Identification of Two Novel Peanut Genotypes Resistant to Aflatoxin Production and Their SNP Markers Associated with Resistance
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation for AFB1 and AFB2 in Chinese Mini-Mini Core
2.2. SNP Genotyping and Genetic Diversity
2.3. Population Structure and Relative Kinship
2.4. Association Analysis and Candidate Genes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Peanut Plant Materials
5.2. Phenotyping for Aflatoxin Production
5.3. RAD-Seq and SNP Calling
5.4. Population Structure, Relative Kinship, Phylogenetic Tree Construction, and Linkage Disequilibrium
5.5. GWAS for Aflatoxin Production in Peanut Seeds
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Toomer, O.T. Nutritional chemistry of the peanut (Arachis hypogaea L.). Crit. Rev. Food Sci. Nutr. 2018, 58, 3042–3053. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Yield of Major Crop Products in China. Available online: http://data.stats.gov.cn/easyquery.htm?cn=C01 (accessed on 2 November 2019).
- Perrone, G.; Gallo, A. Mycotoxigenic Fungi: Methods and Protocols; Springer International Publishing: New York, NY, USA, 2017; pp. 33–49. [Google Scholar]
- Payne, G.A.; Brown, M.P. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 1998, 36, 329–362. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, A.; Walkiewicz, K.; Koziel, P.; Muc-Wierzgon, M. Aflatoxins: Characteristics and impact on human health. Postepy Hig Med. Dosw. 2017, 71, 315–327. [Google Scholar] [CrossRef]
- Moretti, A.; Logrieco, A.F.; Susca, A. Mycotoxins: An underhand food problem. Methods Mol. Biol. 2016, 1542, 3–12. [Google Scholar]
- Chulze, S.N.; Palazzini, J.M.; Torres, A.M.; Barros, G.; Ponsone., M.L.; Geisen, R.; Schmidt-Heydt, M.; Kohl, J. Biological control as a strategy to reduce the impact of mycotoxins in peanuts, grapes and cereals in Argentina. Food Addit Contam, A 2015, 32, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, J.D.; Baker, J.L.; Mahoney, N.E. Isolation of bacterial antagonists of Aspergillus flavus from almonds. Microb Ecol. 2006, 52, 45–52. [Google Scholar] [CrossRef]
- Nesci, A.V.; Bluma, R.V.; Etcheverry, M.G. In vitro selection of maize rhizobacteria to study potential biological control of aspergillus section flavi and aflatoxin production. Eur. J. Plant. Pathol. 2005, 113, 159–171. [Google Scholar] [CrossRef]
- Dorner, J.W. Management and prevention of mycotoxins in peanuts. Food Addit Contam. 2008, 25, 203–208. [Google Scholar] [CrossRef]
- Pandey, M.K.; Kumar, R.; Pandey, A.K.; Soni, P.; Gangurde, S.S.; Sudini, H.K.; Fountain, J.C.; Liao, B.; Desmae, H.; Okori, P.; et al. Mitigating aflatoxin contamination in groundnut through a combination of genetic resistance and post-harvest management practices. Toxins 2019, 11, 315. [Google Scholar] [CrossRef]
- Asare-Bediako, K.; Ofori, K.; Offei, S.K.; Dzidzienyo, D.; Asibuo, J.Y.; Adu-Amoah, R. Aflatoxin contamination of groundnut (Arachis hypogaea L.): Predisposing factors and management interventions. Food Control 2019, 98, 61–67. [Google Scholar] [CrossRef]
- Liao, B.; Zhuang, W.; Tang, R.; Zhang, X.; Shan, S.; Jiang, H.; Huang, J. Peanut aflatoxin and genomics research in China: Progress and perspectives. Peanut Sci. 2009, 36, 21–28. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, L.; Ren, X.; Pandey, M.K.; Wu, B.; Chen, Y.; Zhou, X.; Chen, W.; Xia, Y.; Li, Z.; et al. Genetic variation and association mapping of seed-related traits in cultivated peanut (Arachis hypogaea L.) using single-locus simple sequence repeat markers. Front. Plant. Sci. 2017, 8, 2105. [Google Scholar] [CrossRef] [PubMed]
- Mahanti, N.; Bhatnagar, D.; Cary, J.W.; Joubran, J.; Linz, J.E. Structure and function of fas-1A, a gene encoding a putative fatty acid synthetase directly involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ. Microbiol. 1996, 62, 191–195. [Google Scholar] [CrossRef]
- Minto, R.E.; Townsend, C.A. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 1997, 97, 2537–2556. [Google Scholar] [CrossRef]
- Roze, L.V.; Hong, S.Y.; Linz, J.E. Aflatoxin biosynthesis: Current frontiers. Annu. Rev. Food Sci. Technol. 2013, 4, 293–311. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Rajasekaran, K.; Payne, G.; Brown, R.; Yu, J.; Cleveland, T. The ‘omics’ tools: Genomics, proteomics, metabolomics and their potential for solving the aflatoxin contamination problem. World MycotoxinJ. 2008, 1, 3–12. [Google Scholar] [CrossRef]
- Bhatnagarmathur, P.; Sunkara, S.; Bhatnagarpanwar, M.; Waliyar, F.; Sharma, K.K. Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant. Sci. 2015, 234, 119–132. [Google Scholar] [CrossRef]
- Huang, L.; Liu, X.; Pandey, M.K.; Ren, X.; Chen, H.; Xue, X.; Liu, N.; Huai, D.; Chen, Y.; Zhou, X.; et al. Genome-wide expression quantitative trait locus analysis in a recombinant inbred line population for trait dissection in peanut. Plant. Biotechnol. J. 2019. [Google Scholar] [CrossRef]
- Luo, H.; Pandey, M.K.; Khan, A.W.; Wu, B.; Guo, J.; Ren, X.; Zhou, X.; Chen, Y.; Chen, W.; Huang, L.; et al. Next-generation sequencing identified genomic region and diagnostic markers for resistance to bacterial wilt on chromosome B02 in peanut (Arachis hypogaea L.). Plant. Biotechnol. J. 2019, 17, 2356–2369. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Ren, X.; Huang, L.; Luo, H.; Chen, Y.; Chen, W.; Liu, N.; Liao, B.; Lei, Y.; et al. A major and stable QTL for bacterial wilt resistance on chromosome B02 Identified using a high-density SNP-based genetic linkage map in cultivated peanut Yuanza 9102 derived population. Front. Genet. 2018, 9, 652. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Pandey, M.K.; Khan, A.W.; Guo, J.; Wu, B.; Cai, Y.; Huang, L.; Zhou, X.; Chen, Y.; Chen, W.; et al. Discovery of genomic regions and candidate genes controlling shelling percentage using QTL-seq approach in cultivated peanut (Arachis hypogaea L.). Plant. Biotechnol. J. 2019, 17, 1248–1260. [Google Scholar] [CrossRef]
- Wan, L.; Li, B.; Lei, Y.; Yan, L.; Huai, D.; Kang, Y.; Jiang, H.; Tan, J.; Liao, B. Transcriptomic profiling reveals pigment regulation during peanut testa development. Plant. Physiol. Biochem. 2018, 125, 116–125. [Google Scholar] [CrossRef]
- Davey, J.W.; Blaxter, M.L. RADSeq: Next-generation population genetics. Brief. Funct Genom. 2010, 9, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xia, Y.; Ren, X.; Chen, Y.; Huang, L.; Huang, S.; Liao, B.; Lei, Y.; Yan, L.; Jiang, H.; et al. Construction of a SNP-based genetic linkage map in cultivated peanut based on large scale marker development using next-generation double-digest restriction-site-associated DNA sequencing (ddRADseq). Bmc Genom. 2014, 15, 351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, C.; Chen, H.; Yuan, M.; Nipper, R.; Prakash, C.S.; Zhuang, W.; He, G. QTL mapping for bacterial wilt resistance in peanut (Arachis hypogaea L.). Mol. Breed. 2016, 36, 13. [Google Scholar] [CrossRef]
- Liu, N.; Guo, J.; Zhou, X.; Wu, B.; Huang, L.; Luo, H.; Chen, Y.; Chen, W.; Lei, Y.; Huang, Y.; et al. High-resolution mapping of a major and consensus quantitative trait locus for oil content to a ~ 0.8-Mb region on chromosome A08 in peanut (Arachis hypogaea L.). Appl. Genet. 2019. [Google Scholar] [CrossRef]
- Yu, B.; Huai, D.; Huang, L.; Kang, Y.; Ren, X.; Chen, Y.; Zhou, X.; Luo, H.; Liu, N.; Chen, W.; et al. Identification of genomic regions and diagnostic markers for resistance to aflatoxin contamination in peanut (Arachis hypogaea L.). BMC Genet 2019, 20, 32. [Google Scholar] [CrossRef]
- Jiang, H.; Huang, L.; Ren, X.; Chen, Y.; Zhou, X.; Xia, Y.; Huang, J.; Lei, Y.; Yan, L.; Wan, L.; et al. Diversity characterization and association analysis of agronomic traits in a Chinese peanut (Arachis hypogaea L.) mini-core collection. J. Integr. Plant. Biol 2014, 56, 159–169. [Google Scholar] [CrossRef]
- Fountain, J.C.; Scully, B.T.; Ni, X.; Kemerait, R.C.; Lee, R.D.; Chen, Z.Y.; Guo, B. Environmental influences on maize-Aspergillus flavus interactions and aflatoxin production. Front. Microbiol. 2014, 5, 40. [Google Scholar] [CrossRef]
- Arunyanark, A.; Jogloy, S.; Wongkaew, S.; Akkasaeng, C.; Vorasoot, N.; Wright, G.C.; Rachaputi, R.C.N.; Patanothai, A. Association between aflatoxin contamination and drought tolerance traits in peanut. Field Crop. Res. 2009, 114, 14–22. [Google Scholar] [CrossRef]
- Girdthai, T.; Jogloy, S.; Vorasoot, N.; Akkasaeng, C.; Wongkae, S.; Holbrook, C.C.; Patanothai, A. Associations between physiological traits for drought tolerance and aflatoxin contamination in peanut genotypes under terminal drought. Plant. Breed. 2010, 129, 693–699. [Google Scholar] [CrossRef]
- Kebede, H.; Abbas, H.K.; Fisher, D.K.; Bellaloui, N. Relationship between Aflatoxin contamination and physiological responses of corn plants under drought and heat stress. Toxins 2012, 4, 1385–1403. [Google Scholar] [CrossRef] [PubMed]
- Yabe, K.; Nakajima, H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2004, 64, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, J.; He, X.; Wang, Y.; Ma, X.; Yin, D. Genome-wide association study of major agronomic traits related to domestication in peanut. Front. Plant. Sci. 2017, 8, 1611. [Google Scholar] [CrossRef]
- Wang, X.; Xu, P.; Yin, L.; Ren, Y.; Li, S.; Shi, Y.; Alcock, T.D.; Xiong, Q.; Qian, W.; Chi, X.; et al. Genomic and transcriptomic analysis identified gene clusters and candidate genes for oil content in peanut (Arachis hypogaea L.). Plant. Mol. Biol. Rep. 2018, 36, 518–529. [Google Scholar] [CrossRef]
- Nelson, D.; Sommers, L. Methods of Soil Analysis; American Society of Agronomy: Madison, WI, USA, 1982; pp. 539–577. Available online: https://dl.sciencesocieties.org/publications/books/pdfs/agronomymonogra/methodsofsoilan2/frontmatter (accessed on 2 February 2020).
- ISSCAS (Institute of Soil Science, Chinese Academy of Science). Physical and Chemical Analysis Methods of Soils; Shanghai Science and Technology Press: Shanghai, China, 1978; pp. 7–59, (In Chinese). Available online: http://www.irgrid.ac.cn/handle/1471x/112649# (accessed on 2 February 2020).
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat.Genet. 2016, 48, 438. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A map reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genet. 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Jian, H.; Lu, K.; Filardo, F.; Yin, N.; Liu, L.; Qu, C.; Li, W.; Du, H.; Li, J. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant. Biotechnol. J. 2016, 14, 1368–1380. [Google Scholar] [CrossRef] [PubMed]
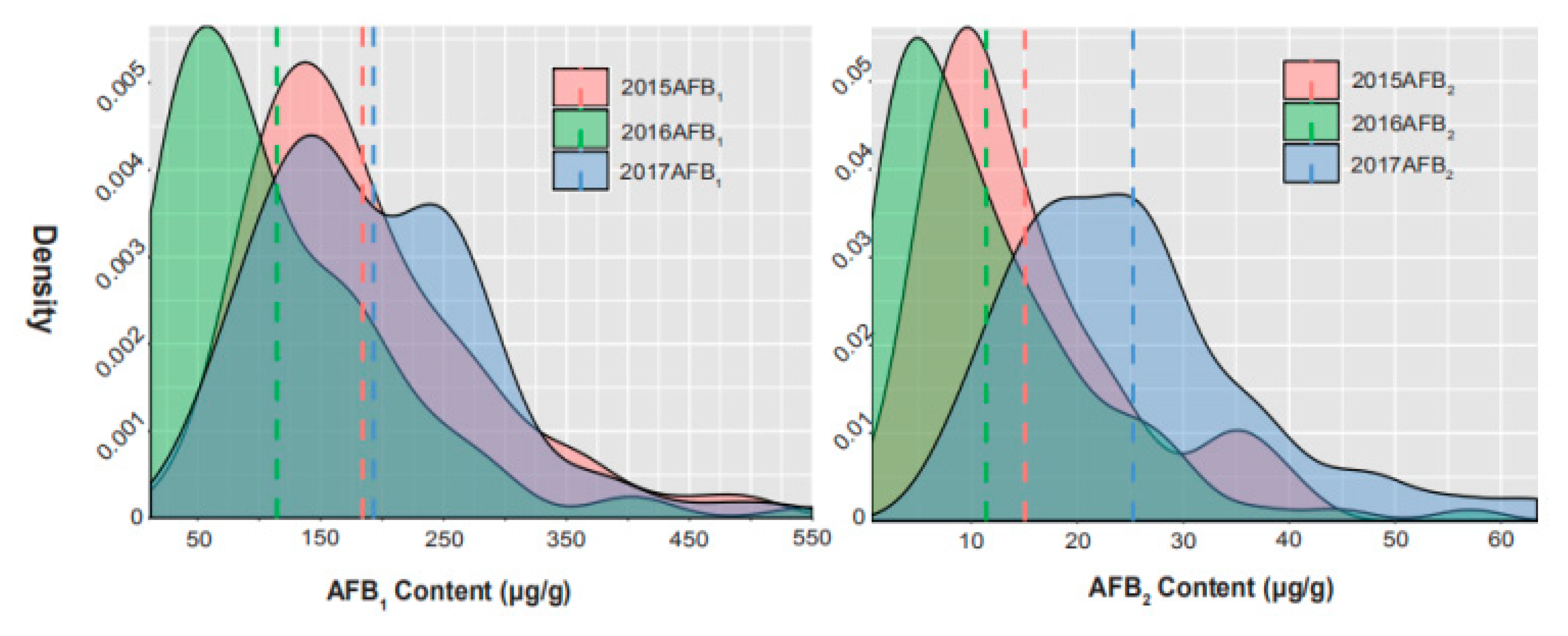
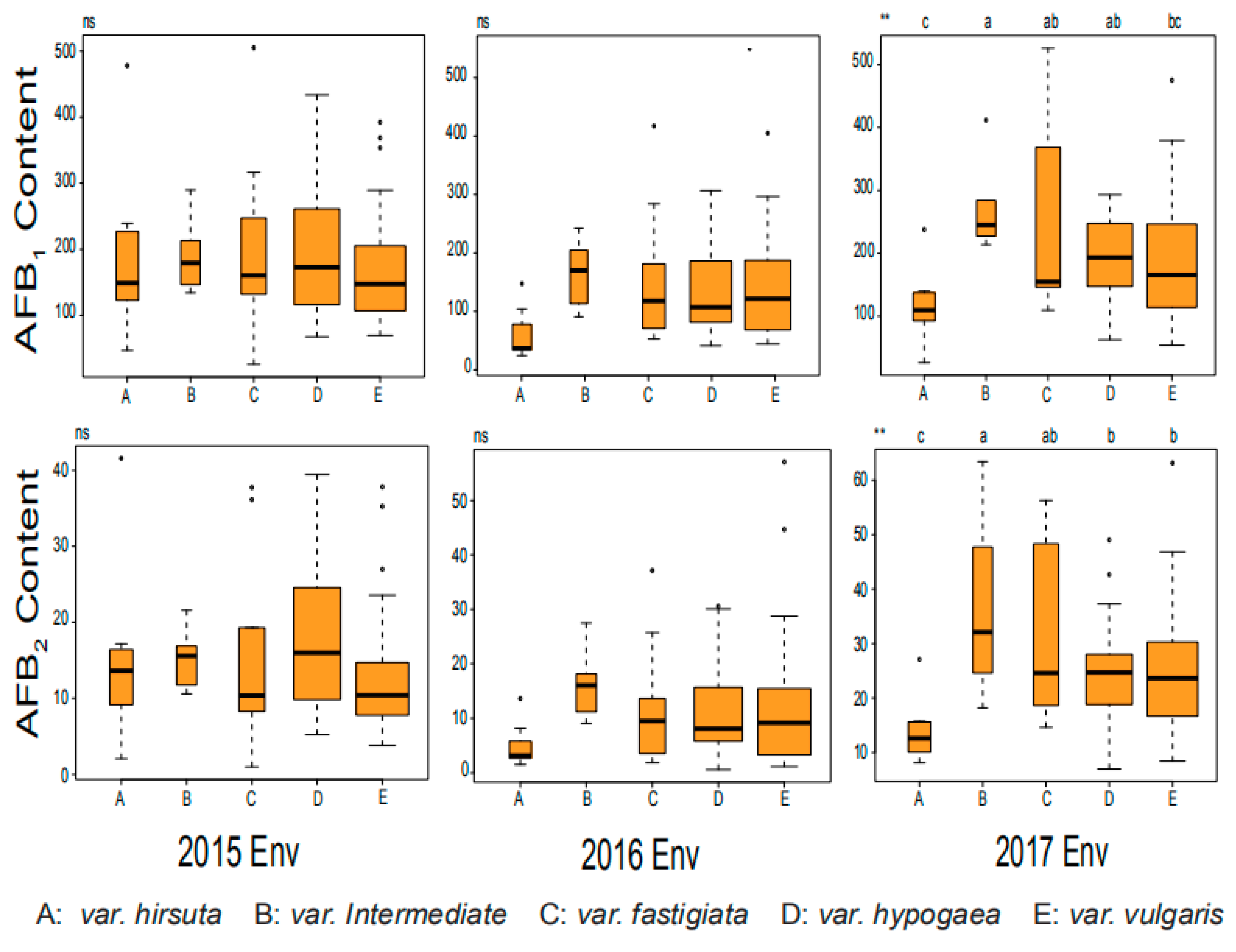
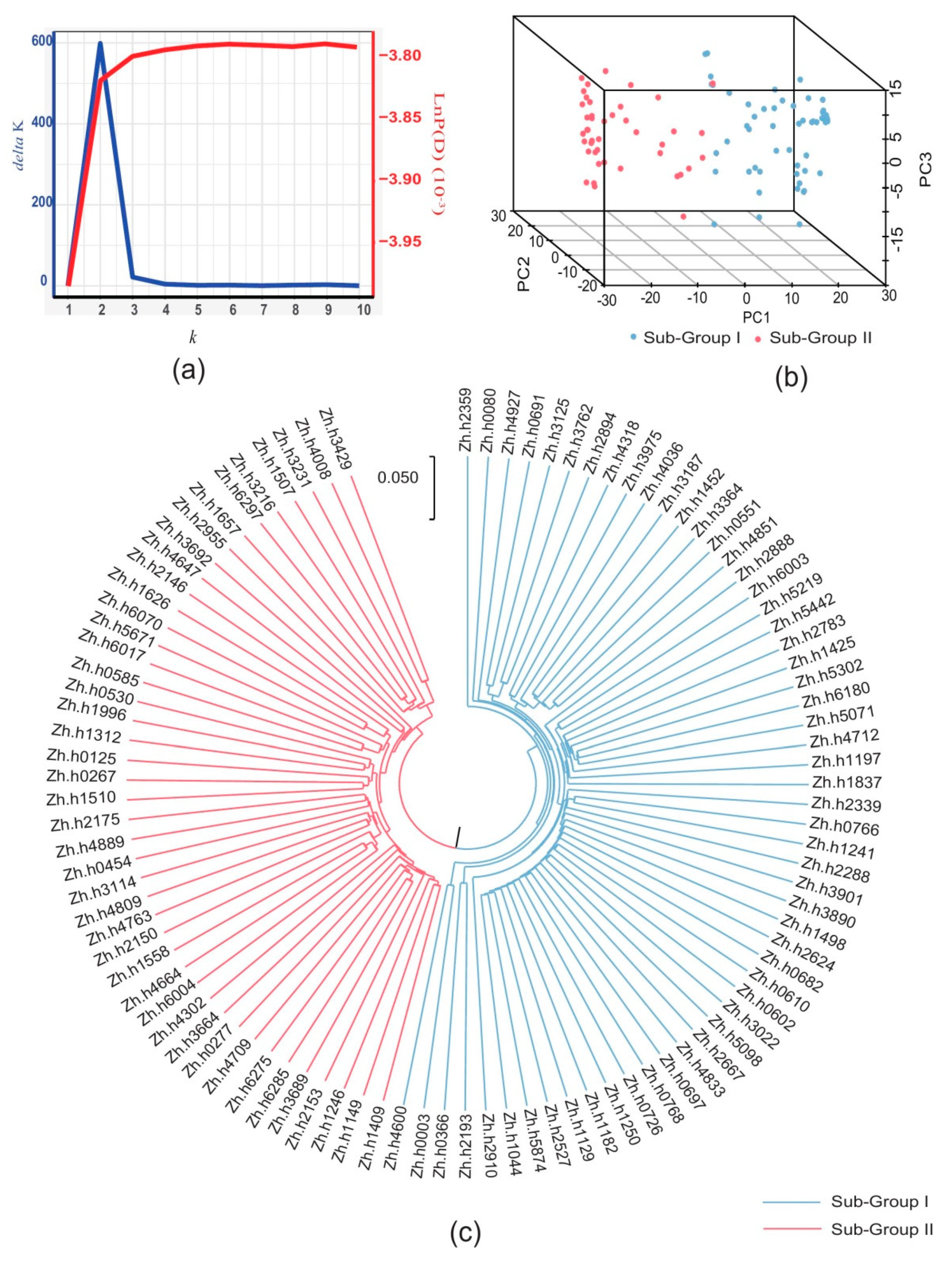
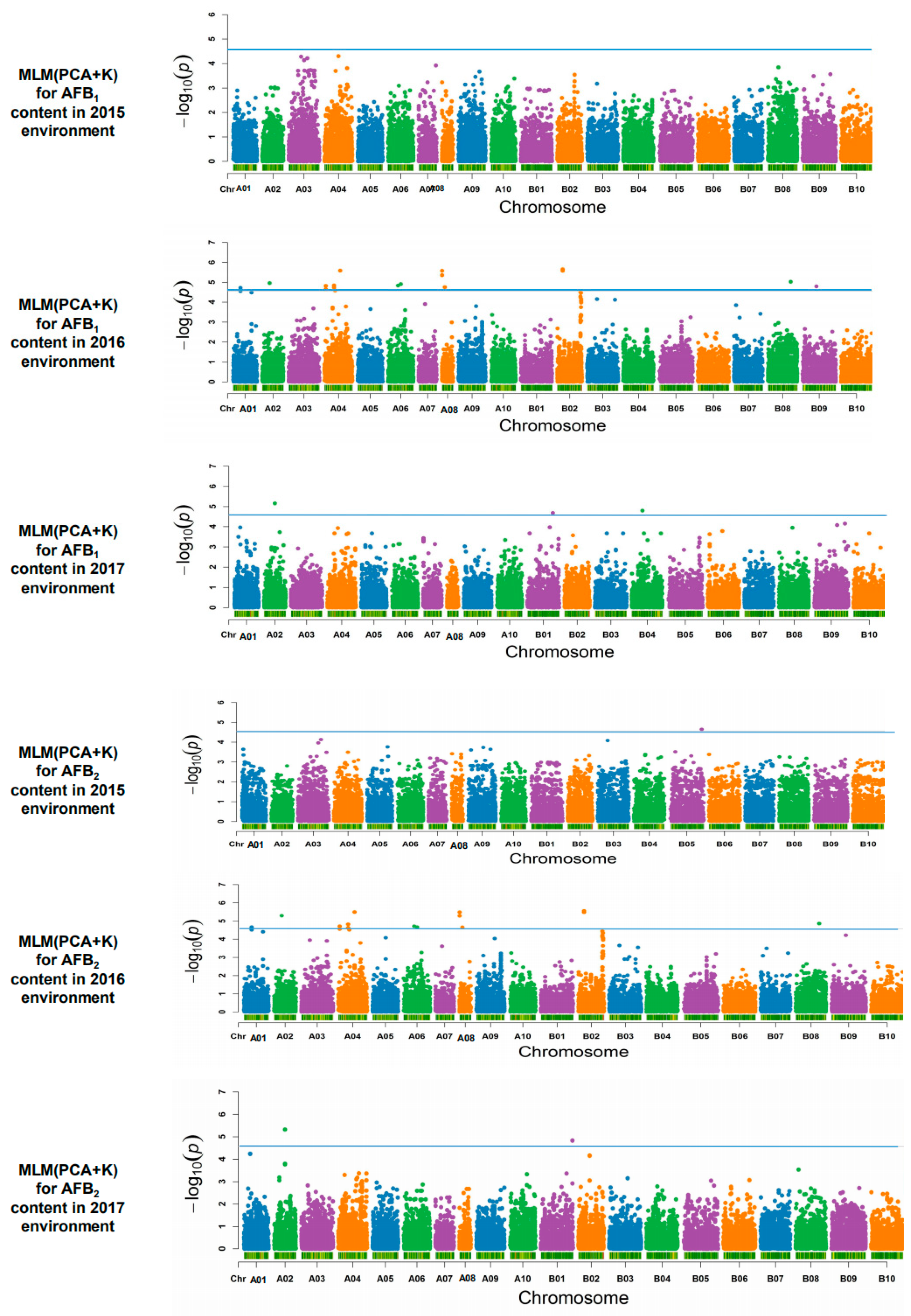
| Traits | Env c | Range | Mean | SD d | CV e |
|---|---|---|---|---|---|
| AFB1 (μg/g) a | 2015 | 25.92–550.17 | 184.08 | 93.14 | 0.51 |
| 2016 | 11.69–505.01 | 114.45 | 94.39 | 0.82 | |
| 2017 | 26.08–526.21 | 193.01 | 91.84 | 0.48 | |
| AFB2 (μg/g) b | 2015 | 0.98–41.60 | 15.06 | 9.42 | 0.63 |
| 2016 | 0.58–57.08 | 11.40 | 9.99 | 0.88 | |
| 2017 | 7.00–63.42 | 25.25 | 11.88 | 0.47 |
| Traits | Source | DF c | SS d | MS e | F Value | p Value | б2f | h2g |
|---|---|---|---|---|---|---|---|---|
| AFB1 a | Genotype | 98 | 4106986 | 41908 | 9.18 | <0.001 | 2667.69 | 0.57 |
| Environment | 2 | 1077196 | 539598 | 117.97 | <0.001 | 1756.56 | ||
| Genotype × Environment | 196 | 3508164 | 17899 | 3.92 | <0.001 | 4444.76 | ||
| Error | 594 | 2711315 | 4565 | 4564.50 | ||||
| AFB2 b | Genotype | 98 | 48863 | 499 | 5.96 | <0.001 | 28.52 | 0.51 |
| Environment | 2 | 30454 | 15227 | 182.19 | <0.001 | 50.45 | ||
| Genotype × Environment | 196 | 47409 | 242 | 1.20 | <0.001 | 52.77 | ||
| Error | 594 | 49644 | 84 | 83.58 |
| Env a | Pearson Correlation between AFB1 and AFB2 | p-Value |
|---|---|---|
| 2015 | 0.88 | <0.01 |
| 2016 | 0.99 | <0.01 |
| 2017 | 0.78 | <0.01 |
| Traits | Group | Accession Number | Var Type | 2015Env c | 2016Env | 2017Env | Average |
|---|---|---|---|---|---|---|---|
| AFB1(μg/g) a | Susceptible control | Zh.h4600 | var.vulgaris | 392.31 | 450.17 | 310.45 | 384.31 |
| Zh.h3231 | var.fastigiata | 247.43 | 409.57 | 368.59 | 341.86 | ||
| Low content | Zh.h0551 | var.hirsuta | 39.00 | 21.42 | 33.08 | 31.17 | |
| Zh.h2150 | var.vulgaris | 49.74 | 20.71 | 31.10 | 33.85 | ||
| AFB2(μg/g) b | Susceptible control | Zh.h4600 | var.vulgaris | 37.83 | 45.08 | 25.55 | 36.15 |
| Zh.h3231 | var.fastigiata | 28.27 | 32.13 | 44.32 | 34.91 | ||
| Low content | Zh.h0551 | var.hirsuta | 8.33 | 5.19 | 6.32 | 6.61 | |
| Zh.h2150 | var.vulgaris | 5.27 | 4.13 | 9.57 | 6.32 |
| Chromosome | SNP Number | Marker Start Loci (Kb) | Marker End Loci (Mb) | Reference Length (Mb) | Density of Markers (kb/SNP) |
|---|---|---|---|---|---|
| A01 | 1856 | 1621.08 | 106.90 | 105.28 | 56.72 |
| A02 | 1512 | 165.40 | 93.53 | 93.36 | 61.75 |
| A03 | 2387 | 69.80 | 134.58 | 134.51 | 56.35 |
| A04 | 2317 | 277.02 | 123.31 | 123.03 | 53.10 |
| A05 | 2019 | 252.20 | 109.66 | 109.41 | 54.19 |
| A06 | 1774 | 302.81 | 112.63 | 112.32 | 63.32 |
| A07 | 1116 | 201.97 | 79.09 | 78.89 | 70.69 |
| A08 | 599 | 519.89 | 49.09 | 48.57 | 81.09 |
| A09 | 2125 | 276.41 | 120.36 | 120.08 | 56.51 |
| A10 | 2184 | 156.37 | 109.21 | 109.05 | 49.93 |
| B01 | 1560 | 136.00 | 137.19 | 137.05 | 87.85 |
| B02 | 1635 | 137.78 | 108.93 | 108.79 | 66.54 |
| B03 | 1967 | 101.11 | 135.70 | 135.60 | 68.94 |
| B04 | 2000 | 176.67 | 133.52 | 133.35 | 66.67 |
| B05 | 1757 | 3440.92 | 149.75 | 146.31 | 83.27 |
| B06 | 1743 | 124.59 | 135.87 | 135.75 | 77.88 |
| B07 | 1608 | 247.60 | 126.20 | 125.95 | 78.33 |
| B08 | 1864 | 578.78 | 129.60 | 129.02 | 69.22 |
| B09 | 2373 | 135.20 | 146.85 | 146.72 | 61.83 |
| B10 | 1689 | 34.68 | 135.89 | 135.86 | 80.44 |
| SNP Marker | Genotype | n | AFB1 | AFB2 |
|---|---|---|---|---|
| SNP02686 | AA | 3 | 312.57 ± 21.93 a | 38.15 ± 1.59 a |
| GG | 6 | 195.35 ± 37.99 b | 19.20 ± 7.24 b | |
| GA | 80 | 159.88 ± 62.86 b | 16.57 ± 6.54 b | |
| SNP19994 | AA | 9 | 223.56 ± 38.64 a | 25.37 ± 9.12 a |
| GG | 3 | 160.75 ± 34.74 b | 16.62 ± 4.76 b | |
| GA | 79 | 145.08 ± 31.12 b | 14.25 ± 2.67 b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, B.; Jiang, H.; Pandey, M.K.; Huang, L.; Huai, D.; Zhou, X.; Kang, Y.; Varshney, R.K.; Sudini, H.K.; Ren, X.; et al. Identification of Two Novel Peanut Genotypes Resistant to Aflatoxin Production and Their SNP Markers Associated with Resistance. Toxins 2020, 12, 156. https://doi.org/10.3390/toxins12030156
Yu B, Jiang H, Pandey MK, Huang L, Huai D, Zhou X, Kang Y, Varshney RK, Sudini HK, Ren X, et al. Identification of Two Novel Peanut Genotypes Resistant to Aflatoxin Production and Their SNP Markers Associated with Resistance. Toxins. 2020; 12(3):156. https://doi.org/10.3390/toxins12030156
Chicago/Turabian StyleYu, Bolun, Huifang Jiang, Manish K. Pandey, Li Huang, Dongxin Huai, Xiaojing Zhou, Yanping Kang, Rajeev K. Varshney, Hari K. Sudini, Xiaoping Ren, and et al. 2020. "Identification of Two Novel Peanut Genotypes Resistant to Aflatoxin Production and Their SNP Markers Associated with Resistance" Toxins 12, no. 3: 156. https://doi.org/10.3390/toxins12030156
APA StyleYu, B., Jiang, H., Pandey, M. K., Huang, L., Huai, D., Zhou, X., Kang, Y., Varshney, R. K., Sudini, H. K., Ren, X., Luo, H., Liu, N., Chen, W., Guo, J., Li, W., Ding, Y., Jiang, Y., Lei, Y., & Liao, B. (2020). Identification of Two Novel Peanut Genotypes Resistant to Aflatoxin Production and Their SNP Markers Associated with Resistance. Toxins, 12(3), 156. https://doi.org/10.3390/toxins12030156








