Abstract
Mycotoxin contamination causes significant economic loss to food and feed industries and seriously threatens human health. Aflatoxins (AFs) are one of the most harmful mycotoxins, which are produced by Aspergillus flavus, Aspergillus parasiticus, and other fungi that are commonly found in the production and preservation of grain and feed. AFs can cause harm to animal and human health due to their toxic (carcinogenic, teratogenic, and mutagenic) effects. How to remove AF has become a major problem: biological methods cause no contamination, have high specificity, and work at high temperature, affording environmental protection. In the present research, microorganisms with detoxification effects researched in recent years are reviewed, the detoxification mechanism of microbes on AFs, the safety of degrading enzymes and reaction products formed in the degradation process, and the application of microorganisms as detoxification strategies for AFs were investigated. One of the main aims of the work is to provide a reliable reference strategy for biological detoxification of AFs.
Keywords:
aflatoxin; biological detoxification; detoxification mechanism; degradation products; probiotics Key Contribution:
The mechanism, advantages and disadvantages of microorganisms and enzymes to detoxification of aflatoxins are reviewed; A reliable reference strategy for biological detoxification of aflatoxins is provided.
1. Introduction
Mycotoxins are metabolites of fungi that are ubiquitous in cereal crops and animal forage [1]. One group of well-known mycotoxins, aflatoxins (AFs), are secondary metabolites produced mainly by Aspergillus flavus, which produces both aflatoxin B1 (AFB1) and aflatoxin B2 (AFB2), and by Aspergillus parasiticus, which produces aflatoxin G1 (AFG1) and aflatoxin G2 (AFG2) [2]. They have a high degree of hepatotoxicity, nephrotoxicity, and immunotoxicity [3]. Among them, AFB1 is the most toxic and is well known for its toxic carcinogenic and teratogenic mutation effects [4,5]. As a result, it was categorized as a Class I carcinogen by the World Health Organization in 1993 [6,7].
The long-term consumption of food contaminated with AFs can induce inflammatory damage to hepatocytes [8]. Furthermore, the AF-DNA adducts can result in the production of cancer cells [9], leading to liver cancer [10,11]. In addition, AFB1 can induce the apoptosis of CASP3 and BAX, and shows extensive cytotoxicity to neuronal cells, including ROS accumulation, DNA damage, S-phase arrest, and apoptosis [12]. AFs can also destroy the metabolic pathways of a variety of intestinal flora. This may affect energy supply and lead to certain metabolic diseases [13,14]. Today, South-East Asia remains a high-risk area for acute AF poisoning [15]. Molecular structures of four naturally occurring AFs are illustrated in Figure 1.
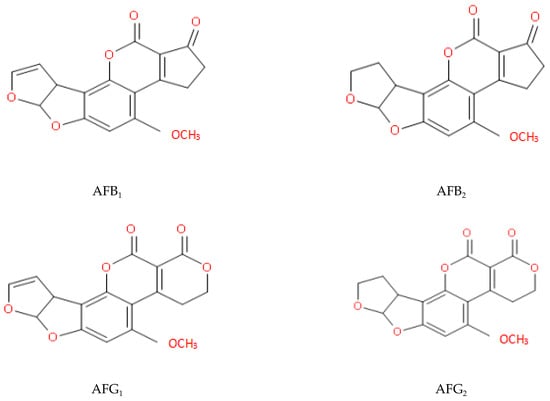
Figure 1.
Structures of some natural AFs (Aflatoxins B1 and G1 have double bonds at positions 8–9; aflatoxins B2 and G2 do not).
AFs are often detected in grains, nuts, and spices [16,17]. Contamination occurs readily when feed and food are exposed to high temperature and high humidity [18]. The toxic effects of AFs are not only manifested in feeding. Animals that consume contaminated feed are likely to be poisoned [19]. However, the toxins found in the animal by-products (e.g., milk and milk products) will enter other animals in the food chain, which can result in further serious consequences and spread the contamination more widely [20]. Finding ways to safely and efficiently detoxify food has thus become a focus of research [21]. Contamination of AFs in food and feed samples in some countries is displayed in Table 1.

Table 1.
Contamination of AFs in food and feed.
AFs can be detoxified using physical, chemical, and biological detoxification methods, and a great deal of research has been carried out using these methods in the past few decades [31,32]. Physical methods are those most commonly used; for example, adsorbents are employed to undertake physical adsorption to control toxin contamination [33]. Although adsorbent products can reduce the bioavailability of mycotoxins, in practice, the toxins cannot be completely adsorbed [34]. In recent years, after continuous improvement, nanotechnology has been applied to adsorbents, such as magnetic adsorbents, whose adsorption capacity has been much improved [35]. However, physical methods show many disadvantages, e.g., limited applicability, poor detoxification effect, and limited detox product status [36]. Chemical methods involve treatment with acid, alkali, or oxidizing agent [37]. The use of chemical substances such as chlorine dioxide to disinfect toxins [38] may impair the appearance and taste of food. After chemical treatment, chemical residues in food may be harmful to humans [39]. Neither approach is the better option for detoxification. Biological detoxification also has certain drawbacks, such as the difficulty of controlling microbial performance and the safety of the newly formed product to the body [39]; however, biological detoxification has high specificity, produces harmless products, and can even completely detoxify samples under appropriate conditions [37,40]. Thus, biological detoxification is gradually becoming the most suitable detoxification approach [41,42].
Beneficial intestinal bacteria have many important functions. They produce various nutrients for the host, prevent infections caused by intestinal pathogens, and regulate the immune response [43]. At the same time, the life activity metabolites of microorganisms (such as exogenous antioxidant compounds) can induce activity among genes related to the oxidative stress toxicity of AFs, restore the oxidative balance destroyed by mycotoxins, and prevent the production of ROS and RNS [44]. Therefore, the use of microorganisms to detoxify AFs is a promising new technology with broad application prospects; as such, their use is a research hotspot both for the beneficial effects and AF detoxification [41,45].
2. Microorganisms with Detoxification Effects
Different microorganisms exert detoxification effects toward AFs [46]. The microorganisms that exert detoxification effects on AFs are listed in Table 2.

Table 2.
Microorganisms that can be used for the detoxification of AFs.
3. Decontamination Mechanism of AFs
3.1. Microorganisms Inhibit the Production of AFs
Mixed populations of microorganisms coexist in the ecosystem, thus forming a complex microbial community [86]. Soil is the natural habitat of Aspergillus flavus, and soil ecotoxicology has gradually become a safety hotspot [87]. The high complexity and heterogeneity of the soil environment make it difficult to analyze the ecological functions of secondary metabolites such as AFs in the soil [87]. Therefore, co-cultivation research has become an effective means to control or reduce specific contaminants in grain, feed, and the environment [88].
Competitive interactions between pathogenic and beneficial microorganisms include both exploitation and interference competition [89]. When Aspergillus flavus and Aspergillus parasiticus are co-cultured with Salmonella, the colony diameter and spore formation of Aspergillus flavus and Aspergillus parasiticus are decreased, and the contents of AFs (AFB1, AFB2, AFG1, and AFG2) are reduced [86]. After 24 h of co-cultivation of Aspergillus flavus and Aspergillus niger, the growth of Aspergillus flavus was inhibited and the production of AFB1 was also reduced by 42.8% [80]. Further studies implied that, during co-cultivation, the life activities of other microorganisms can cause gene mutations or activate silent gene clusters, thereby reducing the production of AFs [90]. The biosynthetic processes that generated AFB1 in Aspergillus flavus were interrupted when the A. flavus was co-cultured with Streptomyces roseolus. More specifically, the interruption to the biosynthetic pathway occurred at an early stage before the synthesis of norsolorinic acid, so the first toxic AFB1 precursor could not be synthesized normally and the concentration of AFB1 was decreased to an undetectable level [73]. The inhibitory compounds secreted by Aspergillus oryzae and a non-aflatoxigenic A. flavus can inhibit the production of AFB1 and the growth and reproduction of Aspergillus flavus. Transcriptome sequencing has shown that some genes such as AflS, FarB, and MtfA are involved in the biosynthetic pathway of AFs. The synthetic gene cluster was significantly down-regulated, and the two conidial transcription factors BrlA and AbaA were significantly down-regulated, which may down-regulate conidia-specific genes (such as the conidial hydrophobin genes RodA and RodB) [91].
Toxins will exist for a long time after polluting the soil. In planting on contaminated land, toxins will be transferred from the soil to the grain, and then transferred to fodder whereafter they are accumulated. If beneficial microorganisms can multiply in the contaminated soil, the toxin content will be greatly reduced. In short, co-cultivation can indeed provide new insights for controlling the synthesis of AFs and the proliferation of Aspergillus flavus. The exact molecular mechanism of this process remains to be studied.
3.2. Microbial Adsorption of AFs
Adsorption means that due to the special structure on the microbial cell wall, AFs interact with non-covalent bonds (the main effect is that of Van der Waals forces), which makes it easier to bind, reducing the bioavailability of mycotoxins in the gastrointestinal tract, and protecting the body from toxin infringement [92,93]. For biosorption, the most often studied strains are Lactobacillaceae and Saccharomyces, which can effectively bind AFs through polysaccharides (such as peptidoglycan and teichoic acid) on the bacterial wall [94,95]. The adsorption mechanisms thereof are illustrated in Figure 2.
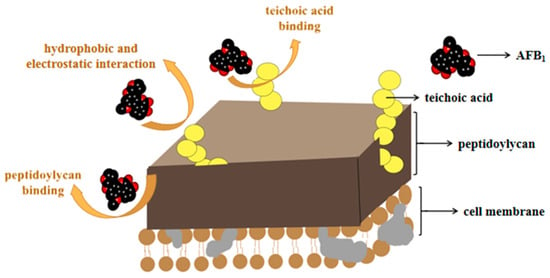
Figure 2.
The adsorption of AFs by microorganisms (taking AFB1 as an example). Microorganisms can adsorb AFs through peptidoglycan or phosphoric acid in the cytoderm, and hydrophobic and electrostatic interaction.
Lactobacillaceae and Saccharomyces are the most commonly used microorganisms in fermentation: Lactobacillus delbrueckii, Lactobacillus kefiri, and Lactobacillus rhamnosus strain (LGG) are used for the fermentation of yogurt or cheese; Saccharomyces cerevisiae can be used for brewing beer [76,96,97]. The excellent adsorption capacity and natural fermentation function make the use of Lactobacillaceae and Saccharomyces essential in the process of detoxifying food. LGG is an excellent biosorption species. The combination of heat-treatment and anaerobic solid fermentation can remove 100% of AFB1 [41]. Of course, this is the result of adsorption under simulated laboratory conditions. Recent research has shown that LGG can adsorb 90% of AFs in pistachio nuts subjected to heat treatment (from an initial concentration of 20 ppb), and it had no effect on the qualitative characteristics of the pistachios, e.g., color, texture, and peroxide value [58].
Not only for food, LGG has outstanding stability with respect to stomach acid and bile, and can therefore enter the intestines of the body in vivo. It is also an excellent species to use in fermentation as it has favorable degradability (so it is safe to use during the fermentation process) and does not affect the palatability of the product [57]. It is worth noting that, although LGG is resistant to the environment in the intestine, its binding to toxins is unstable. The stability of the combination of species and toxins depends on various parameters, such as pH, temperature, sorbate ion concentration, and mixing rate [98]; therefore, careful optimization is required before application. Unlike Lactobacillus, however, Saccharomyces results in adsorption products that are more stable (i.e., less likely to re-release the toxin). The combined product forms a complex that is not readily adsorbed by the body and is mostly excreted. Hence, Saccharomyces species are relatively stable mycotoxin adsorbents (mainly because the toxins form a specific complementary structure with the mannose on the cell walls). A study has concluded that the adsorption capacity of Saccharomyces lysate with respect to AFs can reach 2.5 μg/mg [99]. The problem of how to improve the adsorption capacity of Saccharomyces is also a hot research topic.
In addition, L. plantarum not only exerts a detoxifying effect on AFs but is a biological preservative. It can inhibit the decay of animal manure and residual feed in the middle and late stages of animal breeding, reducing the amount of chemicals required and the cost of breeding. It is therefore very important in production practices [75,76].
3.3. Microbial Degradation of AFs
Degradation involves the microorganisms producing certain substances during their life activities that change the original structure of the mycotoxins and convert them into substances that are low in toxicity or even completely non-toxic. AFs are metabolites of difurans and the double bond in the furan ring is the main site leading to genetic mutations and carcinogenic teratogenic effects [100]. The main toxic structure present in AFs is the coumarin lactone ring, which is readily hydrolyzed [101,102,103]. During the degradation process, the active substances secreted by the microorganisms are mainly enzymes that convert the AFs into other substances. Main degrading enzymes of AFs are displayed in Table 3.

Table 3.
AF-degrading enzymes and their sources.
AFO, as an intracellular enzyme, is a typical member of the dipeptidyl peptidase III (DPP III) enzyme family [112] and was extracted from Armillariella tabescens. It can act on the dilute ether bond of the furan ring of AFB1 and convert it to epoxide. Hydrolysis to generate AFB1-8,9-dihydrodiol was undertaken to achieve the purpose of detoxification [113,114]. Armillariella tabescens is a Chinese edible fungus, and AFO is a new choice in practical applications preventing biodegradation of food and detoxification of AF in feed. The reaction mechanism of AFO is demonstrated in Figure 3.
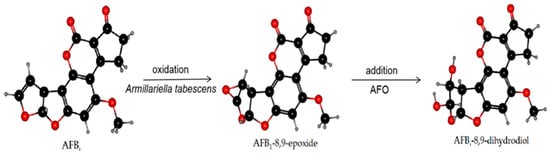
Figure 3.
The mechanism of AFB1 degradation. Armillariella tabescens and the AFO produced therewith can act on the dilute ether bond of the furan ring to activate AFB1 transforming it into an epoxide. The hydrolysis reaction was conducted to generate a new compound with significantly reduced toxicity: AFB1-8,9-dihydrodiol.
Laccase is an extracellular enzyme that contains four copper ions and can be extracted from some microorganisms (e.g., white rot fungi) [115]. Many in vitro experiments have been conducted to ascertain the stability of laccases. In vitro degradation experiments using recombinant fungal laccase found that AFB1, AFB2, AFG1, and AFG2 can interact with the laccase (near the T1 copper center of the enzyme) via hydrogen bonds and hydrophobic interactions with amino acid residues. The binding capacity of the interaction was also shown to decrease in the order AFB1 > AFG2 > AFG1 > AFB2 and the maximum degradation rates were 90.33%, 74.23%, 85.24%, and 87.58%, respectively [116]. The latest research by Zhou et al. found that a new type of laccase that catalyzes the degradation of AFB1 could be purified and identified in white-rot fungus Cerrena unicolor. The half-life of AFB1 degradation catalyzed thereby was 5.16 h, and the degradation product was AFQ1 [85]. These findings are expected to lead to the use of laccase as a new AFO able to degrade AFB1 in food and feed. The reaction mechanism of laccase is displayed in Figure 4.
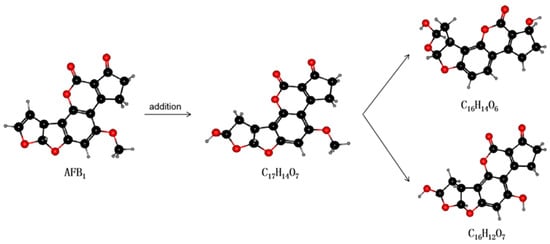
Figure 4.
The mechanism by which laccase degrades AFB1. Laccase can act on the double bond of the furan ring to undergo an addition reaction. As shown, the degradation product with molecular formula C17H14O7 (unstable structure) is first produced, then the elimination reaction occurs to generate two degradation products with different structures: C16H14O6 and C16H12O7.
There are also some newly discovered enzymes that also have detoxification capability for AFs. The alternative oxidase, which is ubiquitous in the plant kingdom, affects the penultimate intermediate of AFB1 biosynthesis [117], but after analysis of the genome sequence, alternative oxidase also has expressed genes in A. clavatus, A. flavus, A. fumigatus, A. nidulans, and A. niger [118]. Alternative oxidase may be used as a target to control the reproduction of Aspergillus flavus and contamination by AFs.
MSMEG-5998 is an AF-degrading enzyme produced by Mycobacterium smegmatis (F. smegmatis), which can reduce AFB1-induced cytotoxicity in HepG2 cells by ameliorating DNA damage and p53-mediated apoptosis. Thioredoxin affected the rate of degradation of MSMEG-5998 to AFB1 as it increased from 31% to 63% [108,119]. The MSMEG-5998 connected by thioredoxin shows great application prospects, but the toxicity of the product remains to be considered.
CotA laccase, a new aflatoxin oxidase in Bacillus licheniformis, can convert AF into AFQ1 and epi-AFQ1. In vitro experiments have found that AFQ1 and epi-AFQ1 do not inhibit the viability of human hepatocytes and induce apoptosis [120]. These findings are expected to allow use of CotA laccase as a new AFO to degrade AFB1 in food.
The two key sites that affect the toxicity of AFs are the furan and lactone rings and the detoxification process mainly involves changes in the structures of these rings. After many years of research, the metabolites of AFs that have been identified fall into the following three categories: (i) hydroxylated metabolites, e.g., AFM1, aflatoxin P1 (AFP1), and aflatoxin Q1 (AFQ1); (ii) epoxides, e.g., AFB1-8,9-epoxide; and (iii) metabolites of microorganisms or animals, e.g., AFG2a, AFB2a, and aflatoxicol (AFL) [44,45,79]. The molecular structures of some of these metabolites are shown in Figure 5 [121,122,123,124].
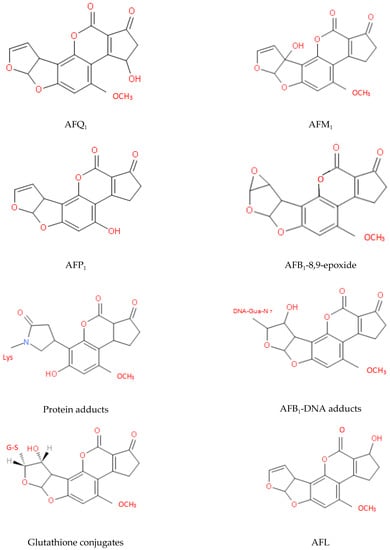
Figure 5.
Molecular structures of some key AF metabolites.
Unlike adsorption, degradation changes the structure of toxins. The toxicity of degradation products is the most important indicator of whether degrading enzymes can be used to detoxify the body. If the degradation product is of low toxicity or even non-toxic, this degradation enzyme is applicable. On the contrary, there is no applied research value otherwise. Melvin et al. found that Pseudomonas putida MTCC 1274 and 2445 can tolerate AFB1 in the medium, break the furan and lactone rings in the AFB1 molecule within 24 h of incubation, and convert it into new products: a non-toxic compound, AFD1 and two compounds, AFD2 and AFD3, of low toxicity [66]. Bacillus velezensis, Lysinibacillus fusiformis, Staphylococcus warneri, and other species can also degrade AFs into new substances with significantly reduced cytotoxicity [54,125]; however, the degradation process is often accompanied by many intermediate metabolites, and it is not enough to analyze only the toxicity of the final degradation products. Tetragenococcus halophilus CGMCC 3792 can produce six non-toxic metabolites in the process of AFB1 degradation, and there are two completely different degradation pathways [63]. The end products of the two pathways are non-toxic C14H20O2 compounds [63]. The high degradation rate of AFB1 achieved using T. halophilus CGMCC 3792 and the non-toxicity of its degradation products suggest it has detoxification applications, both in vivo and in vitro, and huge application potential in the processing of fermented oriental seasonings.
AF degradation results obtained using representative microorganisms and the degradation products formed are displayed in Table 4. Separating and purifying degradation enzymes and determining the toxicity of degradation products are problems that must be faced in any clinical application of biodegradation. The degrading enzyme can be amplified and expressed according to its gene sequence, and has a good degradation effect, laying a solid foundation for its actual clinical application. The toxicity of the product is a reference indicator for the use of degrading enzymes. How to isolate degrading enzymes from a species that can degrade AFs into non-toxic metabolites will be the focus of future research.

Table 4.
Microbial localization of AF-degrading substances and degradation products.
4. Application of Microbial Detoxification
4.1. Compound Probiotics Increase the Ability to Detoxify AFs
Although many microorganisms can detoxify AFs, probiotics are the first choice for detoxification. Adding probiotics during the breeding process can help prevent AFs causing tissues lesions, especially in the liver [10]. The detoxification of AFs using probiotics often involves multiple effects; multiple species can therefore be used together to acquire a better detoxification effect. The saccharomyces-containing mixture present in kombucha can adsorb AFB1 and convert it into four products of low toxicity. Poisoning tests using brine shrimp showed that the mortality rates of these AFB1 degradation products were between 20% and 80%, whereas the mortality rate with AFB1 was up to 100% under the same conditions [128]. This result proved that this mixed yeast product can adsorb part of the toxin while converting another part into less toxic products, thus reducing the impact of AFs on cell tissues and even the body as a whole.
Chen et al. found that Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus can completely remove AFB1 and AFG1 in peanuts subjected to anaerobic, high-temperature, solid fermentation conditions (to the extent that no obvious toxicity was observed in the final products) [58]. In this case, the two species facilitated excellent biotransformation under specific conditions. In general, this research was conducted under optimal growth conditions specific to the strain; however, it is necessary to ascertain the detoxification ability of strains to AFs under specific conditions.
The use of probiotics compound not only improves the rate of degradation of AFs, but also makes the intestinal epithelial barrier more resistant to mycotoxins and toxins from other pathogenic microorganisms [128]. Cavaglieri et al. showed that probiotics of certain bacteria (Pediococcus pentosaceus RC006) and yeasts (Kluyveromyces marxianus VM003) have the ability to adsorb and degrade AFM1 in milk to fewer toxic derivatives when used in combination [129].
The probiotic mixture used by Barati et al. (consisting of Bacillus and Lactobacillus species and cell walls of Saccharomyces cerevisiae) was found to reduce the inhibitory effect that AFs have on the humoral and cellular immune systems of broiler chicks. This mixture was therefore able to weaken the anti-nutritional effects of the AFs. Furthermore, it also improved the synthesis of proteins in the chicks. Thus, the mixture could control the impact of AFs on the chicks and improve their immune functions and biochemical pathways [130,131]. The combined use of probiotics to detoxify AFs in recent years is displayed in Table 5.

Table 5.
Detoxification effects of probiotic compounds on AFs.
4.2. Microbial Preparations Can Remove AFs in Food and Feed
The detoxification method of AFs has attracted increasing attention; however, the in vivo detoxification reaction is difficult due to the problem of the activity of biological factors. Therefore, the in vitro detoxification study of bacterial fermentation broth is warranted. The degrading enzyme activity of Bacillus subtilis BCC 42005 was stable and non-toxic at IC 50.4 mg/mL. Its fermentation broth was mixed with water as a corn-soaking agent. After 2 h of contact, the content of AFB1 was decreased by 54% [136]. The 39 volatile organic compounds produced by Streptomyces philanthi RL-1-178 could replace toxic chemical fungicides as biological fumigants and control the production of AFB1, AFB2, and AFG2 in stored soybean seeds [137]. Therefore, microorganisms can be used as a new biological agent to reduce the contamination of AFs in food and feed.
4.3. Microbes Ameliorate the Damage Caused by AFs to the Body
Fan et al. researched the ability of Bacillus subtilis ANSB060 to detoxify AFs. Their results showed that B. subtilis improved the growth performance and meat quality of broilers [138]. The levels of AF residues in the livers of broilers consuming naturally moldy peanut meal were also decreased [134]. Chen et al. found that oral Lactobacillus bulgaricus or Lactobacillus rhamnosus ingestion can significantly prevent liver injury induced by AFB1, and reduce histopathological changes and inflammation by elevating the expression of NF-κB p65 [138]. Feeding with Lactobacillus plantarum 299v can decrease the contents of serum lactate dehydrogenase and alanine aminotransferase in the liver and increase the body weight of broilers by about 20%-55%, bringing economic benefits [139]. Therefore, microorganisms can ameliorate damage to the body induced by AFs by adjusting related pathways, or they can preferentially combine with AFs to prevent AFs from exerting their toxic effects. The oral administration of microorganisms may be a new treatment for AF poisoning.
4.4. Combined Use of Probiotics, Biological Agents, and Degrading Enzymes
As probiotics are safe to use and have superior detoxification ability, the combined use of compound probiotics and degrading enzymes has also been explored in recent years. For example, when a 1:1:1 mixture of Bacillus subtilis, Lactobacillus casei, and Candida utilis was mixed with Aspergillus oryzae degrading enzyme in the ratio of 3:2, the degradation rate of AFB1 was found to be 63.95% [135]. Another study found that using licorice extract, Protexin probiotic, toxin binder (Agrabound), and poultry litter biochar as additives, during mixed feeding of broiler chickens, can reduce the effects of AFB1 on broiler chickens, improving blood indicators, and immunity to good effect [140].
Evaluating food and feed to identify its safety will also need to be a top priority in future research. In short, the combined use of probiotics, biological agents, and degrading enzymes is another innovative strategy for mycotoxin degradation.
4.5. Detoxification of Mixed Mycotoxins by Microorganisms
The pollution caused by mycotoxins is often not of a single type, but of mixed types: for instance, AFs and zearalenone, etc. Beneficial microorganisms can simultaneously detoxify multiple toxins. Lactic acid bacteria have detoxification effects on AFs, Ochratoxin A, and zearalenone [141]. B. subtilis and B. velezensis have high degradation efficiency when applied to AFs and zearalenone, and the degradation products have also been studied [129]. Based on more thorough research into the mechanisms of detoxification, the joint action of multiple microorganisms and the combined use of multiple degrading enzymes will be the focus of future research.
5. Conclusions
The use of microorganisms (especially microorganisms with probiotic properties) is a specific, effective, environmentally friendly, cheap, and safe strategy. The pleasant harvest produced by microbial detoxification is the elimination of chemical pesticides and pollutants in food and feed, and an absence of toxic residues. At the moment, biological detoxification technology is far from perfect and the determination and purification of metabolites is incomplete in many cases. Therefore, more research is needed to reveal the mechanism, dosage, time of microbial detoxification, and how to use these new microbial preparations to maximize the prevention and beneficial effects on toxins. As the technology develops, the mechanisms by which these probiotics detoxify AFs will gradually become well known and their use as feed/food additives will be mastered and perfected.
It is, therefore, just a matter of time before the production of enzymes and microbial preparations (and other biological additives) are taken to the stage where large-scale industrialization is realized.
Author Contributions
Y.G. wrote the paper; J.C. revised; E.N., M.L., W.W. and K.K. revised and supervised. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (grants No. 31972746; grants No. 31772809; grants No. 31872538), Liaoning Provincial Natural Fund Guidance Program Project (2019-ZD-0708), China Postdoctoral Science Foundation (grants No. 2016T90477), PAPD, Project UHK VT2019-2021.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Jiang, K.; Huang, Q.; Fan, K.; Wu, L.; Nie, D.; Guo, W.; Wu, Y.; Han, Z. Reduced graphene oxide and gold nanoparticle composite-based solid-phase extraction coupled with ultra-high-performance liquid chromatography-tandem mass spectrometry for the determination of 9 mycotoxins in milk. Food Chem. 2018, 264, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, R.; Ortega-Beltran, A.; Akande, A.; Mutegi, C.; Atehnkeng, J.; Kaptoge, L.; Senghor, A.L.; Adhikari, B.N.; Cotty, P.J. Biological control of aflatoxins in Africa: Current status and potential challenges in the face of climate changes. World Mycotoxin J. 2016, 9, 771–789. [Google Scholar] [CrossRef]
- Yang, J.; Wang, T.; Lin, G.; Li, M.; Zhu, R.; Yiannikouris, A.; Zhang, Y.; Mai, K. The assessment of diet contaminated with aflatoxin B1 in juvenile turbot (Scophthalmus maximus) and the evaluation of the efficacy of mitigation of a yeast cell wall extract. Toxins 2020, 12, 597. [Google Scholar] [CrossRef] [PubMed]
- Alshannaq, A.; Yu, J.H. Occurrence, toxicity, and analysis of major mycotoxins in food. Public Health 2017, 14, 632. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.H.; Phillips, T.D.; Jolly, P.E.; Stiles, J.K.; Jolly, C.M.; Aggarwal, D. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 2004, 80, 1106–1122. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y. Codex alimentarius commission and food safety. Shokuhinseigaku Zasshi J. Food Hyg. Soc. Jpn. 2002, 43, 249–253. [Google Scholar]
- Cupid, B.C.; Lightfoot, T.J.; Russell, D. The formation of AFBl-macromolecular adducts in rats and humans at dietary levels of exposure. Food Chem. Toxicol 2004, 42, 559–560. [Google Scholar] [CrossRef]
- Zhang, L.; Zhan, D.; Chen, Y. Aflatoxin B1 enhances pyroptosis of hepatocytes and activation of Kupffer cells to promote liver inflammatory injury via dephosphorylation of cyclooxygenase-2: An in vitro, ex vivo and in vivo study. Arch. Toxicol. 2019, 93, 3305–3320. [Google Scholar] [CrossRef]
- Hathout, A.S.; Aly, S.E. Biological detoxification of mycotoxins: A review. Ann. Microbiol. 2014, 64, 905–919. [Google Scholar] [CrossRef]
- Śliżewska, K.; Cukrowska, B.; Smulikowska, S.; Cielecka-Kuszyk, J. The effect of probiotic supplementation on performance and the histopathological changes in liver and kidneys in broiler chickens fed diets with aflatoxin B1. Toxins 2019, 11, 112. [Google Scholar] [CrossRef]
- Wu, F. Global impacts of aflatoxin in maize: Trade and human health. World Mycotoxin J. 2015, 8, 137–142. [Google Scholar] [CrossRef]
- Huang, B.; Chen, Q.; Wang, L.; Gao, X.; Zhu, W.; Mu, P.; Deng, Y. Aflatoxin B1 Induces Neurotoxicity through Reactive Oxygen Species Generation, DNA Damage, Apoptosis, and S-Phase Cell Cycle Arrest. Int. J. Mol. Sci. 2020, 21, 6517. [Google Scholar] [CrossRef] [PubMed]
- Akinrinmade, F.J.; Akinrinde, A.S. Changes in serum cytokine levels, hepatic and intestinal morphology in afatoxin B1-induced injury: Modulatory roles of melatonin and favonoid-rich fractions from Chromolena odorata. Mycotoxin Res. 2016, 32, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, R.; Sricastaba, A.K.; Rastogi, A.K. Long term effect of aflatoxin B1 on lipid peroxidation in rat liver and kidney: Effect of picroliv and silymarin. Phytother Res. 2001, 15, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Benkerroum, N. Aflatoxins: Producing-Molds, Structure, Health Issues and Incidence in Southeast Asian and Sub-Saharan African Countries. Int. J. Environ. Res. Public Health 2020, 17, 1215. [Google Scholar] [CrossRef] [PubMed]
- Riba, A.; Bouras, N.; Mokrane, S.; Mathieu, F.; Lebrihi, A.; Sabaou, N. Aspegillus section flavi and aflatoxins in Algerian wheat and derived products. Food Chem. Toxicol. 2010, 48, 2772–2777. [Google Scholar] [CrossRef]
- Plaz Torres, M.C.; Bodini, G.; Furnari, M.; Marabotto, E.; Zentilin, P.; Giannini, E.G. Nuts and Non-Alcoholic Fatty Liver Disease: Are Nuts Safe for Patients with Fatty Liver Disease? Nutrients 2020, 12, 3363. [Google Scholar] [CrossRef]
- Zuo, R.Y.; Chang, J.; Yin, Q.Q.; Wang, P.; Yang, Y.R.; Wang, X.; Wang, G.Q.; Zheng, Q.H. Effect of the combined probiotics with aflatoxin B1-degrading enzyme on aflatoxin detoxification, broiler production performance and hepatic enzyme gene expression. Food Chem. Toxicol. 2013, 59, 470–475. [Google Scholar] [CrossRef]
- Fang, L.Q.; Chen, H.; Ying, Y.; Jin-Ming, L. Micro–plate chemiluminescence enzyme immunoassay for aflatoxin B1 in agricultural products. Talanta 2011, 84, 216–222. [Google Scholar] [CrossRef]
- Cherkani-Hassani, A.; Ghanname, I.; Zinedine, A.; Sefrioui, H.; Qmichou, Z.; Mouane, N. Aflatoxin M1 prevalence in breast milk in Morocco: Associated factors and health risk assessment of newborns "CONTAMILK study". Toxicon 2020, 187, 203–208. [Google Scholar] [CrossRef]
- Fandohan, P.; Gnonlonfin, B.; Hell, K.; Marasas, W.F.; Wingfield, M.J. Natural occurrence of Fusarium and subsequent fumonisin contamination in preharvest and stored maize in Benin, West Africa. Int. J. Food Microbiol. 2005, 99, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Namulawa, V.T.; Mutiga, S.; Musimbi, F.; Akello, S.; Ngángá, F.; Kago, L.; Kyallo, M.; Harvey, J.; Ghimire, S. Assessment of fungal contamination in fish feed from the Lake Victoria Basin, Uganda. Toxins 2020, 12, 233. [Google Scholar] [CrossRef] [PubMed]
- Omara, T.; Nassazi, W.; Omute, T.; Awath, A.; Laker, F.; Kalukusu, R.; Musau, B.; Nakabuye, B.V.; Kagoya, S.; Otim, G.; et al. Aflatoxins in Uganda: An Encyclopedic Review of the Etiology, Epidemiology, Detection, Quantification, Exposure Assessment, Reduction, and Control. Int. J. Microbiol. 2020, 2020, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Tsafack Takadong, J.J.; Mouafo, H.T.; Manet, L.; Baomog, A.M.B.; Adjele, J.J.B.; Medjo, E.K.; Medoua, G.N. Assessment of the Presence of Total Aflatoxins and Aflatoxin B1 in Fish Farmed in Two Cameroonian Localities. Int. J. Food Sci. 2020, 2020, 1–6. [Google Scholar] [CrossRef]
- Dada, T.A.; Ekwomadu, T.I.; Mwanza, M. Multi mycotoxin determination in dried beef using liquid chromatography coupled with triple quadrupole mass spectrometry (LC-MS/MS). Toxins 2020, 12, 357. [Google Scholar] [CrossRef]
- Carvajal-Moreno, M.; Vargas-Ortiz, M.; Hernández-Camarillo, E.; Ruiz-Velasco, S.; Rojo-Callejas, F. Presence of unreported carcinogens, Aflatoxins and their hydroxylated metabolites, in industrialized Oaxaca cheese from Mexico City. Food Chem. Toxicol. 2019, 124, 128–138. [Google Scholar] [CrossRef]
- Norlia, M.; Jinap, S.; Nor-Khaizura, M.A.R.; Radu, S.; Samsudin, N.I.P.; Azri, F.A. Aspergillus section Flavi and Aflatoxins: Occurrence, Detection, and Identification in Raw Peanuts and Peanut-Based Products Along the Supply Chain. Front. Microbiol. 2019, 10, 2602. [Google Scholar] [CrossRef]
- Jayaratne, W.M.S.C.; Abeyratne, A.H.M.A.K.; De Zoysa, H.K.S.; Dissanayake, D.M.R.B.N.; Bamunuarachchige, T.C.; Waisundara, V.Y.; Chang, S. Detection and quantification of Aflatoxin B1 in corn and corn-grown soils in the district of Anuradhapura, Sri Lanka. Heliyon 2020, 6, e05319. [Google Scholar] [CrossRef]
- Wenndt, A.; Sudini, H.K.; Pingali, P.; Nelson, R. Exploring aflatoxin contamination and household-level exposure risk in diverse Indian food systems. PLoS ONE 2020, 15, e0240565. [Google Scholar] [CrossRef]
- Pongpraket, M.; Poapolathep, A.; Wongpanit, K.; Phanwimol, T.; Poapolathep, S. Exposure assessment of multiple mycotoxins in black and white sesame seeds consumed in Thailand. Food Prot. 2020, 83, 1198–1207. [Google Scholar] [CrossRef]
- Karlovsky, P.; Suman, M.; Berthiller, F.; De Meester, J.; Eisenbrand, G.; Perrin, I.; Oswald, I.P.; Speijers, G.; Chiodini, A.; Recker, T.; et al. Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res. 2016, 32, 179–205. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tang, L.; Wang, J.; Wang, J.S. Aflatoxin B1 disrupts gut-microbial metabolisms of short-chain fatty acids, long-chain fatty acids, and bile acids in male F344 rats. Toxicol. Sci. 2018, 164, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Jaćević, V.; Dumanović, J.; Lazarević, M.; Nepovimova, E.; Resanović, R.; Milovanović, Z.; Wu, Q.; Kuča, K. Antidotal Potency of the Novel, Structurally Different Adsorbents in Rats Acutely Intoxicated with the T-2 Toxin. Toxins (Basel) 2020, 12, 643. [Google Scholar] [CrossRef] [PubMed]
- Battilani, P. Food mycology-a multifaceted approach to fungi and food. World Mycotoxin J. 2008, 1, 223–224. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, P.P.; Gupta, V.; Prakash, B. Assessing the antifungal and aflatoxin B1 inhibitory efficacy of nanoencapsulated antifungal formulation based on combination of Ocimum spp. essential oils. Int. J. Food Microbiol. 2020, 330, 108766. [Google Scholar] [CrossRef]
- Ji, J.; Xie, W. Removal of aflatoxin B1 from contaminated peanut oils using magnetic attapulgite. Food Chem. 2021, 339, 128072. [Google Scholar] [CrossRef]
- Čolović, R.; Puvača, N.; Cheli, F.; Avantaggiato, G.; Greco, D.; Đuragić, O.; Kos, J.; Pinotti, L. Decontamination of Mycotoxin-Contaminated Feedstuffs and Compound Feed. Toxins (Basel) 2019, 11, 617. [Google Scholar] [CrossRef]
- Yu, Y.; Shi, J.; Xie, B.; He, Y.; Qin, Y.; Wang, D.; Shi, H.; Ke, Y.; Sun, Q. Detoxification of aflatoxin B1 in corn by chlorine dioxide gas. Food Chem. 2020, 328, 127121. [Google Scholar] [CrossRef]
- Conte, G.; Fontanell, i.M.; Galli, F.; Cotrozzi, L.; Pagni, L.; Pellegrini, E. Mycotoxins in Feed and Food and the Role of Ozone in Their Detoxification and Degradation: An Update. Toxins (Basel) 2020, 12, 486. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Gbashi, S.; Nwinyi, O.C.; Mavumengwana, V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 2017, 57, 3208–3217. [Google Scholar] [CrossRef]
- Zychowski, K.E.; Hoffmann, A.R.; Ly, H.J.; Pohlenz, C.; Buentello, A.; Romoser, A.; Gatlin, D.M.; Phillips, T.D. The effect of aflatoxin-B1 on red drum (Sciaenops ocellatus) and assessment of dietary supplementation of NovaSil for the prevention of aflatoxicosis. Toxins (Basel) 2013, 5, 1555–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Chen, Y.; Kong, Q.; Ma, Y.; Liu, Y. Detoxification of Aflatoxin B1 by Zygosaccharomyces rouxii with Solid State Fermentation in Peanut Meal. Toxins (Basel) 2017, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Markowiak, P.; Śliżewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Shokrzadeh, M.; Raeisi, S.N.; Ghorbani-HasanSaraei, A.; Nasiraii, L.R. Aflatoxins biodetoxification strategies based on probiotic bacteria. Toxicon 2020, 178, 50–58. [Google Scholar] [CrossRef]
- Asurmendi, P.; Gerbaldo, G.; Pascual, L.; Barberis, L. Lactic acid bacteria with promising AFB1 binding properties as an alternative strategy to mitigate contamination on brewers’ grains. J. Environ. Sci. Health B 2020, 20, 1–7. [Google Scholar]
- Ren, X.; Zhang, Q.; Zhang, W.; Mao, J.; Li, P. Control of Aflatoxigenic Molds by Antagonistic Microorganisms: Inhibitory Behaviors, Bioactive Compounds, Related Mechanisms, and Influencing Factors. Toxins (Basel) 2020, 12, 24. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Loots, D.T.; Babalola, O.O. Bacillus velezensis: Phylogeny, useful applications, and avenues for exploitation. Appl. Microbiol. Biotechnol. 2019, 103, 3669–3682. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.Q.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 degradation by Bacillus subtilis, UTBSP1 isolated from pistachio nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.Q.; Ahmadzadeh, M.; Hu, L.B.; Ghassempour, A. Inhibition of the Aspergillus flavus growth and aflatoxin B1 contamination on pistachio nut by fengycin and surfactin-producing Bacillus subtilis UTBSP1. Plant. Pathol. J. 2016, 32, 209–215. [Google Scholar] [CrossRef]
- Gao, X.; Ma, Q.; Zhao, L. Isolation of Bacillus subtilis: Screening for aflatoxins B1, M and G1, detoxification. Eur. Food Res. Technol. 2011, 232, 957–962. [Google Scholar] [CrossRef]
- Fan, Y.; Zhao, L.; Ma, Q. Effects of Bacillus subtilis ANSB060 on growth performance, meat quality and aflatoxin residues in broilers fed moldy peanut meal naturally contaminated with aflatoxins. Food Chem. Toxicol. 2013, 59, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Sun, J.; Cui, Y.; Wang, X.; Sang, Y. Biological degradation of aflatoxin M1 by Bacillus pumilus E-1-1-1. Microbiologyopen 2019, 8, 663. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.R.; Vipin, A.V.; Hariprasad, P.; Appaiah, K.A.; Venkateswaran, G. Biological detoxification of aflatoxin b1 by bacillus licheniformis cfr1. Food Control. 2016, 71, 234–241. [Google Scholar]
- Adebo, O.A.; Njobeh, P.B.; Mavumengwana, V. Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control. 2016, 68, 92–96. [Google Scholar] [CrossRef]
- Jebali, R.; Abbèss, S.; Salah-Abbès, J.B.; Younes, R.B.; Haous, Z.; Oueslati, R. Ability of Lactobacillus plantarum Mon03 to mitigate aflatoxins (B1 and M1) immunotoxicities in mice. J. Immunot. 2015, 12, 290–299. [Google Scholar] [CrossRef]
- Huang, L.; Duan, C.; Zhao, Y.; Gao, L.; Niu, C.; Xu, J.; Li, S. Reduction of aflatoxin B1 toxicity by Lactobacillus plantarum C88: A potential probiotic strain isolated from Chinese traditional fermented food “tofu”. PLoS ONE 2017, 12, e0170109. [Google Scholar] [CrossRef]
- Lahtinen, S.J.; Haskard, C.A.; Ouwehand, A.C.; Salminen, S.J.; Ahokas, J.T. Binding of aflatoxin B1 to cell wall components of Lactobacillus rhamnosus strain GG. Food Addit. Contam. 2004, 21, 158–164. [Google Scholar] [CrossRef]
- Rahaie, S.; Emam-Djomeh, Z.; Razavi, S.H. Evaluation of aflatoxin decontaminating by two strains of Saccharomyces cerevisiae and Lactobacillus rhamnosus strain GG in pistachio nuts. Int. J. Food Sci. Tech. 2012, 47, 1647–1653. [Google Scholar] [CrossRef]
- Chen, Y.; Kong, Q.; Chi, C.; Shan, S.; Guan, B. Biotransformation of aflatoxin B1 and aflatoxin G1 in peanut meal by anaerobic solid fermentation of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus. Int. J. Food Microbiol. 2015, 211, 1–5. [Google Scholar] [CrossRef]
- Kumara, S.S.; Gayathri, D.; Hariprasad, P.; Venkateswaran, G.; Swamy, C.T. In vivo AFB1 detoxification by Lactobacillus fermentum LC5-a with chlorophyll and immunopotentiating activity in albino mice. Toxicon 2020, 187, 214–222. [Google Scholar] [CrossRef]
- Topcu, A.; Bulat, T.; Wishah, R.; Boyacı, I.H. Detoxification of aflatoxin B1 and patulin by Enterococcus faecium strains. Int. J. Food Microbiol. 2010, 139, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, J.; Liu, Z.; Yutao, S.; Jinqiu, L.; Xiaofan, H.; Peiqiang, M.; Fengru, D.; Yiqun, D. Aflatoxin B1 degradation and detoxification by Escherichia coli CG1061 isolated from chicken cecum. Front. Pharmacol. 2019, 9, 1548. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, J.; Jin, Y.; Wu, C.; Shen, D.; Zhang, S.; Zhou, R. Aflatoxin B1 degradation by salt tolerant Tetragenococcus halophilus CGMCC 3792. Food Chem. Toxicol. 2018, 121, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Sangare, L.; Zhao, Y.; Folly, Y.M.E.; Chang, J.; Li, J.; Selvaraj, J.N.; Xing, F.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B1 degradation by a Pseudomonas strain. Toxins 2014, 6, 3028–3040. [Google Scholar] [CrossRef]
- Singh, J.; Mehta, A. Protein-mediated degradation of aflatoxin B1 by Pseudomonas putida. Braz. J. Microbiol. 2019, 50, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.S.; Sivaramakrishna, A.; Alka, M. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeter. Biodegr. 2014, 86, 202–209. [Google Scholar] [CrossRef]
- Mengyu, C.; Yingying, Q.; Chen, N.; Tiejun, L.; Jingjing, W.; Hong, J.; Xu, W.; Kezong, Q.; Yu, Z. Detoxification of aflatoxin B1 by Stenotrophomonas sp. CW117 and characterization the thermophilic degradation process. Environ. Pollut. 2020, 6, 114178. [Google Scholar]
- Yang, X.; Chen, X.; Song, Z. Antifungal, plant growth-promoting, and mycotoxin detoxication activities of Burkholderia sp. strain XHY-12. 3 Biotech 2020, 10, 158. [Google Scholar] [CrossRef]
- Wu, Q.; Jezkova, A.; Yuan, Z.; Pavlikova, L.; Dohnal, V.; Kuca, K. Biological degradation of aflatoxins. Drug Metab. Rev. 2009, 41, 1–7. [Google Scholar] [CrossRef]
- Hormisch, D.; Brost, I.; Kohring, G.W.; Giffhorn, F.; Kroppenstedt, R.M.; Stackebrandt, E.; Färber, P.; Holzapfel, W.H. Mycobacterium fluoranthenivorans sp. nov. a fluoranthene and aflatoxin B1 degrading bacterium from contaminated soil of a former coal gas plant. Syst. Appl. Microbiol. 2004, 27, 653–660. [Google Scholar] [CrossRef]
- Ibrahim, S.; Abdul, K.K.; Zahr, i.K.N.M.; Gomez-Fuentes, C.; Convey, P.; Zulkharnain, A.; Sabri, S.; Alias, S.A.; González-Rocha, G.; Ahmad, S.A. Biosurfactant production and growth kinetics studies of the waste canola oil-degrading bacterium Rhodococcuserythropolis AQ5-07 from Antarctica. Molecules 2020, 25, 3878. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Castaneda, Z.I.; Avila-Gonzalez, E.; Casaubon-Huguenin, M.T.; Cervantes-Olivares, R.A.; Vasquez-Pelaez, C.; Hernandez-Baumgarten, E.M.; Moreno-Martinez, E. Bio-detoxification of aflatoxin-contaminated chick feed. Poult. Sci. 2008, 87, 1569–1576. [Google Scholar] [CrossRef]
- Caceres, I.; Snini, S.P.; Puel, O.; Mathieu, F. Streptomyces roseolus, a promising biocontrol agent against Aspergillus flavus, the main aflatoxin B1 producer. Toxins 2018, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Peltonen, K.; El-Nezami, H.; Haskard, C.; Ahokas, J.; Salminen, S. Aflatoxin B1 binding by dairy strains of lactic acid bacteria and bifidobacteria. J. Dairy Sci. 2001, 84, 2152–2156. [Google Scholar] [CrossRef]
- Smiley, R.D.; Draughon, F.A. Preliminary evidence that degradation of aflatoxin B1 by Flavobacterium aurantiacum is enzymatic. J. Food Protect. 2020, 63, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Taheur, F.B.; Fedhila, K.; Chaieb, K.; Kouidhi, B.; Bakhrouf, A.; Abrunhosa, L. Adsorption of aflatoxin B1, zearalenone and ochratoxin A by microorganisms isolated from Kefir grains. Int. J. Food Microbiol. 2017, 251, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Xing, M.; Fei, X.; Zhang, J.H.; Tian, S.L.; Li, M.H.; Liu, S.D. Identification of a novel PSR as the substrate of an SR protein kinase in the true slime mold. J. Biochem. 2011, 149, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.H.; Guan, S.; Gao, X.; Ma, Q.G.; Lei, Y.P.; Bai, X.M.; Ji, C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2010, 110, 147–155. [Google Scholar] [CrossRef]
- Wochner, K.F.; Becker-Algeri, T.A.; Colla, E. The action of probiotic microorganisms on chemical contaminants in milk. Crit Rev. Microbiol. 2018, 44, 112–123. [Google Scholar] [CrossRef]
- Zhang, W.; Xue, B.; Li, M.; Mu, Y.; Chen, Z.; Li, J.; Shan, A. Screening a strain of Aspergillus niger and optimization of fermentation conditions for degradation of aflatoxin B1. Toxins 2014, 6, 3157–3172. [Google Scholar] [CrossRef]
- Li, J.; Huang, J.; Jin, Y.; Wu, C.; Shen, D.; Zhang, S.; Zhou, R. Mechanism and kinetics of degrading aflatoxin B1 by salt tolerant Candida versatilis CGMCC 3790. J. Hazard. Mater. 2018, 359, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Kusumaningtyas, E.; Widiastuti, R.; Maryam, R. Reduction of aflatoxin B1 in chicken feed by using Saccharomyces cerevisiae, Rhizopus oligosporus, and their combination. Mycopathologia 2006, 162, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Chlebicz, A.; Śliżewska, K. In vitro detoxification of aflatoxin B1, deoxynivalenol, fumonisins, T-2 toxin and zearalenone by probiotic bacteria from genus Lactobacillus and Saccharomyces cerevisiae yeast. Probiotics Antimicro. Prot. 2020, 12, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Suresh, G.; Cabezudo, I.; Pulicharla, R.; Cuprys, A.; Rouissi, T.; Brar, S.K. Biodegradation of aflatoxin B1 with cell-free extracts of Trametes versicolor and Bacillus subtilis. Res. Vet. Sci. 2020, 133, 85–91. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, R.; Ng, T.B.; Lai, Y.; Yang, J.; Ye, X. A New Laccase of Lac 2 from the White Rot Fungus Cerrena unicolor 6884 and Lac 2-Mediated Degradation of Aflatoxin B1. Toxins (Basel) 2020, 12, 476. [Google Scholar] [CrossRef]
- Von Hertwig, A.M.; Iamanaka, B.T.; Amorim Neto, D.P.; Rezende, J.B.; Martins, L.M.; Taniwaki, M.H.; Nascimento, M.S. Interaction of Aspergillus flavus and A. parasiticus with Salmonella spp. isolated from peanuts. Int. J. Food Microbiol. 2020, 328, 108666. [Google Scholar] [CrossRef]
- Fouché, T.; Claassens, S.; Maboeta, M. Aflatoxins in the soil ecosystem: An overview of its occurrence, fate, effects and future perspectives. Mycotoxin Res. 2020, 36, 303–309. [Google Scholar] [CrossRef]
- Serrano, R.; González-Menéndez, V.; Rodríguez, L.; Martín, J.; Tormo, J.R.; Genilloud, O. Co-culturing of fungal strains against Botrytis cinerea as a model for the induction of chemical diversity and therapeutic agents. Front. Microbiol. 2017, 8, 649. [Google Scholar] [CrossRef]
- Sarrocco, S.; Mauro, A.; Battilani, P. Use of Competitive Filamentous Fungi as an Alternative Approach for Mycotoxin Risk Reduction in Staple Cereals: State of Art and Future Perspectives. Toxins (Basel) 2019, 11, 701. [Google Scholar] [CrossRef]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Yang, K.; Geng, Q.; Song, F.; He, X.; Hu, T.; Wang, S.; Tian, J. Transcriptome Sequencing Revealed an Inhibitory Mechanism of Aspergillus flavus Asexual Development and Aflatoxin Metabolism by Soy-Fermenting Non-Aflatoxigenic Aspergillus. Int. J. Mol. Sci. 2020, 21, 6994. [Google Scholar] [CrossRef] [PubMed]
- Haskard, C.A.; El-Nezami, H.S.; Kankaanpaa, P.E.; Seppo, S.; Jorma, T.A. Surface binding of aflatoxin B1 by lactic acid bacteria. Appl. Environ. Microb. 2001, 67, 3086–3091. [Google Scholar]
- Kolosova, A.; Stroka, J. Substances for reduction of the contamination of feed by mycotoxins: A review. World Mycotoxin J. 2011, 4, 225–256. [Google Scholar] [CrossRef]
- Kabak, B.; Dobson, A.D.; Var, I.I.L. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Crit. Rev. Food Sci. Nutr. 2006, 46, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Deepak, M.B.; Jhanvi, S.P. Aflatoxin binding and detoxification by non-saccharomyces yeast a new vista for decontamination. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 310–317. [Google Scholar]
- Kim, J.A.; Bayo, J.; Cha, J.; Choi, Y.J.; Jung, M.Y.; Kim, D.H.; Kim, Y. Investigating the probiotic characteristics of four microbial strains with potential application in feed industry. PLoS ONE 2019, 14, e0218922. [Google Scholar] [CrossRef]
- Elghandour, M.M.Y.; Tan, Z.L.; Abu Hafsa, S.H.; Adegbeye, M.J.; Greiner, R.; Ugbogu, E.A.; Cedillo Monroy, J.; Salem, A.Z.M. Saccharomyces cerevisiae as a probiotic feed additive to non and pseudo-ruminant feeding: A review. J. Appl. Microbiol. 2020, 128, 658–674. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Ligas, B.; Skrzypczak, D.; Mikula, K.; Izydorczyk, G.; Witek-Krowiak, A.; Moustakas, K.; Chojnacka, K. Biosorption as a method of biowaste valorization to feed additives: RSM optimization. Environ. Pollut. 2021, 268, 115937. [Google Scholar] [CrossRef]
- Shetty, P.H.; Hald, B.; Jespersen, L. Surface binding of aflatoxin B1 by Saccharomyces cerevisiae strains with potential decontaminating abilities in indigenous fermented foods. Int. J. Food Microbiol. 2007, 113, 41–46. [Google Scholar] [CrossRef]
- Samuel, M.S.; Aiko, V.; Panda, P. Aflatoxin B1 occurrence, biosynthesis and its degradation. J. Pure Appl. Microbiol. 2013, 7, 1–7. [Google Scholar]
- Theumer, M.G.; Henneb, Y.; Khoury, L.; Snini, S.P.; Tadrist, S.; Canlet, C.; Audebert, M. Genotoxicity of aflatoxins and their precursors in human cells. Toxicol. Lett. 2018, 287, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Zuki-Orozco, B.A.; Batres-Esquivel, L.E.; Ortiz-Pérez, M.D.; Juárez-Flores, B.I.; Díaz-Barriga, F. Aflatoxins contamination in maize products from rural communities in San Luis Potosi, Mexico. Ann. Glob. Health 2018, 84, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, Y.; Lv, Y.; Cheng, W.; Guo, P.; Cui, Z. The research process of aflatoxins biodegradation. Agr. Sci. Hubei 2016, 55, 5172–5176. [Google Scholar]
- Cao, H.; Liu, D.; Mo, X.; Xie, C.H.; Yao, D. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011, 166, 474–483. [Google Scholar] [CrossRef]
- Xu, T.; Xie, C.; Yao, D.; Zhou, C.Z.; Liu, J. Crystal structures of aflatoxin-oxidase from armillariella tabescens reveal a dual activity enzyme. Biochem. Biophys. Res. Commun. 2017, 494, 621–625. [Google Scholar] [CrossRef]
- Alberts, J.F.; Gelderblom, W.C.A.; Botha, A.; Van Zyl, W.H. Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 2009, 135, 47–52. [Google Scholar] [CrossRef]
- Zaid, A.M.A. Biodegradation of aflatoxin by peroxidase enzyme produced by local isolate of Pseudomonas sp. Int. J. Sci. Res. Manag. 2017, 5, 7456–7467. [Google Scholar]
- Li, C.H.; Li, W.Y.; Hsu, I.N.; Liao, Y.Y.; Yang, C.Y.; Taylor, M.C.; Liu, Y.F.; Huang, W.H.; Chang, H.H.; Huang, H.L.; et al. Recombinant aflatoxin-degrading F420H2-dependent reductase from mycobacterium smegmatis protects mammalian cells from aflatoxin toxicity. Toxins 2019, 11, 259. [Google Scholar] [CrossRef]
- Karim, G.; Kamkar, A. A study on the effect of lactoperoxidase system (LPS) and LPS plus riboflavin on the aflatoxin M1 in milk. Journal of the Faculty of Veterinary Medicine. Univ. Tehran 2020, 55, 5–7. [Google Scholar]
- Yehia, R.S. Aflatoxin detoxification by manganese peroxidase purified from Pleurotus ostreatus. Braz. J. Microbiol. 2014, 45, 127–134. [Google Scholar] [CrossRef]
- Guan, S.; Zhao, L.; Ma, Q.; Zhou, T.; Wang, N.; Hu, X.; Ji, C. In vitro efficacy of myxococcus fulvus ANSM068 to biotransform aflatoxin B1. Int. J. Mol. Sci. 2010, 11, 4063–4079. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.Z.; Sun, Y.P.; Cai, J.S. The aflatoxin-detoxifizyme specific expression in mouse parotid gland. Transgenic Res. 2015, 24, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Z.; Lu, F.P.; Jiang, H.L.; Tan, C.P.; Yao, D.S.; Xie, C.F.; Liu, D.L. The furofuran-ring selectivity, hydrogen peroxide-production and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 2015, 76, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bai, Y.; Huang, H.; Tu, T.; Wang, Y.; Wang, Y.; Luo, H.; Yao, B.; Su, X. Degradation of aflatoxin B1 and zearalenone by bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators. Toxins (Basel) 2019, 11, 609. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mao, H.; Hu, C.; Tron, T.; Lin, J.; Wang, J.; Sun, B. Molecular docking studies and in vitrodegradation of four aflatoxins (AFB1, AFB2, AFG1, and AFG2) by a recombinant laccase from Saccharomyces cerevisiae. J. Food Sci. 2020, 85, 1353–1360. [Google Scholar] [CrossRef]
- Tomin, M.; Tomić, S. Oxidase or peptidase? A computational insight into a putative aflatoxin oxidase from Armillariella tabescens. Proteins 2019, 87, 390–400. [Google Scholar] [CrossRef]
- Tian, F.; Lee, S.Y.; Woo, S.Y.; Chun, H.S. Alternative Oxidase: A Potential Target for Controlling Aflatoxin Contamination and Propagation of Aspergillus flavus. Front. Microbiol. 2020, 11, 419. [Google Scholar] [CrossRef]
- Li, Q.; Bai, Z.; O-Donnell, A.; Harvey, L.M.; Hoskisson, P.A.; McNeil, B. Oxidative stress in fungal fermentation processes: The roles of alternative respiration. Biotechnol. Lett. 2011, 33, 457–467. [Google Scholar] [CrossRef]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J.; Briggs, L.J.; Lapalikar, G.V.; Campbell, P.M.; Scott, C.; et al. Identification and characterization of two families of F420H2-dependent reductases from mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef]
- Guo, Y.; Qin, X.; Tang, Y.; Ma, Q.; Zhang, J.; Zhao, L. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020, 325, 126877. [Google Scholar] [CrossRef]
- Eaton, D.L.; Gallagher, E.P. Mechanisms of aflatoxin carcinogenesis. Ann. Rev. Pharmacol. Toxicol. 1994, 34, 135–172. [Google Scholar] [CrossRef] [PubMed]
- Massey, T.E.; Stewart, R.K.; Daniels, J.M.; Liu, L. Biochemical and molecular aspects of mammalian susceptibility to aflatoxin B1 carcinogenicity. Proc. Soc. Exp. Biol. Med. 1995, 208, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Ueng, Y.F.; Shimada, T.; Yamazaki, H.; Guengerich, F.P. Oxidation of aflatoxin B1 by bacterial recombinant human cytochrome P450 enzymes. Chem. Res. Toxicol. 1995, 8, 218–225. [Google Scholar] [CrossRef]
- Van Vleet, T.R.; Klein, P.J.; Coulombe, R.A. Metabolism of aflatoxin B1 by normal human bronchial epithelial cells. J. Toxicol. Environ. Health A 2001, 63, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Wang, Y.; Zhou, Q.; Li, M.; Hu, H.; Ma, Y.; Chen, X.; Ni, J.; Zhao, W.; Huang, S.; et al. Biological degradation of aflatoxin B1 by cell-free extracts of Bacillus velezensis DY3108 with broad PH stability and excellent thermostability. Toxins 2018, 10, 330. [Google Scholar] [CrossRef]
- Alberts, J.F.; Engelbrecht, Y.; Steyn, P.S. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [CrossRef]
- Cserhárti, M.; Kriszt, B.; Krifaton, C.; Szoboszlay, S.; Háhn, I.; Tóth, S.; Kukolya, J. Mycotoxin degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 2013, 16, 176–185. [Google Scholar] [CrossRef]
- Peña-Rodas, O.; Martinez-Lopez, R.; Hernandez-Rauda, R. Occurrence of Aflatoxin M1 in cow milk in El Salvador: Results from a two-year survey. Toxicol. Rep. 2018, 5, 671–678. [Google Scholar] [CrossRef]
- Wang, N.; Wu, W.; Pan, J.; Long, M. Detoxification Strategies for Zearalenone Using Microorganisms: A Review. Microorganisms 2019, 7, 208. [Google Scholar] [CrossRef]
- Martínez, M.P.; Magnoli, A.P.; González Pereyra, M.L.; Cavaglieri, L. Probiotic bacteria and yeasts adsorb aflatoxin M1 in milk and degrade it to less toxic AFM1-metabolites. Toxicon 2019, 172, 1–7. [Google Scholar] [CrossRef]
- Barati, M.; Chamani, M.; Mousavi, S.N. Effects of biological and mineral compounds in aflatoxin-contaminated diets on blood parameters and immune response of broiler chickens. J. Appl. Anim. Res. 2018, 46, 707–713. [Google Scholar] [CrossRef]
- Bovo, F.; Corassin, C.H.; Rosim, R.E.; Oliveira, C.A.F. Efficiency of lactic acid bacteria strains for decontamination of aflatoxin M1 in phosphate buffer saline solution and in skimmed milk. Food Bioprocess. Tech. 2009, 6, 2230–2234. [Google Scholar] [CrossRef]
- Ismail, A.; Riaz, M.; Akhtar, S.; Yoo, S.H.; Park, S.; Abid, M.; Ahmad, Z. Seasonal variation of aflatoxin B1 content in dairy feed. J. Anim. Feed Sci. 2017, 26, 33–37. [Google Scholar] [CrossRef]
- Hamad, G.M.; Zahran, E.; Hafez, E.E. The efficacy of bacteria and yeast strain and their combination to bind aflatoxin B1 and B2 in artificially contaminated infants food. J. Food Saf. 2017, 37, e12365. [Google Scholar] [CrossRef]
- Huang, W.; Chang, J.; Wang, P. Effect of the combined compound probiotics with mycotoxin-degradation enzyme on detoxifying aflatoxin B1 and zearalenone. J. Toxicol. Sci. 2018, 43, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Watanakij, N.; Visessanguan, W.; Petchkongkaew, A. Aflatoxin B1-degrading activity from Bacillus subtilis BCC 42005 isolated from fermented cereal products. Food Addit. Contam. A 2020, 37, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Boukaew, S.; Prasertsan, P. Efficacy of volatile compounds from Streptomyces philanthi RL-1-178 as a biofumigant for controlling growth and aflatoxin production of the two aflatoxin-producing fungi on stored soybean seeds. J. Appl. Microbiol. 2020, 129, 652–664. [Google Scholar] [CrossRef]
- Chen, Y.; Li, R.; Chang, Q.; Dong, Z.; Yang, H.; Xu, C. Lactobacillus bulgaricus or Lactobacillus rhamnosus Suppresses NF-κB Signaling Pathway and Protects against AFB₁-Induced Hepatitis: A Novel Potential Preventive Strategy for Aflatoxicosis? Toxins (Basel) 2019, 11, 17. [Google Scholar] [CrossRef]
- Khanian, M.; Karimi-Torshizi, M.A.; Allameh, A. Alleviation of aflatoxin-related oxidative damage to liver and improvement of growth performance in broiler chickens consumed Lactobacillus plantarum 299v for entire growth period. Toxicon 2019, 158, 57–62. [Google Scholar] [CrossRef]
- Rashidi, N.; Khatibjoo, A.; Taherpour, K.; Akbari-Gharaei, M.; Shirzadi, H. Effects of licorice extract, probiotic, toxin binder and poultry litter biochar on performance, immune function, blood indices and liver histopathology of broilers exposed to aflatoxin-B1. Poult. Sci. 2020, 99, 5896–5906. [Google Scholar] [CrossRef]
- Muhialdin, B.J.; Saari, N.; Meor Hussin, A.S. Review on the Biological Detoxification of Mycotoxins Using Lactic Acid Bacteria to Enhance the Sustainability of Foods Supply. Molecules 2020, 7, 2655. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).