups | 6749 |
| Orthogroup inferred | |
| No. of orthogroups | 86,675 |
| No. of species-specific orthogroups | 1539 |
| 35 |
| 36 |
| 201 |
| 581 |
| 686 |
| No. of orthogroups commonly found in STXs-producing species | 3204 |
| No. of orthogroups commonly found in Alexandrium species | 11,136 |
| No. of orthogroups with all species present | 14,338 |
| No. of single-copy orthogroups | 3234 |
| Mean orthogroup size | 4.5 |
| Median orthogroup size | 3 |
| G50 (assigned genes) | 5 |
| G50 (all genes) | 2 |
| O50 (assigned genes) | 20,072 |
| O50 (all genes) | 67,408 |
| CCMP113 | CCMP1888 | ATCI03 | CCMP1937 | K1 | |
|---|---|---|---|---|---|
| CCMP113 | 100 | - | - | - | - |
| CCMP1888 | 95 | 100 | - | - | - |
| ATCI03 | 80 | 80 | 100 | - | - |
| CCMP1937 | 65 | 64 | 53 | 100 | - |
| K1 | 65 | 65 | 73 | 74 | 100 |
| CCMP113 | CCMP1888 | ATCI03 | CCMP1937 | K1 | |
|---|---|---|---|---|---|
| No. of protein identified in SAPEs | |||||
| 180 | 217 | 203 | 218 | 197 |
| ▪ Labeled | 44 | 43 | 23 | 21 | 36 |
| ○ Transporter protein | 4 | 5 | 2 | 1 | 4 |
| ▪ Unlabeled 2 | 136 | 174 | 180 | 197 | 161 |
| 286 | 339 | 309 | 276 | 314 |
| ▪ Labeled | 51 | 25 | 30 | 14 | 79 |
| ▪ Unlabeled | 235 | 314 | 279 | 262 | 235 |
| 154 | 211 | 131 | 210 | 160 |
| 620 | 767 | 643 | 704 | 671 |
| Percentage yield (%) | |||||
| 29.0 | 28.3 | 31.6 | 31.0 | 29.4 |
| ▪ % Yield compared to WL 4 | +6.56 | +2.33 | +6.71 | +5.50 | +4.23 |
| ▪ % Yield compared to ICF 4 | +1.36 | +1.33 | +2.95 | +2.23 | 0 |
| 7.10 | 5.61 | 3.58 | 2.98 | 5.37 |
| Species | Uniprot BLASTp Results # | GO-Terms | Unigene IDs of Protein Identified * | No. of Peptide Species Identified in MS | ||
|---|---|---|---|---|---|---|
| E-Value (Respective Accession No. of Best Match) | Annotation of Homolog | Cellular Component | Biological Process | |||
| CCMP113 | 0 (P499510) 0 (Q2RBN7) | Clathrin heavy chain 1 | Endosome (GO:0005768), Membrane (GO:0016020) | Transferrin transport (GO:0033572), Receptor internalization (GO:0031623), Intracellular protein transport (GO:0006886) | Cluster-16584.55282 Cluster-16584.53449 | 5 4 |
| 4.1 × 10−198 (Q61941) | NAD(P) transhydrogenase, mitochondrial | Membrane (GO:0016020) | Proton transmembrane transport (GO:1902600) | Cluster-16584.47837 | 6 | |
| 7.7 × 10−202 (P80021) | ATP synthase subunit alpha, mitochondrial | Plasma membrane (GO:0005886) | ATP synthesis coupled proton transport (GO:0015986) | Cluster-16584.49485 | 14 | |
| CCMP1888 | 4.1 × 10−198 (Q61941) 1.5 × 10-183 (Q61941) 1.6 × 10−148 (P11024) 1.0 × 10−138 (P11024) | NAD(P) transhydrogenase, mitochondrial | Membrane (GO:0016020) | Proton transmembrane transport (GO:1902600) | Cluster-6994.52752 Cluster-6994.52754 Cluster-6994.53793 Cluster-6994.53135 | 7 5 3 2 |
| 7.7 × 10−202 (P80021) | ATP synthase subunit alpha, mitochondrial | Plasma membrane (GO:0005886) | ATP synthesis coupled proton transport (GO:0015986) | Cluster-6994.53837 | 20 | |
| ATCI03 | 1.8 × 10−138 (Q8DHU4) | Protein translocase subunit SecA | Plasma membrane (GO:0005886) | Protein import (GO:0017038) | Cluster-15238.31582 | 7 |
| 8.4 × 10−190 (P11024) | NAD(P) transhydrogenase, mitochondrial | Membrane (GO:0016020) | Proton transmembrane transport (GO:1902600) | Cluster-15238.36601 | 3 | |
| CCMP1937 | 0 (Q00610) | Clathrin heavy chain 1 | Endolysosome membrane (GO:0036020), Plasma membrane (GO:0005886) | Membrane organization (GO:0061024), Intracellular protein transport (GO:0006886) | Cluster-5567.56526 | 7 |
| K1 | 1.3 × 10−97 (Q13423) 1.5 × 10−97 (Q13423) | NAD(P) transhydrogenase, mitochondrial | Membrane (GO:0016020) | Proton transmembrane transport (GO:1902600), Tricarboxylic acid cycle (GO:0006099) | Cluster-24592.79099 Cluster-24592.45845 | 8 7 |
| 7.8 × 10−198 (Q03265) 3.0 × 10−163 (Q03265) | ATP synthase subunit alpha, mitochondrial | Cell surface (GO:0009986) | ATP synthesis coupled proton transport (GO:0015986) | Cluster-24592.43084 Cluster-24592.74857 | 13 8 | |
| Name of Transporter Homologs | Dinoflagellate Species | ||||
|---|---|---|---|---|---|
| CCMP113 | CCMP1888 | ATCI03 | CCMP1937 | K1 | |
| NAD(P) transhydrogenase, mitochondrial | ✓ | ✓ | ✓ | -- | ✓ |
| ATP synthase subunit alpha, mitochondrial | ✓ | ✓ | -- | -- | ✓ |
| Clathrin heavy chain 1 | ✓ | -- | -- | ✓ | -- |
| Dinoflagellate Species | |||||
|---|---|---|---|---|---|
| CCMP113 | CCMP1888 | ATCI03 | CCMP1937 | K1 | |
| No. of protein identified in SAPE | |||||
| 532 | 648 | 383 | 541 | 384 |
| ▪ Species-specific orthogroups | 1 | 0 | 0 | 1 | 11 |
| ▪ STXs-producing species-specific orthogroup | 7 | 7 | 7 | 7 | N/A |
| 88 | 119 | 119 | 163 | 287 |
| Total | 620 | 767 | 502 | 704 | 671 |
| Species | Uniprot BLASTp Results # | GO-Terms | Unigene IDs of Protein Identified * | No. of Peptide Species Identified in MS | ||
|---|---|---|---|---|---|---|
| E-Value (Respective Accession No. of Best Match) | Uniprot Annotation of Ortholog | Cellular Component | Biological Process | |||
| CCMP113 | 7.9 × 10−277 (P16100) | Isocitrate dehydrogenase [NADP] | Cytoplasm (GO:0005737) | Tricarboxylic acid cycle (GO:0006099) | Cluster-16584.46230 | 1 ^ |
| CCMP1937 | 8.7 × 10−92 (P91622) | Pyruvate dehydrogenase kinase | Mitochondrion (GO:0005739) | Glucose metabolic process (GO:0006006) | Cluster-5567.50671 | 3 |
| K1 | 1.2 × 10−7 (Q39709) | Fucoxanthin-chlorophyll a-c binding protein | Chloroplast thylakoid membrane (GO:0009535) | photosynthesis, light harvesting (GO:0009765) | Cluster-24592.102021 | 4 |
| 3.1 × 10−36 (Q40296) 3.1 × 10−38 (Q40297) 1.3 × 10−34 (Q40296) 7.8 × 10−22 (Q40296) | Fucoxanthin-chlorophyll a-c binding protein B | Chloroplast thylakoid membrane (GO:0009535) | photosynthesis, light harvesting (GO:0009765) | Cluster-24592.122404 Cluster-24592.47428 Cluster-24592.64372 Cluster-24592.96057 | 5 2 2 6 | |
| 2.0 × 10−27 (Q40300) | Fucoxanthin-chlorophyll a-c binding protein F | Chloroplast thylakoid membrane (GO:0009535) | photosynthesis, light harvesting (GO:0009765) | Cluster-24592.21292 | 2 | |
| 2.2 × 10−20 (A0A5C6NSI7) | Dual specificity mitogen-activated protein kinase 4 | protein phosphorylation (GO:0006468) | -- | Cluster-24592.69574 Cluster-24592.107035 | 1 ^ 1 ^ | |
| 5.0 × 10−63 (M2X807) | CAAX amino terminal protease family protein | Integral component of membrane (GO:0016021) | CAAX-box protein processing (GO:0071586) | Cluster-24592.117554 | 2 | |
| Orthogroup No. | Uniprot Annotation of Ortholog # (Accession No.) | GO-Terms | No. of Peptide Species Identified in MS | ||||
|---|---|---|---|---|---|---|---|
| Cellular Component | Biological Process | CCMP113 | CCMP1888 | ATCI03 | CCMP1937 | ||
| OG0004814 | Mitochondrial dicarboxylate/tricarboxylate transporter (Q9C5M0, P0C582) | Mitochondrion inner membrane (GO:0005743) | Oxoglutarate:malate antiporter activity (GO:0015367) | 1 ^ | 2 | 7 | 2 |
| OG0024578 | Glutamine synthetase (P15623, Q54WR9) | phagocytic vesicle (GO:0045335) | Nitrogen compound metabolic process (GO:0006807) | 1 ^ | 1 ^ | 1 ^ | 2 |
| OG0000905 | Fucoxanthin-chlorophyll a-c binding protein E (Q40301) | Chloroplast thylakoid membrane (GO:0009535) | Light-harvesting complex (GO:0030076) | 16 | 10 | 9 | 16 |
| OG0034291 | F-type H+-transporting ATPase subunit beta (A6BM09, Q06J29) | Chloroplast thylakoid membrane (GO:0009535) | Proton-transporting ATP synthase activity (GO:0046933) | 22 | 28 | 32 | 25 |
| OG0034855 * | F-type H+-transporting ATPase subunit alpha (Q9MUT2, Q1ACM8, Q85X67) | Chloroplast thylakoid membrane (GO:0009535) | Proton-transporting ATP synthase activity (GO:0046933) | 13 | 17 | 22 | 19 |
| OG0035644 | Pentatricopeptide repeat-containing protein (Q9SV46, Q9SZ52, Q9M907, Q9LVQ5) | Chloroplast (GO:0009507) | RNA stabilization (GO:0043489) Translation (GO:0006412) | 10 | 7 | 11 | 14 |
| Primer | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Pair 1 | TCCGTAGGTGAACCTGCGG | TCCTCCGCTTATTGATATGC |
| Pair 2 | TGAACCTTAYCACTTAGAGGAAGGA | GCTRAGCWDHTCCYTSTTCATTC |
| Parameters | DDA-MS1 | DDA-MS2 |
|---|---|---|
| Resolution | 60,000 | 15,000 |
| Scan range (m/z) | 350–1500 | Auto |
| Max. injection time (ms) | 20 | 30 |
| AGC target | 4.0 × 105 | 5.0 × 104 |
| HCD collision energy (%) | N/A | 30 |
| Intensity threshold | 1.0 × 104 | N/A |
| Variable Modification | ΔM.W. (Da) | Targeted Amino Acid |
|---|---|---|
| Carbamidomethyl | +57.021 | C |
| Oxidation | +15.994 | M |
| 3-[(Carbamoylmethyl)sulfanyl]propanoyl # | +145.019 | K, Protein N-term |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, K.K.-y.; Kong, H.-k.; Tse, S.P.-k.; Chan, Z.; Lo, P.-y.; Kwok, K.W.H.; Lo, S.C.-l. Finding Species-Specific Extracellular Surface-Facing Proteomes in Toxic Dinoflagellates. Toxins 2021, 13, 624. https://doi.org/10.3390/toxins13090624
Chan KK-y, Kong H-k, Tse SP-k, Chan Z, Lo P-y, Kwok KWH, Lo SC-l. Finding Species-Specific Extracellular Surface-Facing Proteomes in Toxic Dinoflagellates. Toxins. 2021; 13(9):624. https://doi.org/10.3390/toxins13090624
Chicago/Turabian StyleChan, Kenrick Kai-yuen, Hang-kin Kong, Sirius Pui-kam Tse, Zoe Chan, Pak-yeung Lo, Kevin W. H. Kwok, and Samuel Chun-lap Lo. 2021. "Finding Species-Specific Extracellular Surface-Facing Proteomes in Toxic Dinoflagellates" Toxins 13, no. 9: 624. https://doi.org/10.3390/toxins13090624
APA StyleChan, K. K.-y., Kong, H.-k., Tse, S. P.-k., Chan, Z., Lo, P.-y., Kwok, K. W. H., & Lo, S. C.-l. (2021). Finding Species-Specific Extracellular Surface-Facing Proteomes in Toxic Dinoflagellates. Toxins, 13(9), 624. https://doi.org/10.3390/toxins13090624





