Hitchhiking with Nature: Snake Venom Peptides to Fight Cancer and Superbugs
Abstract
Abstract
Key Contribution
1. Introduction
2. Antibacterial and Antitumoral Activity of Snake Venoms (SVs)
2.1. Snake Venom-Derived Antimicrobial and Anticancer Peptides (SV-AMPs and ACPs)
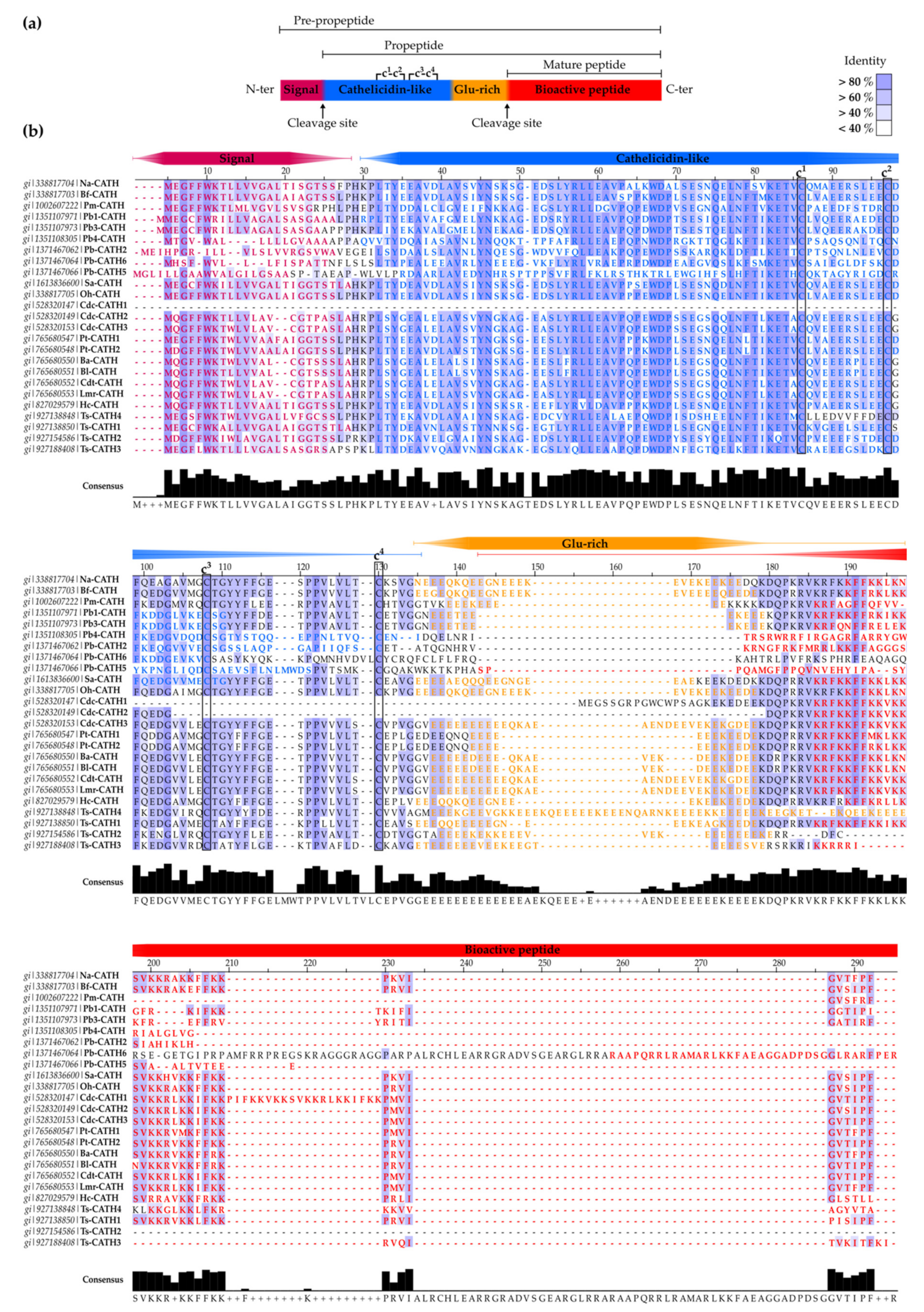
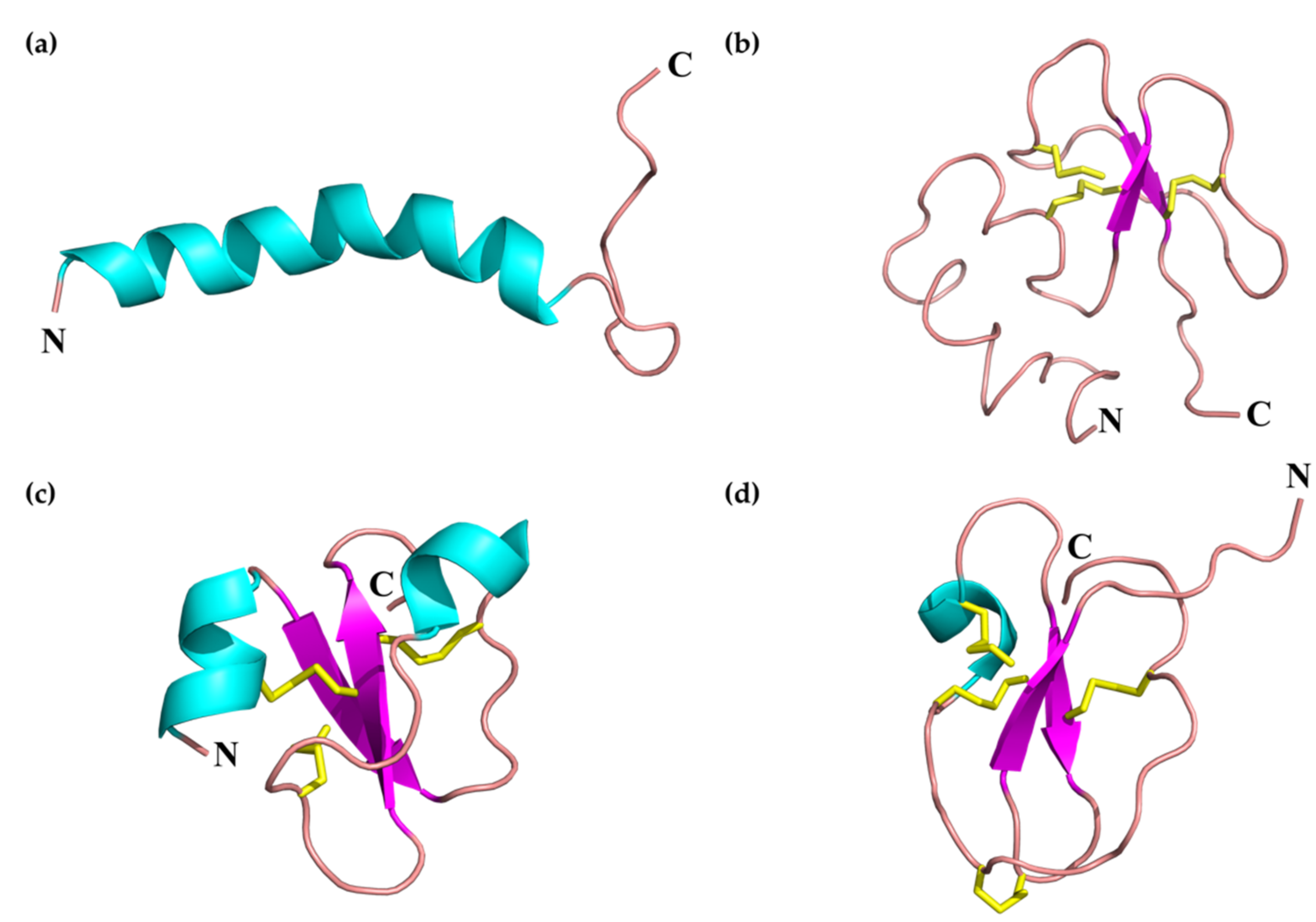
2.1.1. SV-Cathelicidins (SV-CATHs)
- Oh-CATH: identified in the venom gland of the king cobra (O. hannah), this was the first predicted SV-CATH to be synthesized and experimentally validated as an AMP [76] (Table 2). Oh-CATH exerts strong salt-resistant, antibacterial activity against Gram-positive and Gram-negative bacteria (MICs in the 1–20 µg/mL range) with weak hemolysis (~10% hemolysis observed at 200 µg/mL) [76,79]. It is apparently membrane-active and is an inhibitor of ATP-synthase [80,81]. A collection of analogs designed by Zhang et al., to study the structure-function relationships of Oh-CATH, suggested that the four N-terminal aa residues are responsible for cytotoxicity toward eukaryotic cells, while the C-terminal 10 strongly influence antimicrobial activity [79]. Accordingly, OH-CATH30, the most promising analog, lacking the four N-terminal aa, also named OH-CATH(5-34), was further characterized and optimized. Then, OH-CATH30 was tested against a panel of 584 clinical isolates of 14 different species, showing antibacterial activity against 85% of them and overall higher efficacy against Gram-positive strains [82].
- Na-CATH: this CATH from the venom gland of the Chinese cobra N. atra [76] also demonstrated powerful, salt-resistant, antimicrobial activity against Gram-positive and Gram-negative bacteria, including Francisella novicida (the non-virulent strain in humans related to Francisella tularensis, the causative agent of tularemia) [98], E.coli, Aggregatibacter actinomycetemcomitans, Bacillus cereus [99], P. aeruginosa [100], and S. aureus [101] at low concentrations (EC50 < 3 µg/mL). Na-CATH is also active against Mycobacterium smegmatis [102], Burkholderia thailandensis (closely related to B. pseudomallei, the causative agent of melioidosis) [103] and Bacillus anthracis (anthrax) [104]. The last two strains are of particular relevance due to their potential use as biological weapons. Indeed, in vivo studies using wax moth larvae demonstrated that Na-CATH was able to rescue 100% of waxworms after B. anthracis Sterne infection at low peptide concentrations [104]. In addition, Na-CATH is not only active against planktonic bacteria, but also inhibits S. aureus and B. thailandensis biofilm formation [101,102], while inducing minimal hemolysis (<2% at 100 µg/mL) [99]. However, Na-CATH did not inhibit Pseudomonas biofilm formation [100].
- Bf-CATH: unlike other 34-aa AMPs predicted as SV-CATHs, the purified peptide from Bungarus fasciatus venom gland is a 30-aa peptide lacking the four N-terminal residues (Table 2). This difference in length suggests different enzymatic processing rather than the proposed elastase-like protease cleavage at the conserved site (Val) or post-processing of the 34-residue precursor [97]. Expression of Bf-CATH is widespread, including stomach, trachea, skin, muscle, heart, kidney, lung, brain, intestine, spleen, liver, ovary and venom glands [97].
- Cdt-CATH: also named as crotalicidin (Ctn), is a 34-aa peptide from the South American rattlesnake (Crotalus durissus terrificus) venom. Among SV-CATHs identified in pit vipers by Falcão et al. [84] (lutzicidin, lachesicidin, batroxicidin, collectively named vipericidins), Ctn has been the most studied, showing potent bactericidal effects against Gram-negative and Gram-positive bacteria (MICs <10 μM) [84], anti-parasitic (anti-trypanosomatid) activity [109], and activity against opportunistic yeasts and dermatophytes, alone or in combination with conventional antifungals [108]. In addition, Ctn has shown potent anti-tumor activity against different leukemia cell lines (IC50s <5 μM) [107].
- Other snake-derived CATHs: additional pit viper-derived CATHs were predicted by Falcao et al., from the venom gland of Lachesis muta rhombeata (lachesicidin), Bothrops atrox (batroxicidin) and B. lutzi (lutzicidin), as well as two clones from the elapid, Pseudonaja textilis (Pt-CATH1 and Pt-CATH2). Batroxicidin and Pt-CATH1 displayed antibacterial potency comparable to that of Ctn, but were more hemolytic [84]. Moreover, batroxicidin induced T. cruzi cell death by membrane disruption and showed an overall proinflammatory profile [110,111].
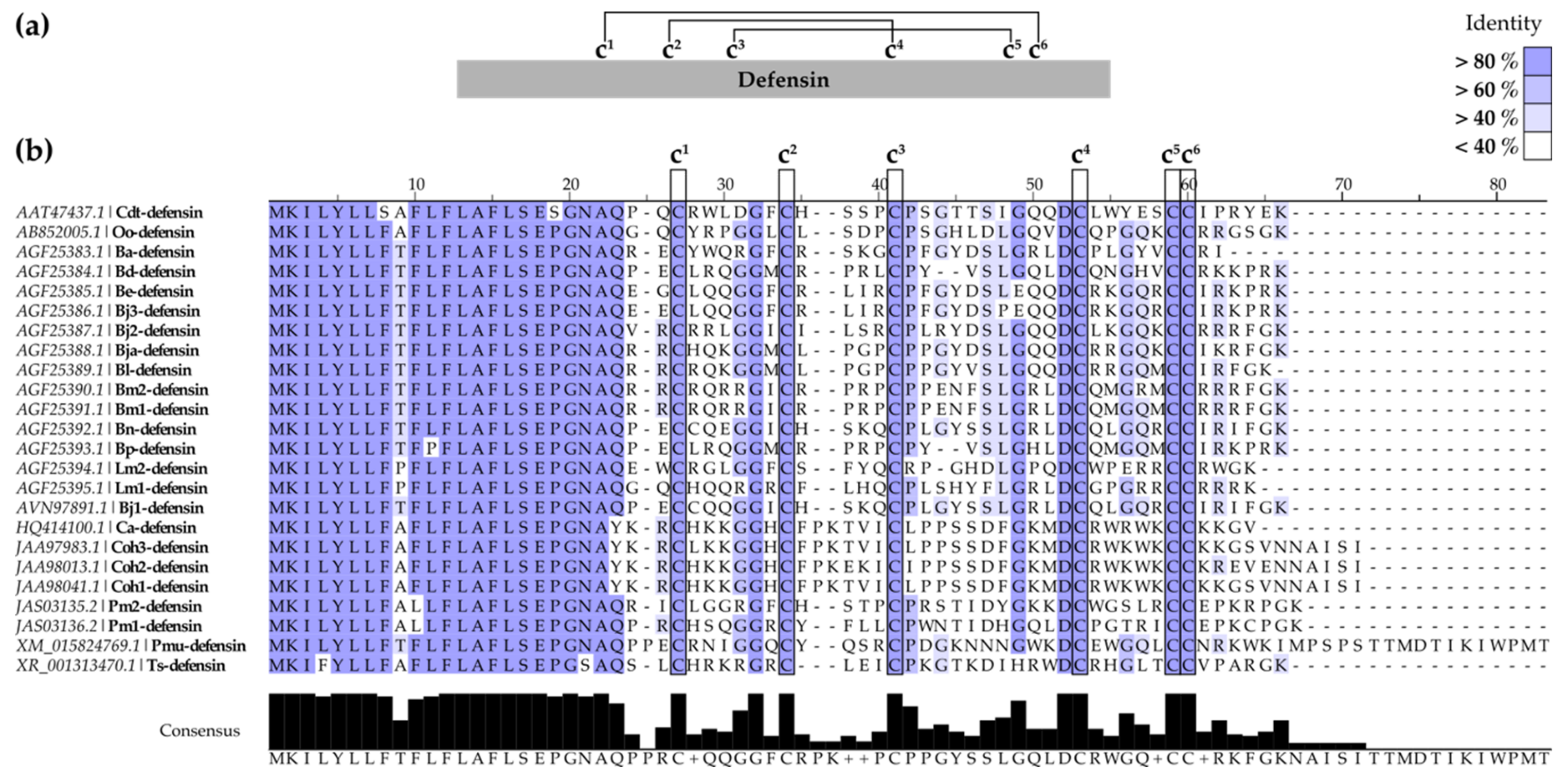
2.1.2. SV-Defensins
2.1.3. Waprins
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACE | angiotensin-converting enzyme |
| ACP | anticancer peptide |
| AMP | antimicrobial peptide |
| CATH | cathelicidin |
| Ctn | crotalicidin |
| EC50 | 50% effective concentration |
| HC10 | 10% hemolytic concentration; |
| HPLC | high-performance liquid chromatography |
| IC50 | 50 % inhibitory concentration |
| L-AAO | L-amino acid oxidases |
| MRSA | methicillin-resistant S. aureus |
| MS | mass spectrometry |
| MSA | multiple sequence analysis |
| Naw | nawaprin |
| NMR | nuclear magnetic resonance |
| Omw | omwaprin |
| PLA2 | phospholipases A2 |
| SV | snake venom |
References
- Waheed, H.; Moin, S.F.; Choudhary, M.I. Snake Venom: From Deadly Toxins to Life-saving Therapeutics. Curr. Med. Chem. 2017, 24, 1874–1891. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.H. A bradykinin-potentiating factor (BPF) present in the venom of bothrops jararaca. Br. J. Pharmacol. Chemother. 1965, 24, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Ondetti, M.A.; Rubin, B.; Cushman, D.W. Design of specific inhibitors of angiotensin-converting enzyme: New class of orally active antihypertensive agents. Science 1977, 196, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. P T Peer-Rev. J. Formul. Manag. 2015, 40, 277–283. [Google Scholar]
- O’Neill, J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist 2014. [Google Scholar]
- O’Neill, J. Tackling drug-resistant infection globally: Final report and recommendations. Rev. Antimicrob. Resist 2016. [Google Scholar]
- WHO. 2018. Cancer Fact Sheets. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 6 August 2019).
- Ferlay, J.E.M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018; Available online: https://gco.iarc.fr/today (accessed on 6 August 2019).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: Globocan sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; Reyes Gil, M. Chapter 164—Diagnostic Use of Venoms. In Transfusion Medicine and Hemostasis, 3rd ed.; Shaz, B.H., Hillyer, C.D., Reyes Gil, M., Eds.; Elsevier: Cambridge, MA, USA, 2019; pp. 969–975. [Google Scholar]
- Koh, D.C.; Armugam, A.; Jeyaseelan, K. Snake venom components and their applications in biomedicine. Cell. Mol. Life Sci. 2006, 63, 3030–3041. [Google Scholar] [CrossRef]
- Takacs, Z.; Nathan, S. Animal Venoms in Medicine. In Encyclopedia of Toxicology, 3rd ed.; Wexler, P., Ed.; Academic Press: Oxford, UK, 2014; pp. 252–259. [Google Scholar]
- King, G.F. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opin. Biol. Ther. 2011, 11, 1469–1484. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Approaching the golden age of natural product pharmaceuticals from venom libraries: An overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr. Pharm. Des. 2007, 13, 2927–2934. [Google Scholar] [CrossRef] [PubMed]
- Vonk, F.J.; Jackson, K.; Doley, R.; Madaras, F.; Mirtschin, P.J.; Vidal, N. Snake venom: From fieldwork to the clinic: Recent insights into snake biology, together with new technology allowing high-throughput screening of venom, bring new hope for drug discovery. Bioessays 2011, 33, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.; Schultz, J. Prospective evaluation of the incidence of wound infection in rattlesnake envenomation in dogs. J. Vet. Emerg. Crit. Care 2015, 25, 546–551. [Google Scholar] [CrossRef]
- De Lima, D.C.; Alvarez Abreu, P.; de Freitas, C.C.; Santos, D.O.; Borges, R.O.; Dos Santos, T.C.; Mendes Cabral, L.; Rodrigues, C.R.; Castro, H.C. Snake Venom: Any Clue for Antibiotics and CAM? Evid. Based Complement. Altern. Med. 2005, 2, 39–47. [Google Scholar] [CrossRef]
- Talan, D.A.; Citron, D.M.; Overturf, G.D.; Singer, B.; Froman, P.; Goldstein, E.J. Antibacterial activity of crotalid venoms against oral snake flora and other clinical bacteria. J. Infect. Dis. 1991, 164, 195–198. [Google Scholar] [CrossRef]
- Perumal Samy, R.; Gopalakrishnakone, P.; Ho, B.; Chow, V.T. Purification, characterization and bactericidal activities of basic phospholipase A2 from the venom of Agkistrodon halys (Chinese pallas). Biochimie 2008, 90, 1372–1388. [Google Scholar] [CrossRef]
- Stiles, B.G.; Sexton, F.W.; Weinstein, S.A. Antibacterial effects of different snake venoms: Purification and characterization of antibacterial proteins from Pseudechis australis (Australian king brown or mulga snake) venom. Toxicon 1991, 29, 1129–1141. [Google Scholar] [CrossRef]
- Perumal Samy, R.; Pachiappan, A.; Gopalakrishnakone, P.; Thwin, M.M.; Hian, Y.E.; Chow, V.T.; Bow, H.; Weng, J.T. In vitro antimicrobial activity of natural toxins and animal venoms tested against Burkholderia pseudomallei. BMC Infect. Dis. 2006, 6, 100. [Google Scholar] [CrossRef]
- Chaim-Matyas, A.; Ovadia, M. Cytotoxic activity of various snake venoms on melanoma, B16F10 and chondrosarcoma. Life Sci. 1987, 40, 1601–1607. [Google Scholar] [CrossRef]
- Da Silva, R.J.; da Silva, M.G.; Vilela, L.C.; Fecchio, D. Antitumor effect of Bothrops jararaca venom. Mediat. Inflamm. 2002, 11, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.; Bhattacharjee, P.; Mishra, R.; Biswas, A.K.; Dasgupta, S.C.; Giri, B. Anticancer potential of animal venoms and toxins. Indian J. Exp. Biol. 2010, 48, 93–103. [Google Scholar] [PubMed]
- Kerkkamp, H.; Bagowski, C.; Kool, J.; van Soolingen, B.; Vonk, F.J.; Vlecken, D. Whole snake venoms: Cytotoxic, anti-metastatic and antiangiogenic properties. Toxicon 2018, 150, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Izidoro, L.F.; Sobrinho, J.C.; Mendes, M.M.; Costa, T.R.; Grabner, A.N.; Rodrigues, V.M.; da Silva, S.L.; Zanchi, F.B.; Zuliani, J.P.; Fernandes, C.F.; et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. BioMed Res. Int. 2014, 2014, 196754. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.R.; Palacios, A.L.V.; Patino, R.S.P.; Mendes, B.; Teixeira, C.A.S.; Gomes, P.; da Silva, S.L. Harnessing snake venom phospholipases A2 to novel approaches for overcoming antibiotic resistance. Drug Dev. Res. 2019, 80, 68–85. [Google Scholar] [CrossRef] [PubMed]
- Modahl, C.M.; Frietze, S.; Mackessy, S.P. Transcriptome-facilitated proteomic characterization of rear-fanged snake venoms reveal abundant metalloproteinases with enhanced activity. J. Proteom. 2018, 187, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Samy, R.P.; Gopalakrishnakone, P.; Chow, V.T.; Ho, B. Viper metalloproteinase (Agkistrodon halys pallas) with antimicrobial activity against multi-drug resistant human pathogens. J. Cell. Physiol. 2008, 216, 54–68. [Google Scholar] [CrossRef]
- Markland, F.S., Jr.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef]
- Sala, A.; Cabassi, C.S.; Santospirito, D.; Polverini, E.; Flisi, S.; Cavirani, S.; Taddei, S. Novel Naja atra cardiotoxin 1 (CTX-1) derived antimicrobial peptides with broad spectrum activity. PLoS ONE 2018, 13, e0190778. [Google Scholar] [CrossRef]
- Samy, R.P.; Stiles, B.G.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; Franco, O.L.; Rowan, E.G.; Kumar, A.P.; Lim, L.H.; Sethi, G. Viperatoxin-II: A novel viper venom protein as an effective bactericidal agent. FEBS Open Bio 2015, 5, 928–941. [Google Scholar] [CrossRef]
- Sampaio, S.C.; Hyslop, S.; Fontes, M.R.; Prado-Franceschi, J.; Zambelli, V.O.; Magro, A.J.; Brigatte, P.; Gutierrez, V.P.; Cury, Y. Crotoxin: Novel activities for a classic beta-neurotoxin. Toxicon 2010, 55, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Samy, R.P.; Kandasamy, M.; Gopalakrishnakone, P.; Stiles, B.G.; Rowan, E.G.; Becker, D.; Shanmugam, M.K.; Sethi, G.; Chow, V.T. Wound healing activity and mechanisms of action of an antibacterial protein from the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). PLoS ONE 2014, 9, e80199. [Google Scholar] [CrossRef] [PubMed]
- Allane, D.; Oussedik-Oumehdi, H.; Harrat, Z.; Seve, M.; Laraba-Djebari, F. Isolation and characterization of an anti-leishmanial disintegrin from Cerastes cerastes venom. J. Biochem. Mol. Toxicol. 2018, 32, e22018. [Google Scholar] [CrossRef] [PubMed]
- Sulca, M.A.; Remuzgo, C.; Cardenas, J.; Kiyota, S.; Cheng, E.; Bemquerer, M.P.; Machini, M.T. Venom of the Peruvian snake Bothriopsis oligolepis: Detection of antibacterial activity and involvement of proteolytic enzymes and C-type lectins in growth inhibition of Staphylococcus aureus. Toxicon 2017, 134, 30–40. [Google Scholar] [CrossRef]
- Nolte, S.; de Castro Damasio, D.; Barea, A.C.; Gomes, J.; Magalhaes, A.; Mello Zischler, L.F.; Stuelp-Campelo, P.M.; Elifio-Esposito, S.L.; Roque-Barreira, M.C.; Reis, C.A.; et al. BJcuL, a lectin purified from Bothrops jararacussu venom, induces apoptosis in human gastric carcinoma cells accompanied by inhibition of cell adhesion and actin cytoskeleton disassembly. Toxicon 2012, 59, 81–85. [Google Scholar] [CrossRef]
- Nunes Edos, S.; de Souza, M.A.; Vaz, A.F.; Santana, G.M.; Gomes, F.S.; Coelho, L.C.; Paiva, P.M.; da Silva, R.M.; Silva-Lucca, R.A.; Oliva, M.L.; et al. Purification of a lectin with antibacterial activity from Bothrops leucurus snake venom. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 159, 57–63. [Google Scholar] [CrossRef]
- Calderon, L.A.; Sobrinho, J.C.; Zaqueo, K.D.; de Moura, A.A.; Grabner, A.N.; Mazzi, M.V.; Marcussi, S.; Nomizo, A.; Fernandes, C.F.; Zuliani, J.P.; et al. Antitumoral activity of snake venom proteins: New trends in cancer therapy. BioMed Res. Int. 2014, 2014, 203639. [Google Scholar] [CrossRef]
- Gomes, V.M.; Carvalho, A.O.; Da Cunha, M.; Keller, M.N.; Bloch, C., Jr.; Deolindo, P.; Alves, E.W. Purification and characterization of a novel peptide with antifungal activity from Bothrops jararaca venom. Toxicon 2005, 45, 817–827. [Google Scholar] [CrossRef]
- Sarzaeem, A.; Zare Mirakabadi, A.; Moradhaseli, S.; Morovvati, H.; Lotfi, M. Cytotoxic effect of ICD-85 (venom-derived peptides) on HeLa cancer cell line and normal LK cells using MTT Assay. Arch. Iran. Med. 2012, 15, 696–701. [Google Scholar]
- Júnior, N.; Cardoso, M.H.; Franco, O.L. Snake venoms: Attractive antimicrobial proteinaceous compounds for therapeutic purposes. Cell. Mol. Life Sci. 2013, 70, 4645–4658. [Google Scholar] [CrossRef]
- Ma, R.; Mahadevappa, R.; Kwok, H.F. Venom-based peptide therapy: Insights into anti-cancer mechanism. Oncotarget 2017, 8, 100908–100930. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.S.; Paidesetty, S.K.; Padhy, R.N. Antibacterial, antifungal and antimycobacterial compounds from cyanobacteria. Biomed. Pharmacother. 2017, 90, 760–776. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Yeung, A.T.; Gellatly, S.L.; Hancock, R.E. Multifunctional cationic host defence peptides and their clinical applications. Cell. Mol. Life Sci. 2011, 68, 2161–2176. [Google Scholar] [CrossRef]
- Gomes, B.; Augusto, M.T.; Felicio, M.R.; Hollmann, A.; Franco, O.L.; Goncalves, S.; Santos, N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef]
- Hancock, R.E.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Wuerth, K.; Hancock, R.E. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 2013, 9, 761–768. [Google Scholar] [CrossRef]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta 2016, 1858, 980–987. [Google Scholar] [CrossRef]
- Xu, N.; Wang, Y.S.; Pan, W.B.; Xiao, B.; Wen, Y.J.; Chen, X.C.; Chen, L.J.; Deng, H.X.; You, J.; Kan, B.; et al. Human alpha-defensin-1 inhibits growth of human lung adenocarcinoma xenograft in nude mice. Mol. Cancer Ther. 2008, 7, 1588–1597. [Google Scholar] [CrossRef]
- Al-Benna, S.; Shai, Y.; Jacobsen, F.; Steinstraesser, L. Oncolytic activities of host defense peptides. Int. J. Mol. Sci. 2011, 12, 8027–8051. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.A.; Lay, F.T.; Poon, I.K.H.; Kvansakul, M.; Hulett, M.D. Tumor cell membrane-targeting cationic antimicrobial peptides: Novel insights into mechanisms of action and therapeutic prospects. Cell. Mol. Life Sci. 2017, 74, 3809–3825. [Google Scholar] [CrossRef] [PubMed]
- Rosca, E.V.; Koskimaki, J.E.; Rivera, C.G.; Pandey, N.B.; Tamiz, A.P.; Popel, A.S. Anti-angiogenic peptides for cancer therapeutics. Curr. Pharm. Biotechnol. 2011, 12, 1101–1116. [Google Scholar] [CrossRef] [PubMed]
- Chavakis, T.; Cines, D.B.; Rhee, J.S.; Liang, O.D.; Schubert, U.; Hammes, H.P.; Higazi, A.A.; Nawroth, P.P.; Preissner, K.T.; Bdeir, K. Regulation of neovascularization by human neutrophil peptides (alpha-defensins): A link between inflammation and angiogenesis. FASEB J. 2004, 18, 1306–1308. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.C.; Watanabe, S.; Watanabe, R.; Hata, K.; Shimazaki, K.; Azuma, I. Bovine lactoferrin and lactoferricin, a peptide derived from bovine lactoferrin, inhibit tumor metastasis in mice. Jpn. J. Cancer Res. 1997, 88, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Brogden, K.A.; Ackermann, M.; McCray, P.B., Jr.; Tack, B.F. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 2003, 22, 465–478. [Google Scholar] [CrossRef]
- Yazici, A.; Ortucu, S.; Taskin, M.; Marinelli, L. Natural-based Antibiofilm and Antimicrobial Peptides from Microorganisms. Curr. Top. Med. Chem. 2018, 18, 2102–2107. [Google Scholar] [CrossRef]
- Hassan, M.; Kjos, M.; Nes, I.F.; Diep, D.B.; Lotfipour, F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef]
- Wong, J.H.; Ye, X.J.; Ng, T.B. Cathelicidins: Peptides with antimicrobial, immunomodulatory, anti-inflammatory, angiogenic, anticancer and procancer activities. Curr. Protein Pept. Sci. 2013, 14, 504–514. [Google Scholar] [CrossRef]
- Agier, J.; Efenberger, M.; Brzezinska-Blaszczyk, E. Cathelicidin impact on inflammatory cells. Cent. Eur. J. Immunol. 2015, 40, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Braff, M.H.; Hawkins, M.A.; Di Nardo, A.; Lopez-Garcia, B.; Howell, M.D.; Wong, C.; Lin, K.; Streib, J.E.; Dorschner, R.; Leung, D.Y.; et al. Structure-function relationships among human cathelicidin peptides: Dissociation of antimicrobial properties from host immunostimulatory activities. J. Immunol. 2005, 174, 4271–4278. [Google Scholar] [CrossRef] [PubMed]
- Tomasinsig, L.; Zanetti, M. The cathelicidins—Structure, function and evolution. Curr. Protein Pept. Sci. 2005, 6, 23–34. [Google Scholar] [CrossRef]
- Gennaro, R.; Zanetti, M. Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers 2000, 55, 31–49. [Google Scholar] [CrossRef]
- Das, H.; Sharma, B.; Kumar, A. Cloning and characterization of novel cathelicidin cDNA sequence of Bubalus bubalis homologous to Bos taurus cathelicidin-4. DNA Seq. 2006, 17, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A.; Kalfa, V.C.; Ackermann, M.R.; Palmquist, D.E.; McCray, P.B., Jr.; Tack, B.F. The ovine cathelicidin SMAP29 kills ovine respiratory pathogens in vitro and in an ovine model of pulmonary infection. Antimicrob. Agents Chemother. 2001, 45, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Nguyen, T.; Boo, L.M.; Hong, T.; Espiritu, C.; Orlov, D.; Wang, W.; Waring, A.; Lehrer, R.I. RL-37, an alpha-helical antimicrobial peptide of the rhesus monkey. Antimicrob. Agents Chemother. 2001, 45, 2695–2702. [Google Scholar] [CrossRef]
- Nagaoka, I.; Tsutsumi-Ishii, Y.; Yomogida, S.; Yamashita, T. Isolation of cDNA encoding guinea pig neutrophil cationic antibacterial polypeptide of 11 kDa (CAP11) and evaluation of CAP11 mRNA expression during neutrophil maturation. J. Biol. Chem. 1997, 272, 22742–22750. [Google Scholar] [CrossRef][Green Version]
- Gallo, R.L.; Kim, K.J.; Bernfield, M.; Kozak, C.A.; Zanetti, M.; Merluzzi, L.; Gennaro, R. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J. Biol. Chem. 1997, 272, 13088–13093. [Google Scholar] [CrossRef]
- Termen, S.; Tollin, M.; Olsson, B.; Svenberg, T.; Agerberth, B.; Gudmundsson, G.H. Phylogeny, processing and expression of the rat cathelicidin rCRAMP: A model for innate antimicrobial peptides. Cell. Mol. Life Sci. 2003, 60, 536–549. [Google Scholar] [CrossRef]
- Xiao, Y.; Cai, Y.; Bommineni, Y.R.; Fernando, S.C.; Prakash, O.; Gilliland, S.E.; Zhang, G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006, 281, 2858–2867. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Gan, T.X.; Liu, X.D.; Jin, Y.; Lee, W.H.; Shen, J.H.; Zhang, Y. Identification and characterization of novel reptile cathelicidins from elapid snakes. Peptides 2008, 29, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Uzzell, T.; Stolzenberg, E.D.; Shinnar, A.E.; Zasloff, M. Hagfish intestinal antimicrobial peptides are ancient cathelicidins. Peptides 2003, 24, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Junior, N.G.O.; Cardoso, M.H.; Candido, E.S.; van den Broek, D.; de Lange, N.; Velikova, N.; Kleijn, J.M.; Wells, J.M.; Rezende, T.M.B.; Franco, O.L.; et al. An acidic model pro-peptide affects the secondary structure, membrane interactions and antimicrobial activity of a crotalicidin fragment. Sci. Rep. 2018, 8, 11127. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, H.; Yu, G.Y.; Liu, X.D.; Shen, J.H.; Lee, W.H.; Zhang, Y. Structure-function relationship of king cobra cathelicidin. Peptides 2010, 31, 1488–1493. [Google Scholar] [CrossRef]
- Azim, S.; McDowell, D.; Cartagena, A.; Rodriguez, R.; Laughlin, T.F.; Ahmad, Z. Venom peptides cathelicidin and lycotoxin cause strong inhibition of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2016, 87, 246–251. [Google Scholar] [CrossRef]
- Li, S.A.; Lee, W.H.; Zhang, Y. Efficacy of OH-CATH30 and its analogs against drug-resistant bacteria in vitro and in mouse models. Antimicrob. Agents Chemother. 2012, 56, 3309–3317. [Google Scholar] [CrossRef]
- Zhao, F.; Lan, X.Q.; Du, Y.; Chen, P.Y.; Zhao, J.; Zhao, F.; Lee, W.H.; Zhang, Y. King cobra peptide OH-CATH30 as a potential candidate drug through clinic drug-resistant isolates. Zool. Res. 2018, 39, 87–96. [Google Scholar]
- Chen, X.-x.; Yu, G.-y.; Zhan, Y.; Zhang, Y.; Shen, J.-h.; Lee, W.-h. Effects of the Antimicrobial Peptide OH-CATH on Escherichia coli. Zool. Res. 2009, 30, 171–177. [Google Scholar] [CrossRef][Green Version]
- Falcao, C.B.; de La Torre, B.G.; Perez-Peinado, C.; Barron, A.E.; Andreu, D.; Radis-Baptista, G. Vipericidins: A novel family of cathelicidin-related peptides from the venom gland of South American pit vipers. Amino Acids 2014, 46, 2561–2571. [Google Scholar] [CrossRef]
- Tajbakhsh, M.; Karimi, A.; Tohidpour, A.; Abbasi, N.; Fallah, F.; Akhavan, M.M. The antimicrobial potential of a new derivative of cathelicidin from Bungarus fasciatus against methicillin-resistant Staphylococcus aureus. J. Microbiol. 2018, 56, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, M.; Akhavan, M.M.; Fallah, F.; Karimi, A. A Recombinant Snake Cathelicidin Derivative Peptide: Antibiofilm Properties and Expression in Escherichia coli. Biomolecules 2018, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Bai, X.; Luan, N.; Yao, H.; Zhang, Z.; Liu, W.; Chen, Y.; Yan, X.; Rong, M.; Lai, R.; et al. A Designed Tryptophan- and Lysine/Arginine-Rich Antimicrobial Peptide with Therapeutic Potential for Clinical Antibiotic-Resistant Candida albicans Vaginitis. J. Med. Chem. 2016, 59, 1791–1799. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Qi, J.; Shan, B.; Gao, R.; Gao, F.; Xie, H.; Yuan, M.; Liu, H.; Jin, S.; Wu, F.; et al. Pretreatment with cathelicidin-BF ameliorates Pseudomonas aeruginosa pneumonia in mice by enhancing NETosis and the autophagy of recruited neutrophils and macrophages. Int. Immunopharmacol. 2018, 65, 382–391. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, B.; Zhang, X.; Wang, X.; Wu, K.; Guan, Q. Effects of cathelicidin-derived peptide from reptiles on lipopolysaccharide-induced intestinal inflammation in weaned piglets. Vet. Immunol. Immunopathol. 2017, 192, 41–53. [Google Scholar] [CrossRef]
- Zhang, H.; Xia, X.; Han, F.; Jiang, Q.; Rong, Y.; Song, D.; Wang, Y. Cathelicidin-BF, a Novel Antimicrobial Peptide from Bungarus fasciatus, Attenuates Disease in a Dextran Sulfate Sodium Model of Colitis. Mol. Pharm. 2015, 12, 1648–1661. [Google Scholar] [CrossRef]
- Yi, H.; Yu, C.; Zhang, H.; Song, D.; Jiang, D.; Du, H.; Wang, Y. Cathelicidin-BF suppresses intestinal inflammation by inhibiting the nuclear factor-kappaB signaling pathway and enhancing the phagocytosis of immune cells via STAT-1 in weanling piglets. Int. Immunopharmacol. 2015, 28, 61–69. [Google Scholar] [CrossRef]
- Song, D.; Zong, X.; Zhang, H.; Wang, T.; Yi, H.; Luan, C.; Wang, Y. Antimicrobial peptide Cathelicidin-BF prevents intestinal barrier dysfunction in a mouse model of endotoxemia. Int. Immunopharmacol. 2015, 25, 141–147. [Google Scholar] [CrossRef]
- Wang, H.; Ke, M.; Tian, Y.; Wang, J.; Li, B.; Wang, Y.; Dou, J.; Zhou, C. BF-30 selectively inhibits melanoma cell proliferation via cytoplasmic membrane permeabilization and DNA-binding in vitro and in B16F10-bearing mice. Eur. J. Pharmacol. 2013, 707, 1–10. [Google Scholar] [CrossRef]
- Zhou, H.; Dou, J.; Wang, J.; Chen, L.; Wang, H.; Zhou, W.; Li, Y.; Zhou, C. The antibacterial activity of BF-30 in vitro and in infected burned rats is through interference with cytoplasmic membrane integrity. Peptides 2011, 32, 1131–1138. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Chen, L.; Guang, H.; Li, Z.; Yang, H.; Li, J.; You, D.; Yu, H.; Lai, R. Cathelicidin-BF, a snake cathelicidin-derived antimicrobial peptide, could be an excellent therapeutic agent for acne vulgaris. PLoS ONE 2011, 6, e22120. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, B.; Zhou, H.; Sun, L.; Dou, J.; Qian, H.; Huang, W.; Mei, Y.; Han, J. Structure-activity relationships of a snake cathelicidin-related peptide, BF-15. Peptides 2011, 32, 2497–2503. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hong, J.; Liu, X.; Yang, H.; Liu, R.; Wu, J.; Wang, A.; Lin, D.; Lai, R. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PLoS ONE 2008, 3, e3217. [Google Scholar] [CrossRef]
- Amer, L.S.; Bishop, B.M.; van Hoek, M.L. Antimicrobial and antibiofilm activity of cathelicidins and short, synthetic peptides against Francisella. Biochem. Biophys. Res. Commun. 2010, 396, 246–251. [Google Scholar] [CrossRef]
- de Latour, F.A.; Amer, L.S.; Papanstasiou, E.A.; Bishop, B.M.; van Hoek, M.L. Antimicrobial activity of the Naja atra cathelicidin and related small peptides. Biochem. Biophys. Res. Commun. 2010, 396, 825–830. [Google Scholar] [CrossRef]
- Dean, S.N.; Bishop, B.M.; van Hoek, M.L. Susceptibility of Pseudomonas aeruginosa Biofilm to Alpha-Helical Peptides: D-enantiomer of LL-37. Front. Microbiol. 2011, 2, 128. [Google Scholar] [CrossRef]
- Dean, S.N.; Bishop, B.M.; van Hoek, M.L. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 2011, 11, 114. [Google Scholar] [CrossRef]
- Gupta, K.; Singh, S.; van Hoek, M.L. Short, Synthetic Cationic Peptides Have Antibacterial Activity against Mycobacterium smegmatis by Forming Pores in Membrane and Synergizing with Antibiotics. Antibiotics 2015, 4, 358–378. [Google Scholar] [CrossRef]
- Blower, R.J.; Barksdale, S.M.; van Hoek, M.L. Snake Cathelicidin NA-CATH and Smaller Helical Antimicrobial Peptides Are Effective against Burkholderia thailandensis. PLoS Negl. Trop. Dis. 2015, 9, e0003862. [Google Scholar] [CrossRef]
- Blower, R.J.; Popov, S.G.; van Hoek, M.L. Cathelicidin peptide rescues G. mellonella infected with B. anthracis. Virulence 2017, 9, 287–293. [Google Scholar] [CrossRef]
- Wei, L.; Gao, J.; Zhang, S.; Wu, S.; Xie, Z.; Ling, G.; Kuang, Y.Q.; Yang, Y.; Yu, H.; Wang, Y. Identification and Characterization of the First Cathelicidin from Sea Snakes with Potent Antimicrobial and Anti-inflammatory Activity and Special Mechanism. J. Biol. Chem. 2015, 290, 16633–16652. [Google Scholar] [CrossRef]
- Carlile, S.R.; Shiels, J.; Kerrigan, L.; Delaney, R.; Megaw, J.; Gilmore, B.F.; Weldon, S.; Dalton, J.P.; Taggart, C.C. Sea snake cathelicidin (Hc-cath) exerts a protective effect in mouse models of lung inflammation and infection. Sci. Rep. 2019, 9, 6071. [Google Scholar] [CrossRef]
- Falcao, C.B.; Perez-Peinado, C.; de la Torre, B.G.; Mayol, X.; Zamora-Carreras, H.; Jimenez, M.A.; Radis-Baptista, G.; Andreu, D. Structural Dissection of Crotalicidin, a Rattlesnake Venom Cathelicidin, Retrieves a Fragment with Antimicrobial and Antitumor Activity. J. Med. Chem. 2015, 58, 8553–8563. [Google Scholar] [CrossRef]
- Cavalcante, C.S.; Falcao, C.B.; Fontenelle, R.O.; Andreu, D.; Radis-Baptista, G. Anti-fungal activity of Ctn[15–34], the C-terminal peptide fragment of crotalicidin, a rattlesnake venom gland cathelicidin. J. Antibiot. 2016, 70, 231–237. [Google Scholar] [CrossRef]
- Bandeira, I.C.J.; Bandeira-Lima, D.; Mello, C.P.; Pereira, T.P.; De Menezes, R.; Sampaio, T.L.; Falcao, C.B.; Radis-Baptista, G.; Martins, A.M.C. Antichagasic effect of crotalicidin, a cathelicidin-like vipericidin, found in Crotalus durissus terrificus rattlesnake’s venom gland. Parasitology 2017, 145, 1059–1064. [Google Scholar] [CrossRef]
- Oliveira-Junior, N.G.; Freire, M.S.; Almeida, J.A.; Rezende, T.M.B.; Franco, O.L. Antimicrobial and proinflammatory effects of two vipericidins. Cytokine 2018, 111, 309–316. [Google Scholar] [CrossRef]
- Mello, C.P.; Lima, D.B.; Menezes, R.R.; Bandeira, I.C.; Tessarolo, L.D.; Sampaio, T.L.; Falcao, C.B.; Radis-Baptista, G.; Martins, A.M. Evaluation of the antichagasic activity of batroxicidin, a cathelicidin-related antimicrobial peptide found in Bothrops atrox venom gland. Toxicon 2017, 130, 56–62. [Google Scholar] [CrossRef]
- Cai, S.; Qiao, X.; Feng, L.; Shi, N.; Wang, H.; Yang, H.; Guo, Z.; Wang, M.; Chen, Y.; Wang, Y.; et al. Python Cathelicidin CATHPb1 Protects against Multidrug-Resistant Staphylococcal Infections by Antimicrobial-Immunomodulatory Duality. J. Med. Chem. 2018, 61, 2075–2086. [Google Scholar] [CrossRef]
- Kim, D.; Soundrarajan, N.; Lee, J.; Cho, H.S.; Choi, M.; Cha, S.Y.; Ahn, B.; Jeon, H.; Le, M.T.; Song, H.; et al. Genome wide analysis of the antimicrobial peptides in Python bivittatus and characterization of cathelicidins with potent antimicrobial activity and low cytotoxicity. Antimicrob. Agents Chemother. 2017, 61, e00530-17. [Google Scholar] [CrossRef]
- Wang, A.; Zhang, F.; Guo, Z.; Chen, Y.; Zhang, M.; Yu, H.; Wang, Y. Characterization of a Cathelicidin from the Colubrinae Snake, Sinonatrix annularis. Zool. Sci. 2019, 36, 68–76. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar]
- Du, H.; Samuel, R.L.; Massiah, M.A.; Gillmor, S.D. The structure and behavior of the NA-CATH antimicrobial peptide with liposomes. Biochim. Biophys. Acta 2015, 1848, 2394–2405. [Google Scholar] [CrossRef][Green Version]
- Juba, M.; Porter, D.; Dean, S.; Gillmor, S.; Bishop, B. Characterization and performance of short cationic antimicrobial peptide isomers. Biopolymers 2013, 100, 387–401. [Google Scholar] [CrossRef]
- Samuel, R.; Gillmor, S. Membrane phase characteristics control NA-CATH activity. Biochim. Biophys. Acta 2016, 1858, 1974–1982. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, L.; Wang, Y. The antimicrobial peptide cathelicidin-BF could be a potential therapeutic for Salmonella typhimurium infection. Microbiol. Res. 2015, 171, 45–51. [Google Scholar] [CrossRef]
- Perez-Peinado, C.; Dias, S.A.; Domingues, M.M.; Benfield, A.H.; Freire, J.M.; Radis-Baptista, G.; Gaspar, D.; Castanho, M.; Craik, D.J.; Henriques, S.T.; et al. Mechanisms of bacterial membrane permeabilization by crotalicidin (Ctn) and its fragment Ctn(15-34), antimicrobial peptides from rattlesnake venom. J. Biol. Chem. 2018, 293, 1536–1549. [Google Scholar] [CrossRef]
- Cavalcante, C.S.P.; de Aguiar, F.L.L.; Fontenelle, R.O.S.; de Menezes, R.; Martins, A.M.C.; Falcao, C.B.; Andreu, D.; Radis-Baptista, G. Insights into the candidacidal mechanism of Ctn[15–34]—A carboxyl-terminal, crotalicidin-derived peptide related to cathelicidins. J. Med Microbiol. 2018, 67, 129–138. [Google Scholar] [CrossRef]
- Zhang, Z.; Mu, L.; Tang, J.; Duan, Z.; Wang, F.; Wei, L.; Rong, M.; Lai, R. A small peptide with therapeutic potential for inflammatory acne vulgaris. PLoS ONE 2013, 8, e72923. [Google Scholar] [CrossRef]
- Hao, Q.; Wang, H.; Wang, J.; Dou, J.; Zhang, M.; Zhou, W.; Zhou, C. Effective antimicrobial activity of Cbf-K16 and Cbf-A7 A13 against NDM-1-carrying Escherichia coli by DNA binding after penetrating the cytoplasmic membrane in vitro. J. Pept. Sci. 2013, 19, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wang, H.; Li, B.; Ke, M.; Wang, J.; Dou, J.; Zhou, C. The cathelicidin-BF Lys16 mutant Cbf-K16 selectively inhibits non-small cell lung cancer proliferation in vitro. Oncol. Rep. 2013, 30, 2502–2510. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; He, X.; Zhang, P.; Shen, C.; Mwangi, J.; Xu, C.; Mo, G.; Lai, R.; Zhang, Z. In Vitro and In Vivo Antimalarial Activity of LZ1, a Peptide Derived from Snake Cathelicidin. Toxins 2019, 11, 379. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Q.; Li, H.; Yuan, M.; Yuan, M. Preparation, characterization, in vitro release and degradation of cathelicidin-BF-30-PLGA microspheres. PLoS ONE 2014, 9, e100809. [Google Scholar] [CrossRef]
- Bao, Y.; Wang, S.; Li, H.; Wang, Y.; Chen, H.; Yuan, M. Characterization, Stability and Biological Activity In Vitro of Cathelicidin-BF-30 Loaded 4-Arm Star-Shaped PEG-PLGA Microspheres. Molecules 2018, 23, 497. [Google Scholar] [CrossRef]
- Luan, C.; Zhang, H.W.; Song, D.G.; Xie, Y.G.; Feng, J.; Wang, Y.Z. Expressing antimicrobial peptide cathelicidin-BF in Bacillus subtilis using SUMO technology. Appl. Microbiol. Biotechnol. 2014, 98, 3651–3658. [Google Scholar] [CrossRef]
- He, Q.; Fu, A.Y.; Li, T.J. Expression and one-step purification of the antimicrobial peptide cathelicidin-BF using the intein system in Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 2015, 42, 647–653. [Google Scholar] [CrossRef]
- Perez-Peinado, C.; Defaus, S.; Sans-Comerma, L.; Valle, J.; Andreu, D. Decoding the human serum interactome of snake-derived antimicrobial peptide Ctn[15–34]: Toward an explanation for unusually long half-life. J. Proteom. 2019, 204, 103372. [Google Scholar] [CrossRef]
- Perez-Peinado, C.; Dias, S.A.; Mendonca, D.A.; Castanho, M.; Veiga, A.S.; Andreu, D. Structural determinants conferring unusual long life in human serum to rattlesnake-derived antimicrobial peptide Ctn[15–34]. J. Pept. Sci. 2019, 25, e3195. [Google Scholar] [CrossRef]
- De Aguiar, F.L.L.; de Paula Cavalcante, C.S.; Dos Santos Fontenelle, R.O.; Falcao, C.B.; Andreu, D.; Radis-Baptista, G. The antiproliferative peptide Ctn[15–34] is active against multidrug-resistant yeasts Candida albicans and Cryptococcus neoformans. J. Appl. Microbiol. 2019, 2, 414–425. [Google Scholar] [CrossRef]
- Vieira-Girao, P.R.N.; Falcao, C.B.; Rocha, I.; Lucena, H.M.R.; Costa, F.H.F.; Radis-Baptista, G. Antiviral Activity of Ctn[15–34], A Cathelicidin-Derived Eicosapeptide, Against Infectious Myonecrosis Virus in Litopenaeus vannamei Primary Hemocyte Cultures. Food Environ. Virol. 2017, 9, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- De Smet, K.; Contreras, R. Human antimicrobial peptides: Defensins, cathelicidins and histatins. Biotechnol. Lett. 2005, 27, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef]
- Correa, P.G.; Oguiura, N. Phylogenetic analysis of beta-defensin-like genes of Bothrops, Crotalus and Lachesis snakes. Toxicon 2013, 69, 65–74. [Google Scholar] [CrossRef]
- Van Hoek, M.L. Antimicrobial peptides in reptiles. Pharmaceuticals 2014, 7, 723–753. [Google Scholar] [CrossRef]
- De Oliveira, Y.S.; Correa, P.G.; Oguiura, N. Beta-defensin genes of the Colubridae snakes Phalotris mertensi, Thamnodynastes hypoconia, and T. strigatus. Toxicon 2018, 146, 124–128. [Google Scholar] [CrossRef]
- Coronado, M.A.; Gabdulkhakov, A.; Georgieva, D.; Sankaran, B.; Murakami, M.T.; Arni, R.K.; Betzel, C. Structure of the polypeptide crotamine from the Brazilian rattlesnake Crotalus durissus terrificus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1958–1964. [Google Scholar] [CrossRef]
- Kerkis, I.; Hayashi, M.A.; Prieto da Silva, A.R.; Pereira, A.; De Sa Junior, P.L.; Zaharenko, A.J.; Radis-Baptista, G.; Kerkis, A.; Yamane, T. State of the art in the studies on crotamine, a cell penetrating peptide from South American rattlesnake. BioMed Res. Int. 2014, 2014, 675985. [Google Scholar] [CrossRef]
- Oguiura, N.; Boni-Mitake, M.; Affonso, R.; Zhang, G. In vitro antibacterial and hemolytic activities of crotamine, a small basic myotoxin from rattlesnake Crotalus durissus. J. Antibiot. 2011, 64, 327–331. [Google Scholar] [CrossRef]
- Yamane, E.S.; Bizerra, F.C.; Oliveira, E.B.; Moreira, J.T.; Rajabi, M.; Nunes, G.L.; de Souza, A.O.; da Silva, I.D.; Yamane, T.; Karpel, R.L.; et al. Unraveling the antifungal activity of a South American rattlesnake toxin crotamine. Biochimie 2013, 95, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Kerkis, A.; Hayashi, M.A.; Pereira, A.S.; Silva, F.S.; Oliveira, E.B.; Prieto da Silva, A.R.; Yamane, T.; Radis-Baptista, G.; Kerkis, I. Crotamine toxicity and efficacy in mouse models of melanoma. Expert Opin. Investig. Drugs 2011, 20, 1189–1200. [Google Scholar] [CrossRef]
- Nascimento, F.D.; Sancey, L.; Pereira, A.; Rome, C.; Oliveira, V.; Oliveira, E.B.; Nader, H.B.; Yamane, T.; Kerkis, I.; Tersariol, I.L.; et al. The natural cell-penetrating peptide crotamine targets tumor tissue in vivo and triggers a lethal calcium-dependent pathway in cultured cells. Mol. Pharm. 2012, 9, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ponnappan, N.; Budagavi, D.P.; Chugh, A. CyLoP-1: Membrane-active peptide with cell-penetrating and antimicrobial properties. Biochim. Biophys. Acta 2017, 1859, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Radis-Baptista, G.; de la Torre, B.G.; Andreu, D. Insights into the uptake mechanism of NrTP, a cell-penetrating peptide preferentially targeting the nucleolus of tumour cells. Chem. Biol. Drug Des. 2012, 79, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Andreu, D.; Santos, N.C. Uptake and cellular distribution of nucleolar targeting peptides (NrTPs) in different cell types. Biopolymers 2015, 104, 101–109. [Google Scholar] [CrossRef]
- Torres, A.M.; Wong, H.Y.; Desai, M.; Moochhala, S.; Kuchel, P.W.; Kini, R.M. Identification of a novel family of proteins in snake venoms. Purification and structural characterization of nawaprin from Naja nigricollis snake venom. J. Biol. Chem. 2003, 278, 40097–40104. [Google Scholar] [CrossRef]
- Nair, D.G.; Fry, B.G.; Alewood, P.; Kumar, P.P.; Kini, R.M. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem. J. 2007, 402, 93–104. [Google Scholar] [CrossRef]
- Banigan, J.R.; Mandal, K.; Sawaya, M.R.; Thammavongsa, V.; Hendrickx, A.P.; Schneewind, O.; Yeates, T.O.; Kent, S.B. Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography. Protein Sci. 2010, 19, 1840–1849. [Google Scholar] [CrossRef]
- Thankappan, B.; Angayarkanni, J. Biological characterization of omw1 and omw2: Antimicrobial peptides derived from omwaprin. 3 Biotech 2019, 9, 295. [Google Scholar] [CrossRef]
- Cooper, M.A.; Shlaes, D. Fix the antibiotics pipeline. Nature 2011, 472, 32. [Google Scholar] [CrossRef] [PubMed]
- Tomaras, A.P.; Dunman, P.M. In the midst of the antimicrobial discovery conundrum: An overview. Curr. Opin. Microbiol. 2015, 27, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Chabner, B.A.; Roberts, T.G., Jr. Timeline: Chemotherapy and the war on cancer. Nat. Rev. Cancer 2005, 5, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Kelderman, S.; Schumacher, T.N.; Haanen, J.B. Acquired and intrinsic resistance in cancer immunotherapy. Mol. Oncol. 2014, 8, 1132–1139. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2015, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2014, 43, D837–D843. [Google Scholar] [CrossRef]
| Commercial Drugs [12,13,14,15,16,17] | ||||
| Drug (Commercial Name) | Source Organism | Target | Therapeutic Use | |
| Captopril (Capoten®) | Bothrops jararaca | Angiotensin-converting enzyme (ACE) | Hypertension | |
| Enalapril (Vasotec®) | Bothrops jararaca | ACE | Hypertension | |
| Tirofiban (Aggrastat®) | Echis carinatus | Glycoprotein IIb/IIIa | Acute coronary syndromes | |
| Eptifibatide (Integrilin®) | Sistrurus miliarius barbouri | Glycoprotein IIb/IIIa | Acute coronary syndromes | |
| Batroxobin (Defibrase®) | Bothrops sp. | Fibrinogen | Infarction / Ischemia / Microcirculation dysfunctions | |
| Platelet gel (Plateltex-Act®) | Bothrops atrox | Fibrinogen | Platelet-induced tissue-healing | |
| Fibrin sealant (Vivostat®) | Bothrops moojeni | Fibrinogen | Autologous fibrin sealant in surgery | |
| Haemocoagulase (Reptilase®) | Bothrops atrox | Fibrinogen Factor X / Prothrombin | Hemorrhage | |
| Ximelagatran (Exanta®)* | Cobra venom | Thrombin | Atrial fibrillation / Blood clotting | |
| Ancrod (Viprinex®)* | Agkistrodon rhodostoma | Fibrinogen | Heparin-induced thrombocytopenia | |
| Drugs in Clinical and Pre-Clinical Trials [12,13,14,15,16,17] | ||||
| Drug Name | Phase | Source Organism | Target | Therapeutic Use |
| Fibrolase (Alfimeprase)* | III | Agkistrodon contortrix | Fibrinogen | Stroke and catheter occlusion |
| Crotoxin | I | Crotalus durissus terrificus | Unknown | Cancer |
| Cenderitide | II | Dendroaspis angusticeps | Natriuretic peptide receptor | Heart failure |
| RPI-MN | I | Naja atra | Nicotinic acetylcholine receptor | HIV / Amyotrophic lateral sclerosis / Herpes simplex keratitis |
| RPI-78M | I/II | Naja atra | Nicotinic acetylcholine receptor | Multiple sclerosis / Herpes simplex infections / Adrenomyeloneuropathy |
| RPI-78 | preclinical | Naja atra | Nicotinic acetylcholine receptor | Pain / Rheumatoid arthritis |
| Prohanin | preclinical | Ophiophagus hannah | Nitric oxide synthase | Chronic pain |
| Oxynor | preclinical | Oxyuranus scutellatus | Unknown | Wound healing |
| Natriuretic peptides | preclinical | Oxyuranus microlepidotus | Natriuretic peptide receptor | Heart failure |
| Textilinin-1TM | preclinical | Pseudonaja textilis | Plasmin | Preoperative bleeding |
| Vicrostatin | preclinical | Chimeric; Echis carinatus / Agkistrodon contortrix | Integrin receptor | Cancer |
| HaempatchTM | preclinical | Pseudonaja textilis | Prothrombin | Blood loss during |
| vascular trauma | ||||
| CoVaseTM | preclinical | Pseudonaja textilis | Factor Xa | Hemorrhage |
| Contortrostatin | preclinical | Agkistrodon contortrix | Integrin | Breast cancer |
| Diagnostic Tools [12,13,14,15,16,17] | ||||
| Product | Source Organism | Target | Clinical Assessment of: | |
| Protac® | Agkistrodon contortix | Protein C activation | Protein C | |
| Reptilase® | Bothrops jararaca | Fibrinogen | Fibrinogen | |
| Ecarin clotting time | Echis carinatus | Prothrombin | Meizothrombin | |
| Textarin®/Ecarin ratio | Pseudonaja textilis | Prothrombin | Lupus anticoagulant | |
| Russell’s viper venom-factor X | Daboia russelii | Factor X | Factor X | |
| Dilute Russell’s Viper Venom Time | Daboia russelii | Factor X, Factor V | Lupus anticoagulant | |
| Taipan Venom Time | Oxyuranus scutellatus | Prothrombin | Lupus anticoagulant | |
| Pefakit® APCR Factor V Leiden | Daboia russelii / Notechis scutatus scutatus | Factor V / Protein C / Prothrombin | Resistance to activated protein C | |
| Botrocetin® | Bothrops sp | Factor VIIIa | von Willebrand Factor | |
| Source Organism | Unified Name | Common name | Mature Peptide Sequence | Length | Hemolysis | Activity | Ref |
|---|---|---|---|---|---|---|---|
| Ophiophagus hannah | Oh-CATH | KF-34 | KRFKKFFKKLKNSVKKRAKKFFKKPRVIGVSIPF | 34 | Medium | G+ and G- bacteria. | [76,82,83,84] |
| Bungarus fasciatus | Bf-CATH | Cath-BF | KRFKKFFRKLKKSVKKRAKEFFKKPRVIGVSIPF | 34 | High | G+ and G- bacteria. | [85,86] |
| Bf-CATH30 | BF-30, cathelicidin-BF, C-BF, cathelicidin-WA, CWA | KFFRKLKKSVKKRAKEFFKKPRVIGVSIPF | 30 | High | G+ and G- bacteria, fungi and tumor cells. Anti-inflamatory. Activation of innate immunity. | [87,88,89,90,91,92,93,94,95,96,97] | |
| Naja atra | Na-CATH | - | KRFKKFFKKLKNSVKKRAKKFFKKPKVIGVTFPF | 34 | Low | G+ and G- bacteria. | [98,99,100,101,102,103,104] |
| Hydrophis cyanocinctus | Hc-CATH | - | KFFKRLLKSVRRAVKKFRKKPRLIGLSTLL | 30 | Low | G+ and G- bacteria and fungi. Anti-inflamatory. Inactive against tumor cells. | [105,106] |
| Crotalus durissus terrificus | Cdt-CATH | crotalicidin, Ctn | KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF | 34 | High | G+ and G- bacteria, fungi, parasites and tumor cells. Overall proinflammatory. | [84,107,108,109,110] |
| Bothrops atrox | Ba-CATH | batroxicidin, BatxC | KRFKKFFKKLKNSVKKRVKKFFRKPRVIGVTFPF | 34 | High | G+ and G- bacteria and parasites. Overall proinflammatory. | [84,110,111] |
| Pseudonaja textilis | Pt-CATH1 | Pt-CRAMP1 | KRFKKFFMKLKKSVKKRVMKFFKKPMVIGVTFPF | 34 | High | G+ and G- bacteria. | [84] |
| Pt-CATH2 | Pt-CRAMP2 | KRFKKFFRKLKKSVKKRVKKFFKKPRVIGVTIPF | 34 | n.d. | n.d. | [84] | |
| Lachesis muta rhombeata | Lmr-CATH | lachesicidin | KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTFPF | 34 | n.d. | n.d. | [84] |
| Bothrops lutzi | Bl-CATH | lutzicidin | KRFKKFFKKLKNNVKKRVKKFFRKPRVIGVTIPF | 34 | n.d. | n.d. | [84] |
| Python bivittatus | Pb-CATH1 | CATHPb1 | KRFKKFFRKIKKGFRKIFKKTKIFIGGTIPI | 31 | Low | G+ and G- bacteria and fungi. Chemotactic. Anti-inflammatory. | [112] |
| ∆Pb-CATH1 | ∆Pb-CATH1 | RVKRFKKFFRKIKKGFRKIFKKTKIFIG | 28 | Medium | G+ and G- bacteria. | [113] | |
| Pb-CATH2 | CATHPb2 | KRNGFRKFMRRLKKFFAGGGSSIAHIKLH | 29 | Low | G+ and G- bacteria and fungi. Chemotactic. Weakly anti-inflammatory. | [112] | |
| ∆Pb-CATH2 | Pb-CATH3 | HRVKRNGFRKFMRRLKKFFAGG | 22 | Medium or low | G+ and G- bacteria. | [113] | |
| Pb-CATH3 | CATHPb3 | KRFQNFFRELEKKFREFFRVYRITIGATIRF | 31 | Low | Inactive against G+ and G- bacteria and fungi. Immunomodulatory inactive. | [112] | |
| Pb-CATH4 | CATHPb4 | TRSRWRRFIRGAGRFARRYGWRIALGLVG | 29 | Medium or high | G+ and G- bacteria and fungi. Weakly anti-inflammatory. | [112] | |
| ∆Pb-CATH4 | ∆Pb-CATH4 | TRSRWRRFIRGAGRFARRYGWRIA | 24 | Medium | G+ and G- bacteria and tumor cells. | [113] | |
| Pb-CATH5 | CATHPb5 | SPPQAMGFPPQVNVEHYIPASYSVAALTVTEEE | 33 | Low | Inactive against G+ and G- bacteria and fungi. Immunomodulatory inactive. | [112] | |
| Pb-CATH6 | CATHPb6 | RAAPQRRLRAMARLKKFAEAGGADPDSGGLRARFPER | 37 | Low | Inactive against G+ and G- bacteria and fungi. Weakly anti-inflammatory. | [112] | |
| Sinonatrix annularis | Sa-CATH | - | KFFKKLKKSVKKHVKKFFKKPKVIGVSIPF | 30 | Low | G+ and G- bacteria and fungi. Anti-inflamatory. | [114] |
| Crotalus durissus cascavella | Cdc-CATH1 | Cas-CATH isoform 1 | KRFKKFFKKVKKSVKKRLKKIFKKPIFKKVKKSVKKRLKKIFKKPMVIGVTIPF | 54 | n.d. | n.d. | NCBI |
| Cdc-CATH2 | Cas-CATH isoform 2 | KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVSIPF | 34 | n.d. | n.d. | NCBI | |
| Cdc-CATH3 | Cas-CATH isoform 3 | KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF | 34 | n.d. | n.d. | NCBI | |
| Thamnophis sirtalis | Ts-CATH1 | - | KRFKKFFKKIKKSVKKRVKKLFKKPRVIPISIPF | 34 | n.d. | n.d. | NCBI |
| Ts-CATH3 | - | KKRRRIRVQITVKITFKI | 18 | n.d. | n.d. | NCBI | |
| Ts-CATH4 | - | KKGLKKLFKRKKVVAGYVTA | 20 | n.d. | n.d. | NCBI | |
| Protobothrops mucrosquamatus | Pm-CATH | - | KRFAGFFQFVVGVSFRF | 17 | n.d. | n.d. | NCBI |
| Microorganism: | Peptide: | Oh-CATH | Bf-CATH | Bf-CATH30 | Hc-CATH | Cdt-CATH | Ba-CATH | Pt-CATH1 | ∆Pb-CATH1 | ∆Pb-CATH2 | ∆Pb-CATH4 | Pb-CATH1 | Pb-CATH2 | Pb-CATH3 | Pb-CATH4 | Pb-CATH5 | Pb-CATH6 | Sa-CATH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hemolysis (HC10) | ||||||||||||||||||
| ~200 | ~31 | 50 - >400 | >200 | ~104 | ~53 | ~13 | ~64 | >64 | ~64 | >100 | >100 | >100 | <100 | >100 | >100 | >200 | ||
| Gram-negative bacteria (MIC) | ||||||||||||||||||
| E. coli ATCC 25922 | 0.25–8 | 8 | 2.3–8 | 2.3 | 0.25–0.78 | 0.25–0.78 | 2 | 2 | 3 | 1 | 9.4 | 37.5 | n.d. | 18.8 | n.d. | n.d. | 18.8 | |
| E. coli (CI) | 2–20 | 0.6–16 | 2.3–9.4 | 16 | 16 | 16 | 4.7 | 9.4 | 75 | n.d. | 18.8 | n.d. | n.d. | 75 | ||||
| Selectivity ratio | 25 | 4 | 6 | >87 | 133 | 68 | 7 | 32 | >21 | 64 | >11 | >3 | - | <5 | - | - | >11 | |
| Gram-positive bacteria (MIC) | ||||||||||||||||||
| S. aureus ATCC 25923 | 4–64 | 16 | 16 > 400 | 4.7–25.8 | 32 | 32 | 32 | >128 | >128 | >128 | 37.5 | n.d. | n.d. | 18.8 | n.d. | n.d. | 75 | |
| S. aureus (CI) | 8–64 | 32–64 | 16 > 400 | 4.7 > 200 | 32 | 32 | 32 | 4.7–37.5 | 75 | n.d. | 18.8 | n.d. | n.d. | |||||
| Selectivity ratio | 3 | 2 | <0.1 | >8 | 3 | 2 | 0.4 | <0.5 | 0.5 | <0.5 | >3 | - | - | <5 | - | - | >3 | |
| Fungi (MIC) | ||||||||||||||||||
| C. albicans ATCC 2002 | 4.7 | |||||||||||||||||
| C. albicans (CI) | 2.3–4.7 | 10–40 | 9.4–18.8 | 18.8–37.5 | n.d. | 9.4–18.8 | n.d. | n.d. | 18.8–37.5 | |||||||||
| Selectivity ratio | 11 | 43* | 3* | >5* | >3* | - | <5* | - | - | >5* | ||||||||
| Tumor cells (IC50) | ||||||||||||||||||
| PC-3 (prostate cancer) | 70.2 | n.d. | ||||||||||||||||
| U937 (leukemia) | <4 | |||||||||||||||||
| MCF-7 (breast cancer) | n.d. | 353 | ~64 | |||||||||||||||
| HepG2 (liver cancer) | n.d. | |||||||||||||||||
| Selectivity ratio | - | 0.7 | - | >26 | 1 | |||||||||||||
| Ref. | [76,84] | [85] | [93,94,96,97] | [105,106] | [84,107,108,122,123] | [84] | [84] | [113] | [113] | [113] | [112] | [112] | [112] | [112] | [112] | [112] | [114] | |
| Color code: | Hemolysis: | Low (>50) | Medium (50–10) | High (>10) | ||||||||||||||
| Biological activity (MIC/IC50): | Low (>50) | Medium (50–10) | High (<10) | |||||||||||||||
| Non-selective (ratio < 1) | Selective (ratio > 1) | |||||||||||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Peinado, C.; Defaus, S.; Andreu, D. Hitchhiking with Nature: Snake Venom Peptides to Fight Cancer and Superbugs. Toxins 2020, 12, 255. https://doi.org/10.3390/toxins12040255
Pérez-Peinado C, Defaus S, Andreu D. Hitchhiking with Nature: Snake Venom Peptides to Fight Cancer and Superbugs. Toxins. 2020; 12(4):255. https://doi.org/10.3390/toxins12040255
Chicago/Turabian StylePérez-Peinado, Clara, Sira Defaus, and David Andreu. 2020. "Hitchhiking with Nature: Snake Venom Peptides to Fight Cancer and Superbugs" Toxins 12, no. 4: 255. https://doi.org/10.3390/toxins12040255
APA StylePérez-Peinado, C., Defaus, S., & Andreu, D. (2020). Hitchhiking with Nature: Snake Venom Peptides to Fight Cancer and Superbugs. Toxins, 12(4), 255. https://doi.org/10.3390/toxins12040255





