Molecular Identification and Toxin Analysis of Alexandrium spp. in the Beibu Gulf: First Report of Toxic A. tamiyavanichii in Chinese Coastal Waters
Abstract
1. Introduction
2. Results
2.1. Sequence Analysis
2.2. Phylogenetic Tree Analysis
2.3. Toxin Analysis
3. Discussion
- (1)
- Low-toxicity algal strains are widely distributed in the Beibu Gulf and survive for long periods. PSTs can thus accumulate in shellfish through time to levels sufficient to exceed regulatory standards. Anderson et al. [62] noted a similar phenomenon in Daya Bay, China where A. pacificum has very low toxicity of 7.2–12.7 fmol/cell, yet PSP poisoning still occasionally occurs there.
- (2)
- Based on reports of A. tamiyavanichii toxin content varying by more than 5 orders of magnitude among isolates from the same locations in Malaysia and by factors of 857 in Thailand and 651 in Japan (Table 3), it seems very likely that other highly toxic A. tamiyavanichii strains occur in the Beibu Gulf that were not isolated. Low-abundance Alexandrium spp. in seawater can be missed due to limitations of small numbers of culture isolations, and thus highly toxic strains could easily be overlooked. Furthermore, because isolates were only established from Weizhou Island in the Beibu Gulf, and not from other highly productive areas of the Gulf, such as Qinzhou Bay and Tieshan Harbor, the presence of highly toxic species in these regions cannot be ruled out. In this regard, it is of note that the PST composition of shellfish from the Beibu Gulf differs significantly from the A. tamiyavanichii isolates analyzed here (Table 3 and Table 4). This could reflect biotransformation (see below) and/or different toxic Alexandrium species or strains from those analyzed here.
- (3)
- Alexandrium in field populations can grow faster and produce more toxins than laboratory cultures, as has been observed by others through variations in temperature, salinity, light, and nutrients [63,64,65,66]. Brosnahan et al. [67] recently reported in situ growth rates for A. catenella that were more than twice those observed in laboratory cultures under similar growth conditions.
- (4)
- Metabolism and bioconversion processes in different shellfish greatly affect the accumulation and potency of PSTs [68,69]. For example, the PSP toxicity of Paphia undulata is over 1000 times higher than that of Meretrix lusoria in the Beibu Gulf [29]. Thus, further study of biotransformation of PSTs in different shellfish from the Beibu Gulf is needed.
- (5)
- Although Alexandrium spp.are the principal causative organisms of PSP in many regions, the presence of other PST-producing microalgae, such as Gymnodinium catenatum and Pyrodinium bahamense [70,71], and atypical toxin-producing organisms such as brackish cyanobacteria, as well as calcareous red macroalgae [72], cannot be ruled out.
4. Conclusions
5. Materials and Methods
5.1. Algal Source and Culture Conditions
5.2. Extraction of DNA, PCR Amplification, and Sequencing
5.3. Sequence Analyses
5.4. Toxin Extraction
5.5. Liquid Chromatography-Mass Spectrometry
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, D.M.; Alpermann, T.J.; Cembella, A.D.; Collos, Y.; Masseret, E.; Montresor, M. The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 2012, 14, 10–35. [Google Scholar] [CrossRef]
- Halim, Y. Alexandrium minutum nov. g. nov. sp. Dinoflagellé provocant des “eaux rouges”. Vie Milieu 1960, 11, 102–105. [Google Scholar]
- Mackenzie, L.; Salas, M.D.; Adamson, J.; Beuzenberg, V. The dinoflagellate genus Alexandrium (Halim) in New Zealand coastal waters: Comparative morphology, toxicity and molecular genetics. Harmful Algae 2004, 3, 71–92. [Google Scholar] [CrossRef]
- Bill, B.D.; Moore, S.K.; Hay, L.R.; Anderson, D.M.; Trainer, V.L. Effects of temperature and salinity on the growth of Alexandrium (Dinophyceae) isolates from the Salish Sea. J. Phycol. 2016, 52, 230–238. [Google Scholar] [CrossRef]
- Liang, Y.; Li, D.; Yao, J.; Jin, W.; Gong, C.; Liu, R. Progresses in investigation and research on phycotoxins and toxic microalgaes in the coastal waters of China. Oceanol. Et Limnol. Sin. 2019, 50, 511–524. [Google Scholar]
- Litaker, R.W.; Fraga, S.; Montresor, M.; Brosnahan, M.; Anderson, D.M.; Hoppenrath, M.; Murray, S.; Wolny, J.; John, U.; Sampedro, N.; et al. A practical guide to new nomenclature for species within the “Alexandrium tamarense species complex”. Harmful Algae News 2018, 61, 13–15. [Google Scholar]
- Balech, E. The Genus Alexandrium Halim (Dinoflagellata); Sherkin Island Marine Station: Cork, Ireland, 1995; p. 452. [Google Scholar]
- Murray, S.A.; Hoppenrath, M.; Orr, R.J.S.; Bolch, C.; John, U.; Diwan, R.; Yauwenas, R.; Harwood, T.; Salas, M.D.; Neilan, B.; et al. Alexandrium diversaporum sp. nov., a new non-saxitoxin producing species: Phylogeny, morphology and sxtA genes. Harmful Algae 2014, 31, 54–65. [Google Scholar] [CrossRef]
- Mertens, K.N.; Adachi, M.; Anderson, D.M.; Band-Schmidt, C.J.; Bravo, I.; Brosnahan, M.L.; Bolch, C.J.S.; Calado, A.J.; Carbonell-Moore, M.C.; Chomérat, N.; et al. Morphological and phylogenetic data do not support the split of Alexandrium into four genera. Harmful Algae 2020, 98, 101902. [Google Scholar] [CrossRef]
- John, U.; Litaker, R.W.; Montresor, M.; Murray, S.; Brosnahan, M.L.; Anderson, D.M. Formal revision of the Alexandrium tamarense species complex (Dinophyceae) taxonomy: The introduction of five species with emphasis on molecular-based (rDNA) classification. Protist 2014, 165, 779–804. [Google Scholar] [CrossRef]
- Fraga, S.; Sampedro, N.; Larsen, J.; Moestrup, Ø.; Calado, A.J. Arguments against the proposal 2302 by John & al. to reject the name Gonyaulax catenella (Alexandrium catenella). Taxon 2015, 64, 634–635. [Google Scholar]
- Prud’homme van Reine, W.F. Report of the nomenclature committee for algae: 15. Taxon 2017, 66, 191–192. [Google Scholar] [CrossRef]
- Lim, A.S.; Jeong, H.J.; Kim, J.H.; Lee, S.Y. Description of the new phototrophic dinoflagellate Alexandrium pohangense sp. nov. from Korean coastal waters. Harmful Algae 2015, 46, 49–61. [Google Scholar] [CrossRef]
- Branco, S.; Oliveira, M.M.M.; Salgueiro, F.; Vilar, M.C.P.; Azevedo, S.M.F.O.; Menezes, M. Morphology and molecular phylogeny of a new PST-producing dinoflagellate species: Alexandrium fragae sp. nov. (Gonyaulacales, Dinophyceae). Harmful Algae 2020, 95, 101793. [Google Scholar] [CrossRef] [PubMed]
- Gu, H. Morphology, phylogenetic position, and ecophysiology of Alexandrium ostenfeldii (Dinophyceae) from the Bohai Sea, China. J. Syst. Evol. 2011, 49, 606–616. [Google Scholar] [CrossRef]
- Gu, H.; Zeng, N.; Liu, T.; Yang, W.; Mueller, A.; Krock, B. Morphology, toxicity, and phylogeny of Alexandrium (Dinophyceae) species along the coast of China. Harmful Algae 2013, 27, 68–81. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, T.; Zhou, J. Historical occurrence of algal blooms in the northern Beibu Gulf of China and implications for future trends. Front. Microbiol. 2019, 10, 451. [Google Scholar] [CrossRef]
- Kang, Z.; Yang, B.; Lai, J.; Ning, Y.; Zhong, Q.; Lu, D.; Liao, R.; Wang, P.; Solomon, F.D.; She, Z.; et al. Phaeocystis globosa bloom monitoring: Based on P. globosa induced seawater viscosity modification adjacent to a nuclear power plant in Qinzhou Bay, China. J. Ocean Univ. China 2020, 19, 1207–1220. [Google Scholar] [CrossRef]
- Zou, C. Genetic Structure, Phylogeography Relationship and PSP Toxin Profile of Alexandrium tamarense Species Complex. Master’s Thesis, Jinan University, Guangzhou, China, 2015. [Google Scholar]
- Lu, X. Biogeological Distribution of Dinoflagellate Cysts in Surface Sediments from Southern Chinese Coast and Their Roles as a Signal of Environmental Changes. Ph.D. Thesis, Jinan University, Guangzhou, China, 2016. [Google Scholar]
- Jiang, F.; Chen, B.; He, B.; Xu, M.; Zhuang, J.; Zhang, R.; Lei, F. Phytoplankton community in coastal waters of Qinzhou Bay in Guangxi. Guangxi Sci. 2012, 19, 268–275. [Google Scholar]
- Wang, Z.; Ma, C.; Xu, Z. Phylogenetic relationship of four Alexandrium tamarense/catenella isolates from Southeast Chinese costal waters using rDNA sequences. Mar. Environ. Sci. 2015, 34, 1–5. [Google Scholar]
- Anderson, D.M.; Stock, C.A.; Keafer, B.A.; Nelson, A.B.; Thompson, B.; Mcgillicuddy, D.J.; Keller, M.; Matrai, P.A.; Martin, J. Alexandrium fundyense cyst dynamics in the Gulf of Maine. Deep Sea Res. Part II Top. Stud. Oceanogr. 2005, 52, 2522–2542. [Google Scholar] [CrossRef]
- Martin, J.L.; Legresley, M.M.; Hanke, A.R. Thirty years—Alexandrium fundyense cyst, bloom dynamics and shellfish toxicity in the Bay of Fundy, eastern Canada. Deep Sea Res. Part II Top. Stud. Oceanogr. 2014, 103, 27–39. [Google Scholar] [CrossRef]
- Wang, D.; Jiang, T.; Hsieh, D.P.H. Toxin composition variations in cultures of Alexandrium species isolated from the coastal waters of southern China. Harmful Algae 2005, 4, 109–121. [Google Scholar] [CrossRef]
- Zou, C.; Ye, R.; Zheng, J.; Luo, Z.; Gu, H.; Yang, W.; Li, H.; Liu, J. Molecular phylogeny and PSP toxin profile of the Alexandrium tamarense species complex along the coast of China. Mar. Pollut. Bull. 2014, 89, 209–219. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, S.; Gu, H.; Chan, L.; Hong, H. Paralytic shellfish toxin profiles and toxin variability of the genus Alexandrium (Dinophyceae) isolated from the Southeast China Sea. Toxicon 2006, 48, 138–151. [Google Scholar] [CrossRef]
- Zhong, X.; Liu, D.; Liang, H. Risk assessment of paralytic shellfish poison in exported shellfish from Beibu Gulf. Enterp. Sci. Technol. Dev. 2010, 22, 314–315. [Google Scholar]
- Zhao, P.; Qin, X.; Hu, K.; Li, Z.; Jiang, N.; Xu, Y. Paralytic shellfish poisoning of bivalves in seafood market, Beibu Gulf of Guangxi. Ecol. Sci. 2016, 35, 73–77. [Google Scholar]
- Dai, Z.; Guo, W.; Zhang, C.; Wang, P.; Huang, M.; Li, C. Detection and safety evaluation of paralytic shellfish toxins in 7 species of Qinzhou Bay economically shellfish. Food Res. Dev. 2018, 39, 120–125. [Google Scholar]
- Guo, L.; Lu, S. A new record of Alexandrium tamiyavanichii Balech from the coast of China. In Proceedings of the 8th Member Conference of the Chinese Phycological Society and 16th Academic Symposium, Shanghai, China, 11 November 2011; p. 191. [Google Scholar]
- Guo, L. Morphological Taxonomy and Ecological Distribution on Alexandrium in Chinese Coastal Waters. Master’s Thesis, South China Normal University, Guangzhou, China, 2012. [Google Scholar]
- Li, D.; Lu, D.; Dai, X.; He, P.; Xia, P.; Wang, H. Morphological-phylogenetic analysis and population dynamics of Alexandrium tamarense in the bloom area of the East China Sea. Oceanol. Et Limnol. Sin. 2014, 45, 1241–1250. [Google Scholar]
- Law, S.P.C.; Lee, F.Y.K. Harmful Marine Microalgae in Hong Kong, 2nd ed.; Agriculture, Fisheries and Conservation Department: Hong Kong, China, 2013; pp. 1–47.
- Hashimoto, T.; Matsuoka, S.; Yoshimatsu, S.A.; Miki, K.; Nishibori, N.; Nishio, S.; Noguchi, T. First paralytic shellfish poison (PSP) infestation of bivalves due to toxic dinoflagellate Alexandrium tamiyavanichii, in the southeast coasts of the Seto Inland Sea, Japan. J. Food Hyg. Soc. Jpn. 2002, 43, 1–5. [Google Scholar] [CrossRef][Green Version]
- Menezes, M.; Varela, D.; Proença, L.A.D.O.; Tamanaha, M.D.S.; Paredes, J. Identification of the toxic alga Alexandrium tamiyavanichi (Dinophyceae) from northeastern brazil: A combined morphological and rDNA sequence (partial LSU and ITS) approach. J. Phycol. 2010, 46, 1239–1251. [Google Scholar] [CrossRef]
- Sagara, T.; Taniyama, S.; Yoshimatsu, S.; Takatani, T.; Hashimoto, T.; Nishibori, N.; Nishio, S.; Arakawa, O. Toxicity and toxin profile of the dinoflagellate Alexandrium tamiyavanichii and toxic mussels in Harima-Nada of Seto Inland Sea, Japan. Shokuhinseigaku Zasshi 2010, 51, 170–177. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hamdan, N.A.; Hassan, M.S.A.; Hamid, S.D.; Mohammad-Noor, N.; Bunnori, N.M. Toxicity profiles between strains of Alexandrium tamiyavanichii. Int. J. Adv. Sci. Eng. Technol. 2018, 6, 2321–9009. [Google Scholar]
- Hernandez-Becerril, D.U.; Lau, W.L.; Hii, K.S.; Leaw, C.P.; Varona-Cordero, F.; Lim, P.T. Abundance and distribution of the potentially toxic thecate dinoflagellate Alexandrium tamiyavanichii (Dinophyceae) in the central Mexican pacific, using the quantitative PCR method. Front. Mar. Sci. 2018, 5, 366. [Google Scholar] [CrossRef]
- Triki, H.Z.; Laabir, M.; Moeller, P.; Chomérat, N.; Daly-Yahia, O.K. First report of goniodomin A production by the dinoflagellate Alexandrium pseudogonyaulax developing in southern Mediterranean (Bizerte Lagoon, Tunisia). Toxicon 2016, 111, 91–99. [Google Scholar] [CrossRef]
- Yang, I.; John, U.; Beszteri, S.; Glöckner, G.; Krock, B.; Goesmann, A.; Cembella, A.D. Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genom. 2010, 11, 248. [Google Scholar] [CrossRef]
- Krock, B.; Tillmann, U.; Wen, Y.; Hansen, P.J.; Larsen, T.O.; Andersen, A.J.C. Development of a LC-MS/MS method for the quantification of goniodomins A and B and its application to Alexandrium pseudogonyaulax strains and plankton field samples of Danish coastal waters. Toxicon 2018, 155, 51–60. [Google Scholar] [CrossRef]
- Harris, C.M.; Reece, K.S.; Stec, D.F.; Scott, G.P.; Jones, W.M.; Hobbs, P.L.M.; Harris, T.M. The toxin goniodomin, produced by Alexandrium spp., is identical to goniodomin A. Harmful Algae 2020, 92, 101707. [Google Scholar] [CrossRef]
- Draredja, M.A.; Frihi, H.; Boualleg, C.; Abadie, E.; Laabir, M. Distribution of dinoflagellate cyst assemblages in recent sediments from a southern Mediterranean lagoon (Mellah, Algeria) with emphasis on toxic species. Environ. Sci. Pollut. Res. Int. 2020, 27, 25173–25185. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Jeong, H.J.; Kang, H.C.; Ok, J.H.; You, J.H.; Park, S.A. Growth rates and nitrate uptake of co-occurring red-tide dinoflagellates Alexandrium affine and A. fraterculus as a function of nitrate concentration under light-dark and continuous light conditions. Algae 2019, 34, 237–251. [Google Scholar] [CrossRef]
- Nguyen-Ngoc, L. An autecological study of the potentially toxic dinoflagellate Alexandrium affine isolated from Vietnamese waters. Harmful Algae 2004, 3, 117–129. [Google Scholar] [CrossRef]
- Kodama, M.; Ogata, T.; Fukuyo, Y.; Ishimaru, T.; Wisessang, S.; Sartanu, K.; Panichyakarn, V.; Piyakarncana, T. Protogonyaulax cohorticula, a toxic dinoflagellate found in the Gulf of Thailand. Toxicon 1988, 26, 707–712. [Google Scholar] [CrossRef]
- Ogata, T.; Pholpunthin, P.; Fukuyo, Y.; Kodama, M. Occurrence of Alexandrium cohorticula in Japanese coastal water. J. Appl. Phycol. 1990, 2, 351–356. [Google Scholar] [CrossRef]
- Usup, G.; Pin, L.C.; Ahmad, A.; Teen, L.P. Alexandrium (Dinophyceae) species in Malaysian waters. Harmful Algae 2002, 1, 265–275. [Google Scholar] [CrossRef]
- Ruiz Sebastián, C.; Etheridge, S.M.; Cook, P.A.; O’Ryan, C.; Pitcher, G.C. Phylogenetic analysis of toxic Alexandrium (Dinophyceae) isolates from South Africa: Implications for the global phylogeography of the Alexandrium tamarense species complex. Phycologia 2005, 44, 49–60. [Google Scholar] [CrossRef]
- Nagai, S.; Suzuki, M.; Matsuyama, Y.; Hakura, S.; Go, J. Molecular and toxicity analysis of toxic dinoflagellate A. tamiyavanichii isolated from Seto Inland Sea. In Proceedings of the Annual Meeting of the Japanese Society of Fisheries Science Conference Abstracts, Tokyo, Japan; 2005; p. 259. [Google Scholar]
- Lim, P.T.; Ogata, T. Salinity effect on growth and toxin production of four tropical Alexandrium species (Dinophyceae). Toxicon 2005, 45, 699–710. [Google Scholar] [CrossRef]
- Lim, P.T.; Leaw, C.P.; Usup, G.; Kobiyama, A.; Koike, K.; Ogata, T. Effects of light and temperature on growth, nitrate uptake, and toxin production of two tropical dinoflagellates: Alexandrium tamiyavanichii and Alexandrium minutum (Dinophyceae). J. Phycol. 2006, 42, 786–799. [Google Scholar] [CrossRef]
- Usup, G.; Cheah, M.Y.; Ng, B.K.; Leaw, C.P.; Ahmad, A. Toxin profile and relative toxicity of three paralytic shellfish poisoning toxin-producing dinoflagellates from Malaysia. Malays. Appl. Biol. 2006, 35, 41–45. [Google Scholar]
- Beppu, R.; Nojima, K.; Tsuruda, S.; Gomez-Delan, G.; Barte-Quilantang, M.; Taniyama, S.; Sagara, T.; Nishio, S.; Takayama, H.; Miyazawa, K.; et al. Occurrence of PSP-producing dinoflagellate Alexandrium tamiyavanichii in Bingo-Nada, the central coastal water of the Seto Inland Sea, Hiroshima Prefecture, Japan. Mar. Pollut. Bull. 2008, 56, 758–763. [Google Scholar] [CrossRef]
- Oh, S.J.; Matsuyama, Y.; Nagai, S.; Itakura, S.; Yoon, Y.H.; Yang, H.S. Comparative study on the PSP component and toxicity produced by Alexandrium tamiyavanichii (Dinophyceae) strains occurring in Japanese coastal water. Harmful Algae 2009, 8, 362–368. [Google Scholar] [CrossRef]
- Mohammad-Noor, N.; Adam, A.; Lim, P.T.; Leaw, C.P.; Lau, W.L.S.; Liow, G.R.; Muhamad-Bunnor, N.; Hamdan, N.A.; Md-Nor, A.; Kemat, N.; et al. First report of paralytic shellfish poisoning (PSP) caused by Alexandrium tamiyavanichii in Kuantan Port, Pahang, East Coast of Malaysia. Phycol. Res. 2018, 66, 37–44. [Google Scholar] [CrossRef]
- Lu, C.; Wu, S.; Zhang, C.; Yu, H.; Zhu, X.; Liu, M. Analysis of paralytic shellfish poison in shellfish in the offshore of the Guangxi. Mar. Environ. Sci. 2005, 24, 47–50. [Google Scholar]
- Jiang, T.; Jiang, T. Detection and analysis of PSP toxins in shellfish in coastal areas of China. Oceanol. Et Limnol. Sin. 2007, 38, 36–41. [Google Scholar]
- Huang, Y.; Huang, G.; Ye, X.; Li, X.; Wu, X.; Pang, Y.; Xie, Z.; Chen, J. Rapid detection of paralytic shellfish poison (PSP) in aquatic products. Jiangsu Agric. Sci. 2012, 40, 255–256. [Google Scholar]
- Du, K.; Lei, F.; Wu, N.; Jiang, T. The pattern of paralytic shellfish poisoning in shellfish cultured in East China Sea and South China Sea. J. Jinan Univ. 2013, 34, 343–346. [Google Scholar]
- Anderson, D.M.; Kulis, D.M.; Qi, Y.Z.; Zheng, L.; Lu, S.; Lin, Y.T. Paralytic shellfish poisoning in southern China. Toxicon 1996, 34, 579–590. [Google Scholar] [CrossRef]
- Etheridge, S.M.; Roesler, C.S. Effects of temperature, irradiance, and salinity on photosynthesis, growth rates, total toxicity, and toxin composition for Alexandrium fundyense isolates from the Gulf of Maine and Bay of Fundy. Deep Sea Res. Part II Top. Stud. Oceanogr. 2005, 52, 2491–2500. [Google Scholar] [CrossRef]
- Murray, S.; John, U.; Kremp, A. Alexandrium spp.: Genetic and ecological factors influencing saxitoxin production and proliferation. In Climate Change and Marine and Freshwater Toxins, 2nd ed.; Botana, L.M., Louzao, M.C., Eds.; Walter de Gruyter GmbH: Berlin, Germany; Boston, MA, USA, 2015; pp. 125–155. [Google Scholar]
- Vandersea, M.W.; Kibler, S.R.; Tester, P.A.; Holderied, K.; Hondolero, D.E.; Powell, K.; Baird, S.; Dorof, A.; Dugan, D.; Litaker, R.W. Environmental factors influencing the distribution and abundance of Alexandrium catenella in Kachemak bay and lower cook inlet, Alaska. Harmful Algae 2018, 77, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Tobin, E.D.; Wallace, C.L.; Crumpton, C.; Johnson, G.; Eckert, G.L. Environmental drivers of paralytic shellfish toxin producing Alexandrium catenella blooms in a fjord system of northern Southeast Alaska. Harmful Algae 2019, 88, 101659. [Google Scholar] [CrossRef]
- Brosnahan, M.L.; Velo-Suárez, L.; Ralston, D.K.; Fox, S.E.; Sehein, T.R.; Shalapyonok, A.; Sosik, H.M.; Olson, R.J.; Anderson, D.M. Rapid growth and concerted sexual transitions by a bloom of the harmful dinoflagellate Alexandrium fundyense (Dinophyceae). Limnol. Oceanogr. 2015, 60, 2059–2078. [Google Scholar] [CrossRef]
- Bricelj, V.M.; Shumway, S.E. Paralytic shellfish toxins in bivalve molluscs: Occurrence, transfer kinetics, and biotransformation. Rev. Fish. 1998, 6, 315–383. [Google Scholar] [CrossRef]
- Xun, X.; Cheng, J.; Wang, J.; Li, Y.; Li, X.; Li, M.; Lou, J.; Kong, Y.; Bao, Z.; Hu, X. Solute carriers in scallop genome: Gene expansion and expression regulation after exposure to toxic dinoflagellate. Chemosphere 2020, 241, 124968. [Google Scholar] [CrossRef]
- Etheridge, S.M. Paralytic shellfish poisoning: Seafood safety and human health perspectives. Toxicon 2010, 56, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.D.; Hatfield, R.G.; Maskrey, B.H.; Algoet, M.; Lawrence, J.F. Evaluation of the new European Union reference method for paralytic shellfish toxins in shellfish: A review of twelve years regulatory monitoring using pre-column oxidation LC-FLD. TrAC Trends Anal. Chem. 2019, 113, 124–139. [Google Scholar] [CrossRef]
- Deeds, J.R.; Landsberg, J.H.; Etheridge, S.M.; Pitcher, G.C.; Longan, S.W. Non-traditional vectors for paralytic shellfish poisoning. Mar. Drugs 2008, 6, 308–348. [Google Scholar] [CrossRef] [PubMed]
- Chomérat, N.; Sellos, D.Y.; Zentz, F.; Nézan, E. Morphology and molecular phylogeny of Prorocentrum consutum sp. nov. (Dinophyceae), a new benthic dinoflagellate from south Brittany (northwestern France). J. Phycol. 2010, 46, 183–194. [Google Scholar]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Adachi, M.; Sako, Y.; Ishida, Y. Restriction fragment length polymorphism of ribosomal DNA internal transcribed spacer and 5.8S regions in Japanese Alexandrium species (Dinophyceae). J. Phycol. 1994, 30, 857–863. [Google Scholar] [CrossRef]
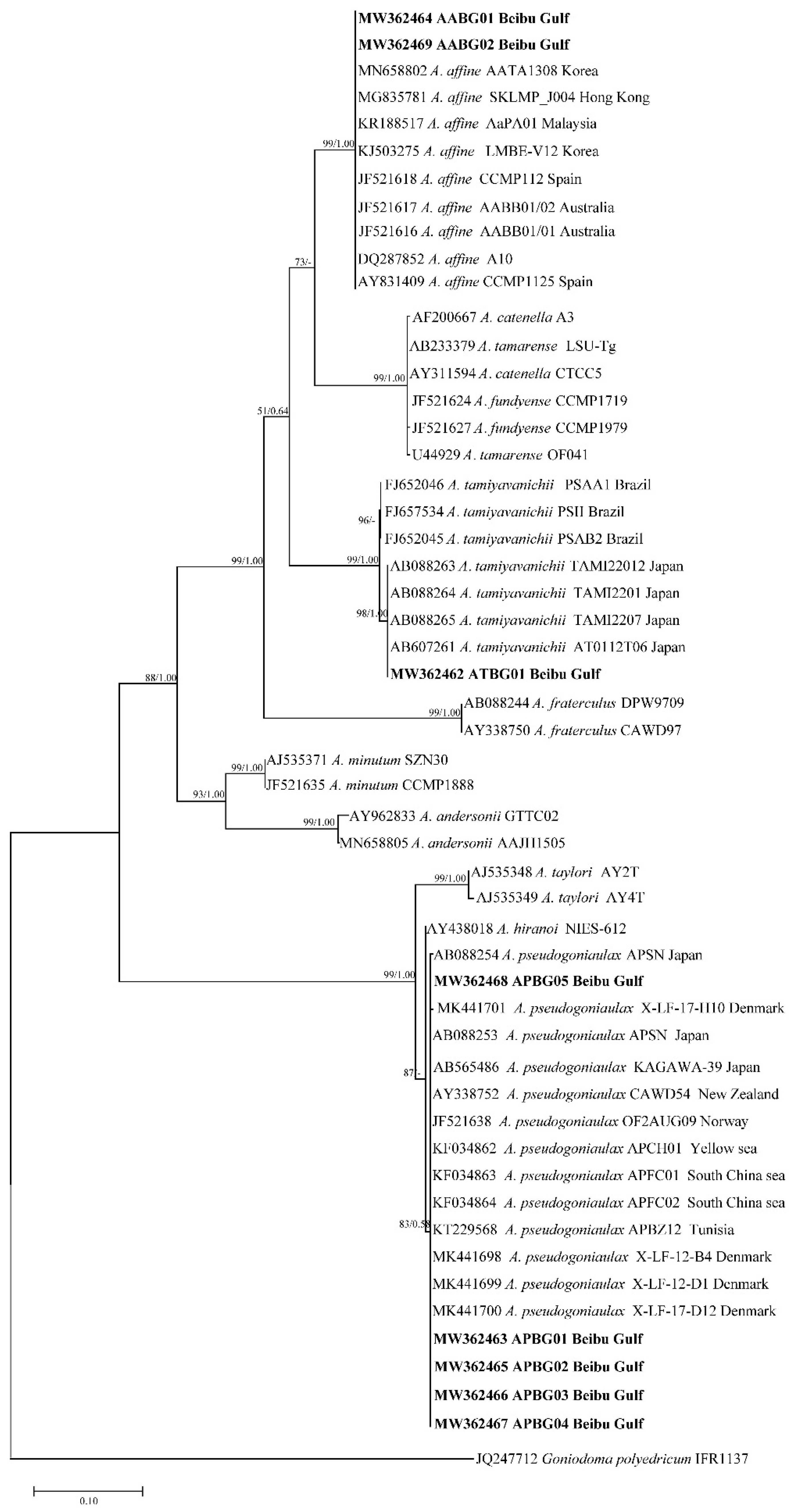
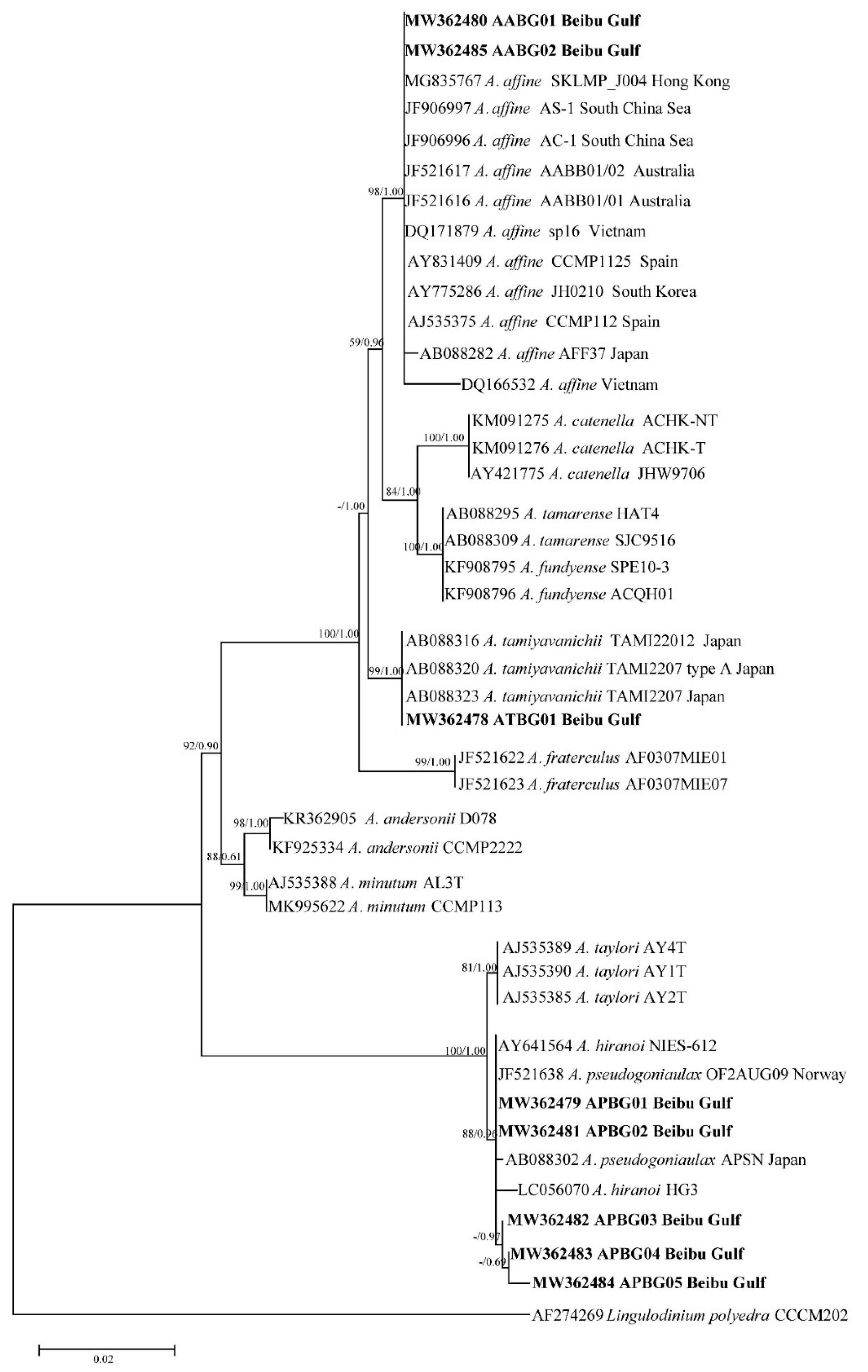
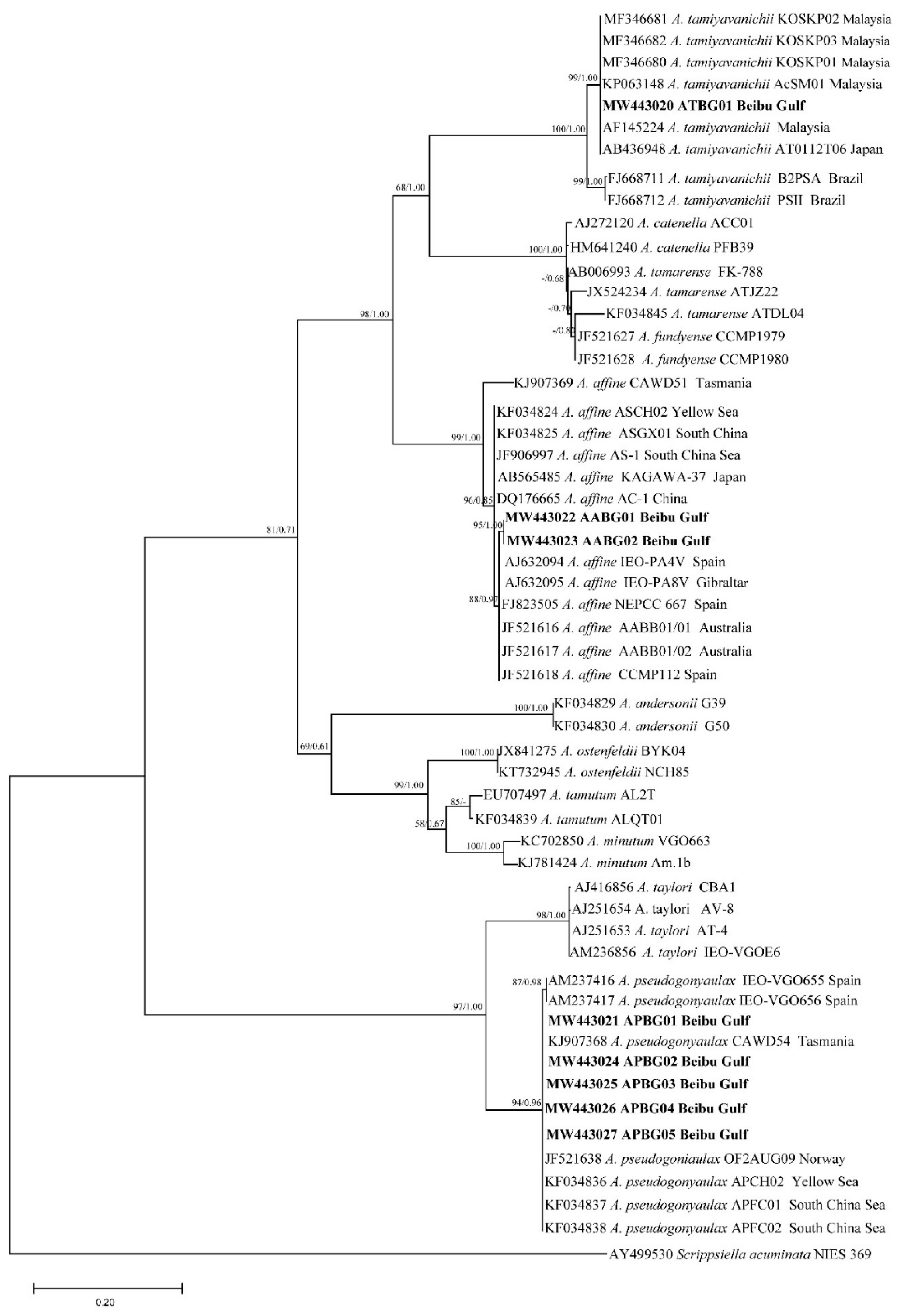
| Gene | Analysis Length | Average Content (%) | Conserved Site | Variable Site | Parsimonious Information Site | Monomorphic Site | Conversion/Transversion Ratio | |||
|---|---|---|---|---|---|---|---|---|---|---|
| A | T | G | C | |||||||
| LSU rDNA | 996 | 26.9 | 31.5 | 25.6 | 16.0 | 416 | 555 | 373 | 182 | 1.0 |
| SSU rDNA | 1769 | 27.7 | 29.4 | 25.2 | 17.7 | 1462 | 275 | 245 | 29 | 1.9 |
| ITS | 666 | 24.7 | 34.0 | 23.9 | 17.4 | 160 | 494 | 449 | 45 | 0.7 |
| Toxins | A. tamiyavanichii | A. pseudogonyaulax |
|---|---|---|
| ATBG01 | APBG04 | |
| GTX1, 4 | 0.88 | ND |
| GTX2, 3 | 1.2 | ND |
| GTX5 | 0.32 | ND |
| dcGTX2 | ND | ND |
| dcGTX3 | ND | ND |
| STX | 2.20 | ND |
| dcSTX | ND | ND |
| neoSTX | ND | ND |
| dcNEO | ND | ND |
| C1 | ND | ND |
| C2 | ND | ND |
| Total PST content | 4.60 | ND |
| Strain | Locality | Toxin Component | Toxin Content (fmol/cell) | References |
|---|---|---|---|---|
| Chula 5 | Gulf of Thailand | C1-2, GTX1-5, STX | 16,200 a | [47] |
| Chula 6 | Gulf of Thailand | GTX1-5, STX | 7500 a | [47] |
| Chula 8 | Gulf of Thailand | C1-3, GTX1-5, STX | 18.9 | [48] |
| MBS8811-1 | Sagami Bay, Japan | C1-4, GTX1-5, STX | 3.7 | [48] |
| MBS8811-3 | Sagami Bay, Japan | C1-2, GTX1, 4, GTX5, STX | 66.3 | [48] |
| — | Seto Island, Japan | C1-2, GTX1-5, STX | 112.5 | [35] |
| — | Malaysia | — | 26 | [49] |
| CTCC23 | South Africa | C1-4, GTX1-4, STX, neoSTX, B1 | 0.26 b | [50] |
| Western Japan | Japan | C1-2, GTX1-5, STX, neoSTX | 40–424 | [51] |
| AcMS01 | Sebatu Malacca, Malaysia | C1-2, GTX1-5, STX | 38–80 | [52] |
| AcMS01 | Sebatu Malacca, Malaysia | C1-2, GTX1-5, STX | 60–180 | [53] |
| — | Malaysia | C2, GTX1-4, dcGTX3, STX | 54 | [54] |
| ATY041106 | Seto Island, Japan | C1-2, GTX1-4 | 38.7 ± 10.9 × 10−6 a | [55] |
| ATY051018 | Seto Island, Japan | — | 111.5 × 10−6 a | [55] |
| Fukuyama Bay | Seto Island, Japan | C1-4, GTX1-5, neoSTX, STX | 244 ± 102 | [56] |
| Kasato Bay | Seto Island, Japan | C1-4, GTX1-5, neoSTX, STX | 307 ± 83.8 | [56] |
| Uchinoumi | Seto Island, Japan | C1-4, GTX1-5, neoSTX, STX | 328 ± 152 | [56] |
| Inokushi Bay | Kyushu Island, Japan | C1-4, GTX1-5, neoSTX, STX | 54.7 ± 5.32 | [56] |
| PSAA1 | Brazil | GTX3-4, dcGTX2-3, neoSTX, STX | 16.85 | [36] |
| At2 | Seto Island, Japan | C1-2, GTX1-6, neoSTX, STX | 2410 | [37] |
| At4 | Seto Island, Japan | C1-2, GTX1-6, neoSTX, STX | 840 | [37] |
| At6-C1 | Seto Island, Japan | C1-2, GTX1-6, neoSTX, STX | 289 | [37] |
| At6-C2 | Seto Island, Japan | C1-2, GTX1-6, neoSTX, STX | 359 | [37] |
| At6-C3 | Seto Island, Japan | C1-2, GTX1-6, neoSTX, STX | 264 | [37] |
| At6-C4 | Seto Island, Japan | C1-2, GTX1-6, neoSTX, STX | 220 | [37] |
| KOSKP01 | Kuantan Port, Malaysia | GTX1-5 | 3070 | [57] |
| KOSKP02 | Kuantan Port, Malaysia | GTX1-5 | 5960.4 | [57] |
| KOSKP03 | Kuantan Port, Malaysia | GTX1-5 | 1027.2 | [57] |
| — | Kuantan Port, Malaysia | GTX1-5 | 3.07 × 106 | [38] |
| — | Sebatu Malacca, Malaysia | GTX1-5 | 1.167 | [38] |
| ATBG01 | Weizhou Island, Beibu Gulf | GTX1-5, STX | 4.6 | This study |
| Sampling Date | Locality | Methodology | Toxins | Detection Rate/Exceedance Rate | References |
|---|---|---|---|---|---|
| 2001.11–2004.04 | Tieshan, Beihai, Fangcheng, Weizhou | MBA, HPLC | C1-2, GTX1-5, STX | exceeding standard 2.15–3.54 times | [58] |
| 2003.03–2004.05 | Beihai | MBA, HPLC | C1-2, GTX1-4, STX | detection rate 8.30% | [59] |
| 2005–2009 | Beibu Gulf | MBA | — | exceedance rate 1.0% | [28] |
| — | Qinzhou, Fangcheng, Beihai | American ABRAXIS kit | — | — a | [60] |
| 2007.11–2008.10 | Beihai | MBA | — | — b | [61] |
| 2014.09–2014.11 | Weizhou, Beihai, Fangcheng, Qinzhou | HPLC | GTX1-5, dcGTX2-3, neoSTX, dcSTX, STX | detection rate 100%, exceedance rate 6.1% | [29] |
| 2015.09 | Qinzhou | MBA, HPLC | GTX1, GTX4, neoSTX, STX | detection rate 86%, no exceedance | [30] |
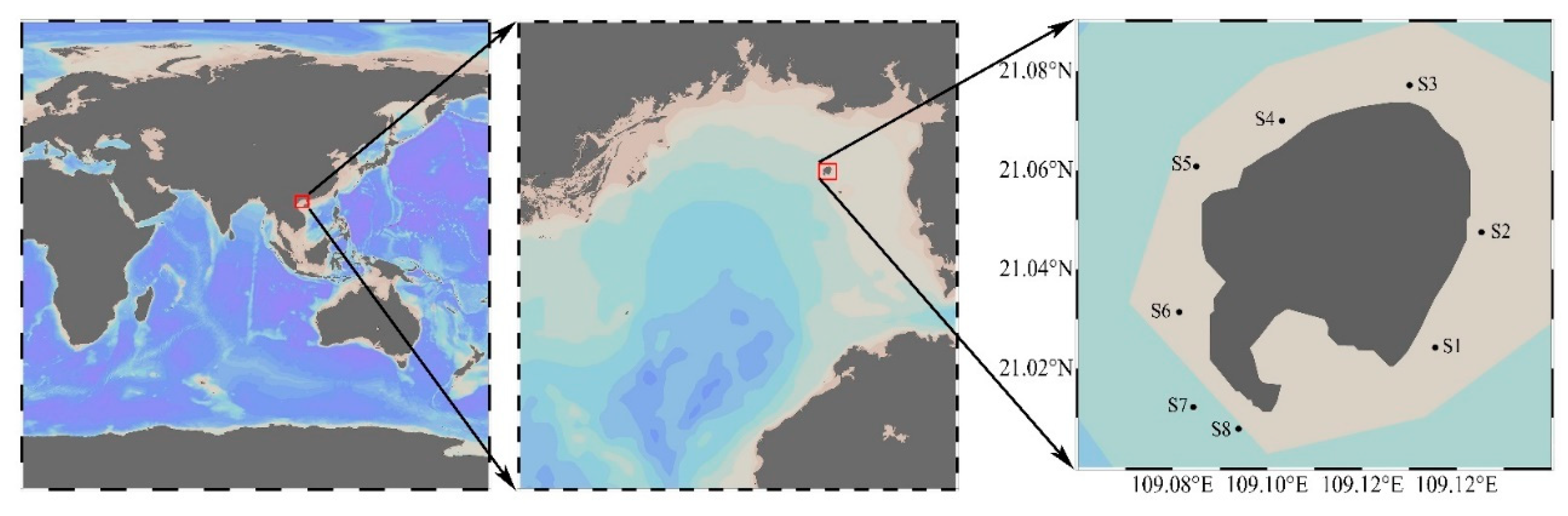
| Strains | Collection Date | Location |
|---|---|---|
| ATBG01 | 2018-06-09 | S2 (21°02′51″ N, 109°08′43″ E) |
| APBG01 | 2018-09-15 | S2 (21°02′51″ N, 109°08′43″ E) |
| AABG01 | 2018-09-15 | S2 (21°02′51″ N, 109°08′43″ E) |
| AABG02 | 2018-12-20 | S8 (21°00′28″ N, 109°05′38″ E) |
| APBG02 | 2019-06-10 | S2 (21°02′51″ N, 109°08′43″ E) |
| APBG03 | 2019-06-10 | S4 (21°04′12″ N, 109°06′11″ E) |
| APBG04 | 2019-06-10 | S5 (21°03′39″ N, 109°05′06″ E) |
| APBG05 | 2019-06-10 | S5 (21°03′39″ N, 109°05′06″ E) |
| Gene | Primers | Primer Sequences | References |
|---|---|---|---|
| LSU rDNA | D1R | 5′-ACCCGCTGAATTTAAGCATA-3′ | [73] |
| D2C | 5′-TGATCCTTCTGCAGGTTCACCTAC-3′ | ||
| SSU rDNA | 1F | 5′-AACCTGGTTGATCCTGCCAGT-3′ | [74] |
| 1528R | 5′-TGATCCTTCYGCAGGTTCAC-3′’ | ||
| ITS | FA | 5′-CCAAGCTTCTAGATCGTAACAAGG(ACT)TCCGTAGGT-3′ | [75] |
| RB | 5′-CCTGCAGTCGACA(TG)ATGCTTAA(AG)TTCAGC(AG)GG-3′ |
| Toxins | Quantitative Transition m/z | Qualitative Transition m/z | Residence Time (ms) | Impact Voltage (V) | Fragmentor Voltage (V) |
|---|---|---|---|---|---|
| GTX1 | 412 > 332.1 | 412 > 314.2 | 100 | 6/20 | 100/100 |
| GTX2 | 396.1 > 316.1 | 396.1 > 297.8 | 100 | 5/15 | 110/110 |
| GTX3 | 396.1 > 316.1 | 396.1 > 298.7 | 100 | 5/15 | 110/110 |
| GTX4 | 412 > 332.1 | 412 > 314.2 | 100 | 6/20 | 100/100 |
| GTX5 | 380.1 > 300 | 380.1 > 282.1 | 100 | 11/15 | 94/105 |
| dcGTX2 | 352.8 > 334.7 | 352.8 > 254.9 | 100 | 5/10 | 120/120 |
| dcGTX3 | 352.8 > 334.7 | 352.8 > 254.9 | 100 | 5/10 | 120/120 |
| STX | 300 > 282 | 300 > 204 | 100 | 10/20 | 100/100 |
| dcSTX | 257 > 222.1 | 257 > 126 | 100 | 10/20 | 80/80 |
| neoSTX | 316.1 > 298 | 316.1 > 220 | 100 | 10/20 | 100/100 |
| dcNEO | 273 > 225.1 | 273 > 126 | 100 | 18/18 | 80/80 |
| C1 | 474.1 > 251 | 474.1 > 122 | 100 | 20/30 | 60/60 |
| C2 | 474.1 > 351 | 474.1 > 122 | 100 | 20/30 | 60/60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; He, X.; Li, H.; Zhang, T.; Lei, F.; Gu, H.; Anderson, D.M. Molecular Identification and Toxin Analysis of Alexandrium spp. in the Beibu Gulf: First Report of Toxic A. tamiyavanichii in Chinese Coastal Waters. Toxins 2021, 13, 161. https://doi.org/10.3390/toxins13020161
Xu Y, He X, Li H, Zhang T, Lei F, Gu H, Anderson DM. Molecular Identification and Toxin Analysis of Alexandrium spp. in the Beibu Gulf: First Report of Toxic A. tamiyavanichii in Chinese Coastal Waters. Toxins. 2021; 13(2):161. https://doi.org/10.3390/toxins13020161
Chicago/Turabian StyleXu, Yixiao, Xilin He, Huiling Li, Teng Zhang, Fu Lei, Haifeng Gu, and Donald M. Anderson. 2021. "Molecular Identification and Toxin Analysis of Alexandrium spp. in the Beibu Gulf: First Report of Toxic A. tamiyavanichii in Chinese Coastal Waters" Toxins 13, no. 2: 161. https://doi.org/10.3390/toxins13020161
APA StyleXu, Y., He, X., Li, H., Zhang, T., Lei, F., Gu, H., & Anderson, D. M. (2021). Molecular Identification and Toxin Analysis of Alexandrium spp. in the Beibu Gulf: First Report of Toxic A. tamiyavanichii in Chinese Coastal Waters. Toxins, 13(2), 161. https://doi.org/10.3390/toxins13020161







